ExCYT: A Graphical User Interface for Streamlining Analysis of High-Dimensional Cytometry Data
Summary
ExCYT is a MATLAB-based Graphical User Interface (GUI) that allows users to analyze their flow cytometry data via commonly employed analytical techniques for high-dimensional data including dimensionality reduction via t-SNE, a variety of automated and manual clustering methods, heatmaps, and novel high-dimensional flow plots.
Abstract
With the advent of flow cytometers capable of measuring an increasing number of parameters, scientists continue to develop larger panels to phenotypically explore characteristics of their cellular samples. However, these technological advancements yield high-dimensional data sets that have become increasingly difficult to analyze objectively within traditional manual-based gating programs. In order to better analyze and present data, scientists partner with bioinformaticians with expertise in analyzing high-dimensional data to parse their flow cytometry data. While these methods have been shown to be highly valuable in studying flow cytometry, they have yet to be incorporated in a straightforward and easy-to-use package for scientists who lack computational or programming expertise. To address this need, we have developed ExCYT, a MATLAB-based Graphical User Interface (GUI) that streamlines the analysis of high-dimensional flow cytometry data by implementing commonly employed analytical techniques for high-dimensional data including dimensionality reduction by t-SNE, a variety of automated and manual clustering methods, heatmaps, and novel high-dimensional flow plots. Additionally, ExCYT provides traditional gating options of select populations of interest for further t-SNE and clustering analysis as well as the ability to apply gates directly on t-SNE plots. The software provides the additional advantage of working with either compensated or uncompensated FCS files. In the event that post-acquisition compensation is required, the user can choose to provide the program a directory of single stains and an unstained sample. The program detects positive events in all channels and uses this select data to more objectively calculate the compensation matrix. In summary, ExCYT provides a comprehensive analysis pipeline to take flow cytometry data in the form of FCS files and allow any individual, regardless of computational training, to use the latest algorithmic approaches in understanding their data.
Introduction
Advances in flow cytometry as well as the advent of mass cytometry has allowed clinicians and scientists to rapidly identify and phenotypically characterize biologically and clinically interesting samples with new levels of resolution, creating large high-dimensional data sets that are information rich1,2,3. While conventional methods for analyzing flow cytometry data such as manual gating have been more straightforward for experiments where there are few markers and those markers have visually discernable populations, this approach can fail to generate reproducible results when analyzing higher-dimensional data sets or those with markers staining on a spectrum. For example, in a multi-institutional study, where intra-cellular staining (ICS) assays were being performed to assess the reproducibility of quantitating antigen-specific T cell responses, despite good inter-laboratory precision, analysis, particularly gating, introduced a significant source of variability4. Furthermore, the process of manually gating population of interests, besides being highly subjective is highly time consuming and labor intensive. However, the problem of analyzing high-dimensional data sets in a robust, efficient, and timely manner is not one new to the research sciences. Gene expression studies often generate extremely high-dimensional data sets (often on the order of hundreds of genes) where manual forms of analysis would be simply infeasible. In order to tackle the analysis of these data sets, there has been much work in developing bioinformatic tools to parse gene expression data5. These algorithmic approaches have just been recently adopted in the analysis of cytometry data as the number of parameters has increased and have proven to be invaluable in the analysis of these high dimensional data sets6,7.
Despite the generation and application of a variety of algorithms and software packages that allow scientists to apply these high-dimensional bioinformatic approaches to their flow cytometry data, these analytical techniques still remain largely unused. While there may be a variety of factors that have limited the widespread adoption of these approaches to cytometry data8, the major hindrance we suspect in use of these approaches by scientists, is a lack of computational knowledge. In fact, many of these software packages (i.e., flowCore, flowMeans, and OpenCyto) are written to be implemented in programming languages such as R that still require substantive programming knowledge. Software packages such as FlowJo have found favor among scientists due to simplicity of use and 'plug-n-play' nature, as well as compatibility with the PC operating system. In order to provide the variety of accepted and valuable analytical techniques to the scientist unfamiliar programming, we have developed ExCYT, a graphical-user interface (GUI) that can be easily installed on a PC/Mac that pulls many of the latest techniques including dimensionality reduction for intuitive visualization, a variety of clustering methods cited in the literature, along with novel features to explore the output of these clustering algorithms with heatmaps and novel high-dimensional flow/box plots.
ExCYT is a graphical user interface built in MATLAB and therefore can either be run within MATLAB directly or an installer is provided that can be used to install the software on any PC/Mac. The software is available at https://github.com/sidhomj/ExCYT. We present a detailed protocol for how to import data, pre-process it, conduct t-SNE dimensionality reduction, cluster data, sort & filter clusters based on user preferences, and display information about the clusters of interest via heatmaps and novel high-dimensional flow/box plots (Figure 1). Axes in t-SNE plots are arbitrary and in arbitrary units and as such as not always shown in the figures for simplicity of the user interface. The coloring of data points in the "t-SNE Heatmaps" is from blue to yellow based on the signal of the indicated marker. In clustering solutions, the color of the data point is based arbitrary on cluster number. All parts of the workflow can be carried out in the single panel GUI (Figure 2 & Table 1). Finally, we will demonstrate the use of ExCYT on previously published data exploring the immune landscape of renal cell carcinoma in the literature, also analyzed with similar methods. The sample dataset we used to create the figures in this manuscript along with the protocol below can be found at https://premium.cytobank.org/cytobank/projects/875, upon registering an account.
Protocol
1. Collecting and Preparing Cytometry Data
- Place all single stains in a folder by themselves and label by the channel name (by fluorophore, not marker).
2. Data Importation & Pre-Processing
- To pause or save throughout this analysis pipeline, use the Save Workspace button at the bottom left of the program to save the workspace as a ‘.MAT’ file that can later be loaded via the Load Workspace button. Do not run more than one instance of the program at a time. Therefore, when loading a new workspace, make sure to check there is no other instance of ExCYT running.
- To begin analysis pipeline, first select type of cytometry (Flow Cytometry or Mass Cytometry – CYTOF), under the File Selection Parameters select number of events to sample from the file (for this example use 2,000). Once data has been successfully imported, a dialogue box will pop up informing the user that the data has been successfully imported.
- Press the Auto-Compensation button to conduct an optional auto-compensation step, as done by Bagwell & Adams9. Select the directory containing single stains. Select the unstained sample within the user interface dialogue.
- Place a forward/side-scatter gate on any of the samples in this directory that will be used to select events to calculate the compensation matrix. It is recommended to use the unstained sample for this purpose. At this point, an algorithm has been implemented to set consistent thresholds at the 99th percentile of the unstained sample to define positive events in each of the single stains to calculate the compensation matrix. When this is finished, a dialogue box will inform the user that the compensation has been performed.
- Next, press Gate Population and select the populations of cells of interest, as is the convention in flow cytometry analyses. When population of cells is selected, enter number of percentage of events downstream analysis (in this 10,000 events).
- Next, select the number channels to be used for analysis in the listbox in the far right of the Pre-Processing box (use the specific channels shown in the example).
3. t-SNE Analysis
- Press the t-SNE button to have the program begin start to compute the reduced dimensionality data set for visualization in the window below the t-SNE button. To save image of t-SNE, press Save TSNE Image. On a machine with 8 CPU @ 3.4 GHz each and 8 GM RAM this step should take about 2 minutes for 10,000 events, 10 minutes for 50,000 events, and 20 minutes for 100,000 events.
- To create a ‘t-SNE heatmap’, as seen in several CYTOF publications10,11, select an option from the Marker-Specific t-SNE pop-up menu (use the specific markers CD64 or CD3 as shown in the example). A figure will pop up showing a heatmap representation of the t-SNE plot that can be saved for figure generation.
- Select areas of interest in the t-SNE plots by the user for further downstream analyses using the Gate t-SNE button.
4. Cluster Analysis
- To begin clustering analysis, select an option in Clustering Method listbox (in this example us DBSCAN with a distance factor of 5 in dialogue box to the right of the listbox). Press the Cluster button.
- Use one of the following options for automated clustering algorithms found in the ‘Automated Clustering Parameters’ panel:
- Hard KMEANS (on t-SNE): Apply k-means clustering to the reduced 2-dimensional t-SNE data and requires the number of clusters to be provided to the algorithm12.
- Hard KMEANS (on HD Data): Apply k-means clustering to the original high-dimensional data that was given to the t-SNE algorithm. Once again, the number of clusters needs to be provided to the algorithm.
- DBSCAN: Apply the clustering method of clustering, called Density-Based Spatial Clustering of Applications with Noise13 that clusters the reduced 2-dimensional t-SNE data and requires a non-dimensional distance factor that determines the general size of the clusters. This type of clustering algorithm is well suited to cluster the t-SNE reduction as it is able to cluster non-spheroidal cluster that are often present in the reduced t-SNE representation. Additionally, due to the fact that it operates on the 2-dimensional data, it is one of the faster clustering algorithms.
- Hierarchical Clustering: Apply the conventional hierarchical clustering method to the high-dimensional data where the entire Euclidean distance matrix is calculated between all events before providing the algorithm a distance factor that sets the size of the cluster.
- Network Graph-Based: Apply a clustering method that has been most recently introduced into analyzing flow cytometry data when there are rare subpopulations that the user wants to detect11,14. This method relies on first creating a graph that determines the connections between all events in the data. This step consists of providing an initial parameter to create the graph, which is the number of k-nearest neighbors. This parameter generally governs the size of the clusters. At this point, another dialogue box pops up asking the user to employ one of 5 clustering algorithms that is applied to the graph. These include 3 options to maximize the modularity of the graph, the Danon Method, and a spectral clustering algorithm14,15,16,17,18. If one wants a generally faster clustering solution, we recommend Spectral Clustering or the Fast Greedy Modularity Maximization. While the Modularity Maximization methods along with the Danon method determine the optimal number of clusters, Spectral Clustering requires the number of clusters to be given to the program.
- Self-Organized Map: Employ an artificial neural network to cluster the high-dimensional data.
- GMM – Expectation Maximization: Create a Gaussian Mixture Model using Expectation Maximization (EM) technique to cluster the high-dimensional data.19 This type of clustering method also requires the user to input the number of clusters.
- Variational Bayesian Inference for GMM: Create a Gaussian Mixture Model but unlike EM, it can automatically determine the number of the mixture components k.20 While the program does require a number of clusters to be given (larger than the expected number of clusters), the algorithm will determine the optimal number on its own.
- To study a particular area of the t-SNE plot, press the Select Cluster Manually button to draw a set of user-defined clusters. Of note, clusters cannot share members (i.e., each event can only belong to 1 cluster).
5. Cluster Filtration
- Set(s) of clusters identified either manually or via one of the automatic methods described above can be filter via as follows.
- To sort clusters (in the Cluster Filter panel) by any of the markers measured in the experiment, select an option from the Sort pop-up menu. To set whether the order is ascending or descending, press the Ascending/Descending button to the right of the Sort pop-up menu. This will update the list of Clusters in the ‘Clusters (Filtration)’ listbox and re-order them in descending order of median cluster expression of that marker. The percentage denoted in the ‘Clusters (Filtration)’ listbox denotes the percent of the population that this cluster represents.
- To set a minimum threshold value for a given cluster across a certain channel, select an option from the Threshold pop-up menu (in this example us the marker CD65 and set a threshold at 0.75). Either type a value in the numerical box below the graph or use the slide-bar to set a threshold. Once threshold is set, press Add Above Threshold or Add Below Threshold to specify the direction of threshold. Once this threshold has been set, it will be listed in the Thresholds box next to the ‘Cluster Filter’ panel where the marker, the threshold value, and the direction will be listed so the user is aware of which thresholds are currently being applied. Finally, the t-SNE plot will update by blurring out clusters that do not meet the requirements of the filtration and the ‘Clusters (Filtration)’ listbox will update to show clusters that meet the filtration requirements.
- To set a minimum threshold for frequency of a cluster, enter a numerical cut-off in the Cluster Frequency Threshold (%) box in the Cluster Filter panel (in this example use 1%).
6. Cluster Analysis & Visualization
- To select clusters for further analysis and visualization, select clusters In Clusters (Filtration) listbox and press the Select à button to move them to the Cluster Analyze listbox.
- To create heatmaps of clusters, select the clusters of interest in the Cluster Analyze listbox and press the HeatMap of Clusters button. When this button is pressed, a figure will pop up containing a heat map along with dendrograms on the cluster and parameter axes. The dendrogram on the vertical axis will group clusters by those that are closely related while the dendrogram on the horizontal axis will group markers that are co-associated. To save heatmap, press File | Export Setup | Export.
- To create a ‘High Dimensional Box Plot’ or ‘High Dimensional Flow Plot,’ select the clusters of interest in the Cluster Analyze listbox and press either the High Dimensional Box Plot button or the High Dimensional Flow Plot button. These plots can be used to visually assess the distribution of given channels of various clusters across all dimensions.
- To show clusters in traditional 2D flow plots, select the transformation (linear, log10, arcsinh) and channel in the Conventional Flow Plot panel and press Conventional Flow Plot.
Representative Results
In order to test the usability of ExCYT, we analyzed a curated data set published by Chevrier et al. titled 'An Immune Atlas of Clear Cell Renal Carcinoma' where the group conducted CyTOF analysis with an extensive immune panel on tumor samples taken from 73 patients11. Two separate panels, a myeloid and lymphoid panel, were used to phenotypically characterize the tumor microenvironment. The objective of our study was to recapitulate the results of their t-SNE and cluster analysis, showing that ExCYT could be used to come to the same conclusions as well as show additional methods of visualization and cluster analysis.
In the original manuscript, the group described 22 T cell clusters identified by the lymphoid panel and 17 cell clusters identified by the myeloid panel. In Figure 3 & Figure 4 of the publication, the group shows heatmaps of clusters, t-SNE plots with color-coded clustering solutions, and t-SNE heatmaps in subpanels A, B, & C. In order to perform the analysis, we obtained the manually gated data from Cytobank and sampled 2,000 events from each file or took the entire file if it had less than 2,000 events, following the analysis pipeline illustrated in the original manuscript. At this point, we sampled a total of 100,000 events via our post-gating subsampling parameter, conducted t-SNE analysis, and used a variety of clustering methods to explore the data in various ways.
First, we examined the myeloid panel by following the same analysis pipeline as the original manuscript by completing the t-SNE analysis and creating heatmaps of the various markers (Figure 3A). While the original manuscript normalized the t-SNE heatmaps to the 99th percentile of each marker, ExCYT does not do this type of normalization for its heatmaps. However, similar distributions of marker co-expression were observed as described in the original manuscript. We then applied a Network Graph-Based method of clustering the data by creating the graph with 100 k-nearest neighbors and clustering the graph via optimizing the modularity of the graph by using the Fast-Greedy implementation within ExCYT, where we found 19 sub-populations of cells (Figure 3B). When comparing the heatmap of these clusters created by ExCYT with the heatmap published in the original manuscript, we noted that we were able to identify similar clusters of myeloid cells (Figure 3C). Of note, the original manuscript identified and contrasted two sub-populations of myeloid cells that we identified in our analysis defined by HLA-DRintCD68intCD64intCD36+CD11b+ (Cluster 13) and HLA-DR+CD4+CD68+CD64+CD36–CD11b– (Cluster 18). Visualization by high-dimensional box plot of these two populations revealed statistically significant differences (Mann-Whitney) in the six markers mentioned (Figure 1D).
Next, we analyzed the lymphoid panel with a more conventional and faster hierarchical clustering approach. This approach yielded similar marker distributions via t-SNE heatmaps (Figure 4A). Furthermore, clustering of the data via hierarchical clustering (Figure 4B), demonstrated similar clusters of lymphoid cells (Figure 4C). Of note, we also identified the unique regulatory T cell population from the original manuscript defined as CD4+CD25+Foxp3+CTLA-4+CD127– (Cluster 17) via our high-dimensional flow plot (Figure 4D).
Finally, we wanted to employ a method within ExCYT to quickly and quantitatively assess co-associations among markers. We began by using a hard k-means clustering algorithm to lay down 5,000 clusters on the two-dimensional t-SNE data (Figure 4E). We then used the median expression of all the markers of all these clusters to create a heatmap from these clusters (Figure 4F). Since these heatmaps cluster rows as well as columns that are similar, this method of abstracting the data by applying a fine mesh of clusters and then creating a heatmap allows us to pick up co-associations easily, such as the co-association of Tim-3, PD-1, CD38, and 4-1BB.
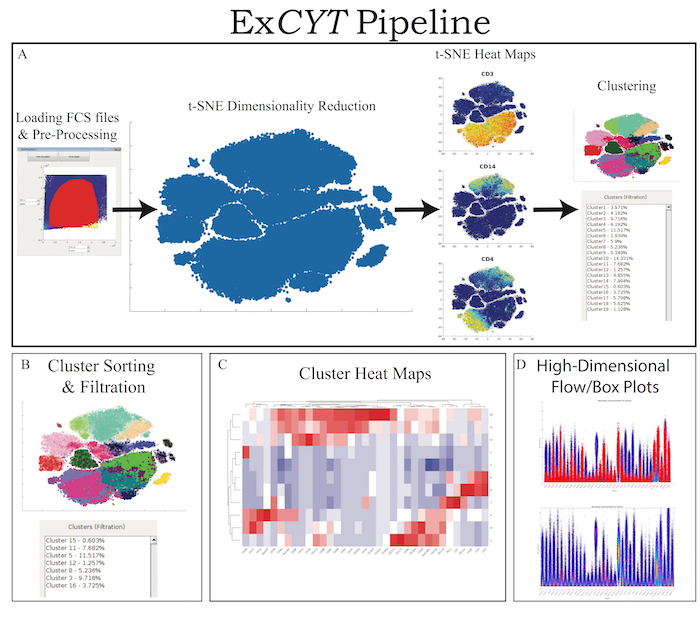
Figure 1: ExCYT Pipeline & Features. (A) ExCYT begins by importing raw FCS data, applying optional compensation, gating, and random subsampling prior to downstream analysis. This ensures all events being analyzed are relevant to the experiment being analyzed. t-SNE dimensionality reduction is then performed to visualize all events and t-SNE heatmaps can be generated to visualize phenotypic distributions. Finally, a variety of clustering algorithms can be applied on either t-SNE transformation or high-dimensional raw data. (B) Novel sorting and thresholding features allow users to quickly sort through possibly hundreds of clusters to find ones of interest. (C) Heatmaps of clusters can be created to examine how multiple clusters compare to each other as well as which markers co-associate. (D) Novel high-dimensional flow/box plots can be generated as a form of back-gating clusters on original data while appreciating the high-dimensional nature of the data. Please click here to view a larger version of this figure.
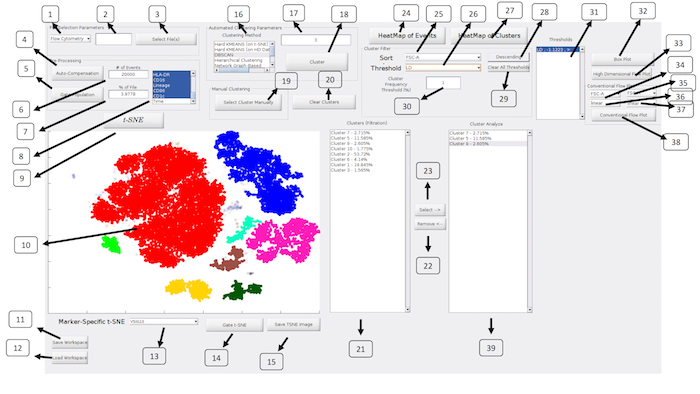
Figure 2: ExCYT Graphical User Interface: The ExCYT graphical user interface allows for a streamline work flow working from the left to right of the panel as the user imports their data, conducts t-SNE dimensionality reduction, clustering, and final cluster analysis and visualization. Please click here to view a larger version of this figure.
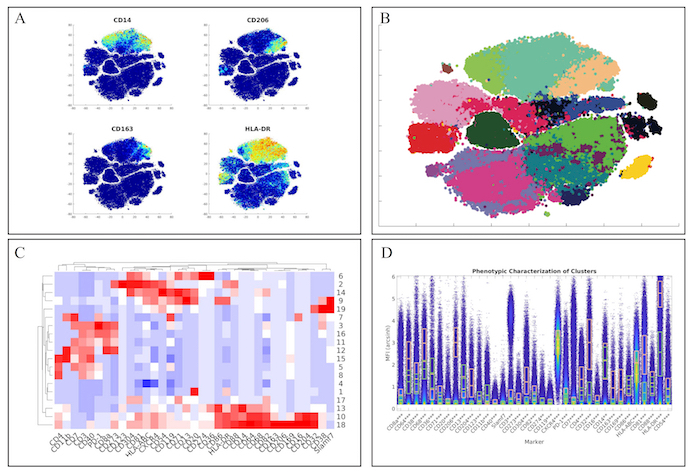
Figure 3: Recapitulation of Myeloid Sub-Populations from Chevrier et al. (A) Token t-SNE heatmaps of myeloid panel (B) t-SNE plot of myeloid panel color coded by Network-Graph clustering algorithm (C) Heatmap of clusters identified by clustering solution on myeloid panel (D) Comparative high dimensional box plot comparing contrasting myeloid subpopulations (Clusters 13 & 18) referenced in original manuscript Please click here to view a larger version of this figure.
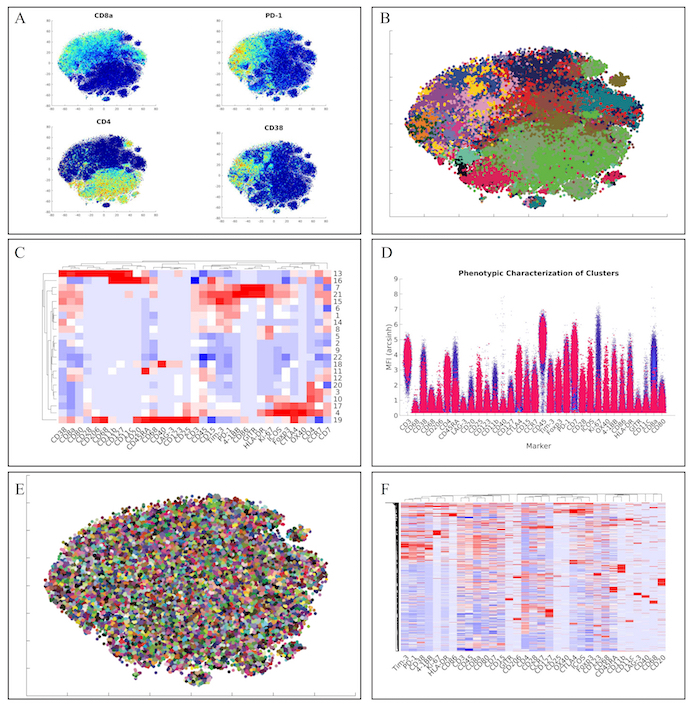
Figure 4: Recapitulation of Lymphoid Sub-Populations from Chevrier et al. (A) Token t-SNE heatmaps of lymphoid panel (B) t-SNE plot of lymphoid panel color coded by hierarchical clustering algorithm (C) Heatmap of clusters identified by clustering solution on lymphoid panel (D) High dimensional flow plot of identified regulatory T cell population (Cluster 17) in original manuscript (E) Clustering solution of 5,000 cluster hard k-means analysis on t-SNE data (F) Heatmap of clusters identified by k-means clustering solution on lymphoid panel showing marker co-associations. Please click here to view a larger version of this figure.
| No. | Description | Name (in GUI) | |
| 1 | Select type of Cytometry | NA | |
| 2 | Random subsampling of raw data | NA | |
| 3 | Select files for analysis | Select File(s) | |
| 4 | Auto-compensation of raw data based on directory of single stains provided to software | Auto-Compensation | |
| 5 | Gating to select events for t-SNE and clustering analysis | Gate Population | |
| 6 | Random subsampling of gated data (absolute number) | NA | |
| 7 | Random subsampling of gated data (percent of gated population) | NA | |
| 8 | Select channels for analysis | NA | |
| 9 | Run t-SNE dimensionality reduction | t-SNE | |
| 10 | t-SNE Window | NA | |
| 11 | Save workspace | Save Workspace | |
| 12 | Load Workspace | Load Workspace | |
| 13 | Create t-SNE heatmap on select marker | NA | |
| 14 | Gate t-SNE to re-do t-SNE analysis of select population | Gate t-SNE | |
| 15 | Save t-SNE window as image | Save TSNE Image | |
| 16 | Select Clustering Algorithm | Clustering Method | |
| 17 | Enter Clustering Parameter for given algorithm | NA | |
| 18 | Cluster Analysis | Cluster | |
| 19 | Draw Clusters Manually | Select Cluster Manually | |
| 20 | Clear All Clusters to redo cluster analysis | Clear Clusters | |
| 21 | Show Clusters under current filter conditions | Clusters (Filtration) | |
| 22 | Remove select clusters from Cluster Analyze listbox | Remove <– | |
| 23 | Add cluster to Cluster Analyze listbox | Select –> | |
| 24 | Create conventional heatmap of all events in analysis | HeatMap of Events | |
| 25 | Sort clusters by select marker | Sort | |
| 26 | Set threshold by select marker | Threshold | |
| 27 | Create conventional heatmap of select clusters from Cluster Analyze listbox | HeatMap of Clusters | |
| 28 | Flip order of sort | Ascending/Descending | |
| 29 | Clear all thresholds | Clear All Thresholds | |
| 30 | Set frequency threshold for clusters | Cluster Frequency Threshold (%) | |
| 31 | List of current thresholds active on 'Clusters (Filtration)' listbox | Thresholds | |
| 32 | High Dimensional Box Plot | High Dimensional Box Plot | |
| 33 | High Dimensional Flow Plot | High Dimensional Flow Plot | |
| 34 | Horizontal axis parameter for conventional flow plot | NA | |
| 35 | Vertical axis parameter for conventional flow plot | NA | |
| 36 | Data transformation for conventional flow plot on horizontal axis | NA | |
| 37 | Data transformation for conventional flow plot on vertical axis | NA | |
| 38 | Create conventional flow plot | Conventional Flow Plot | |
| 39 | Show Clusters for Analysis | NA | |
Table 1: Overview of All Functions Present in the ExCYT GUI
| Name of Software/Package | ExCYT | CYT | FCS Express | flowCore | openCyto | FlowMeans |
| Program Type | Matlab | Matlab | Stand-Alone Application | R | R | R |
| Price to User | Free | Free | $1,000 | Free | Free | Free |
| Graphical User Interface | Yes | Yes | Yes | No | No | No |
| Dimensionality Reduction Techniques | t-SNE | t-SNE,PCA | t-SNE, PCA, SPADE | none | none | none |
| Clustering Algorithms | K-Means DBSCAN Hierarchical Clustering Self-Organized Map Multiple Network-Graph Based Methods GMM – EM GMM – Variational Bayesian Inference |
K-Means GMM – EM Single Network-Graph Based Method (Phenograph) |
K-Means | none | automation of manual gating workflow | K-Means |
| Ability to Sort/Filter Clusters | Yes | No | No | No | No | No |
| High Dimensional Flow Plots | Yes | No | No | No | No | No |
Table 2: Overview of Software-assisted Flow Cytometry Analysis Solutions
Discussion
Here we present ExCYT, a novel graphical user interface running MATLAB-based algorithms to streamline analysis of high-dimensional cytometry data, allowing individuals with no background in programming to implement the latest in high-dimensional data analysis algorithms. The availability of this software to the broader scientific community will allow scientists to explore their flow cytometry data in an intuitive and straightforward workflow. Through conducting t-SNE dimensionality reduction, applying a clustering method, being able to sort/filter through these clusters quickly, and make flexible, customizable heatmaps and high-dimensional flow/box plots, scientists will be able to not only understand the uniquely defined subpopulations in their samples but will be able to create visualizations that are intuitive and easily understood by their colleagues.
While the program is flexible in handling a variety of data types (conventional flow cytometry vs mass cytometry), there are a few considerations for optimal utility of the program. The first of these is regarding the data quality, specifically of flow cytometry data. Proper compensation and resolution of overlapping emission spectra is of paramount importance. Poorly compensated data can inadvertently lead to false co-associations of markers and formation of clusters that are not of true biological significance. Therefore, it is highly advisable that the input data is of sound quality before proceeding with the t-SNE analysis and further downstream analysis. Furthermore, use of the automatic compensation algorithm implemented in ExCYT requires clear single stains for all channels in order to accurately calculate the compensation parameters.
Another important consideration for use of ExCYT is when concatenating multiple FCS files into one analysis (as demonstrated in this manuscript), they must be comparable across all channels. First, this means that the same panel needs to be used across all samples and that there is no drift between samples across all channels. For example, if one were to read two samples on separate days and stained CD8 in FITC on both days but the voltage of the cytometer was set differently on one day resulting in a slightly shifted CD8 population, one could generate false clusters in the downstream analysis, as this shift was generated as a function of instrument variation and not due to biological significance. While future versions of ExCYT may be able to normalize samples to their single stains, at this point, careful consideration must be made that FCS files can be compared to each other before importing them into ExCYT.
Finally, the process of clustering is not one that is absolute/rigid. Different clustering algorithms and parameters can generate different clustering solutions. Whether the solution of the algorithm is appropriate is for the user to determine by synthesizing their understanding of the biology with the clustering solution. For example, when understanding the immune environment of tumors, one may be interested in macroscopic clusters (i.e., T cells vs B cells vs Myeloid cells) while another may be interested in subpopulations of macroscopic clusters. The resolution of the clusters is determined by the user and therefore, no single clustering solution is 'correct.' This is one of the main advantages of using the high dimensional flow plots available in ExCYT. The ability to visualize the distribution of a given cluster across all channels can help the user determine whether they have clustered in not only a biologically relevant way but in a way that is relevant to the scientific question being asked in the experiment. While our goal is to provide a plethora of methods used in the literature to cluster high-dimensional flow cytometry data while providing additional methods of clustering, we recommend using methods such as k-means and DBSCAN to explore the data via quickly iterating on cluster number and size and move towards network-graph and gaussian-mixed model approaches for more robust but more time-consuming approaches.
Given these considerations, ExCYT is still a highly flexible and valuable tool for exploring high dimensional cytometry data, and offers unique/differentiating features than other available packages available to conduct this type of analysis (Table 2). First, ExCYT differentiates itself over most flow cytometry analysis approaches utilizing dimensionality reduction and clustering algorithms by its ability to be used without any scripting/programming knowledge. Additionally, by aggregating many clustering algorithms cited throughout the literature, we believe we provide the most options for clustering data. Finally, our unique feature of cluster filtration and sorting along with display via novel high dimensional flow plots, allows users to explore the characteristics of their clusters quickly and efficiently, making the process of 'discovering' rare subpopulations simple and efficient.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors have no acknowledgements.
Materials
| Desktop | SuperMicro | Custom Build | Computer used to run analysis |
| MATLAB | Mathworks | N/A | Software used to develop ExCYT |
References
- Benoist, C., Hacohen, N. Flow cytometry, amped up. Science. 332 (6030), 677-678 (2011).
- Ornatsky, O., et al. Highly multiparametric analysis by mass cytometry. Journal of immunological methods. 361 (1), 1-20 (2010).
- Tanner, S. D., et al. Flow cytometer with mass spectrometer detection for massively multiplexed single-cell biomarker assay. Pure and Applied Chemistry. 80 (12), 2627-2641 (2008).
- Maecker, H. T., et al. Standardization of cytokine flow cytometry assays. BMC immunology. 6 (1), 13 (2005).
- Brazma, A., Vilo, J. Gene expression data analysis. FEBS letters. 480 (1), 17-24 (2000).
- Pyne, S., et al. Automated high-dimensional flow cytometric data analysis. Proceedings of the National Academy of Sciences. 106 (21), 8519-8524 (2009).
- Ge, Y., Sealfon, S. C. flowPeaks: a fast unsupervised clustering for flow cytometry data via K-means and density peak finding. Bioinformatics. 28 (15), 2052-2058 (2012).
- Venkatesh, V. Determinants of perceived ease of use: Integrating control, intrinsic motivation, and emotion into the technology acceptance model. Information systems research. 11 (4), 342-365 (2000).
- Bagwell, C. B., Adams, E. G. Fluorescence spectral overlap compensation for any number of flow cytometry parameters. Annals of the New York Academy of Sciences. 677 (1), 167-184 (1993).
- Lavin, Y., et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 169 (4), 750-765 (2017).
- Chevrier, S., et al. An immune atlas of clear cell renal cell carcinoma. Cell. 169 (4), 736-749 (2017).
- Hartigan, J. A., Wong, M. A. Algorithm AS 136: A k-means clustering algorithm. Journal of the Royal Statistical Society. Series C (Applied Statistics). 28 (1), 100-108 (1979).
- Ester, M., Kriegel, H. P., Sander, J., Xu, X. Density-based spatial clustering of applications with noise. International Conference Knowledge Discovery and Data Mining. 240, (1996).
- Levine, J. H., et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 162 (1), 184-197 (2015).
- Blondel, V. D., Guillaume, J. L., Lambiotte, R., Lefebvre, E. Fast unfolding of communities in large networks. Journal of statistical mechanics: theory and experiment. 2008 (10), P10008 (2008).
- Le Martelot, E., Hankin, C. Fast multi-scale detection of relevant communities in large-scale networks. The Computer Journal. 56 (9), 1136-1150 (2013).
- Newman, M. E. Fast algorithm for detecting community structure in networks. Physical review E. 69 (6), 066133 (2004).
- Hespanha, J. P. . An efficient matlab algorithm for graph partitioning. , 1-8 (2004).
- Moon, T. K. The expectation-maximization algorithm. IEEE Signal processing. 13 (6), 47-60 (1996).
- Bishop, C. M. . Pattern recognition and machine learning. , (2006).

