Single-cell Quantitation of mRNA and Surface Protein Expression in Simian Immunodeficiency Virus-infected CD4+ T Cells Isolated from Rhesus macaques
Summary
Described is a methodology to quantitate the expression of 96 genes and 18 surface proteins by single cells ex vivo, allowing for the identification of differentially expressed genes and proteins in virus-infected cells relative to uninfected cells. We apply the approach to study SIV-infected CD4+ T cells isolated from rhesus macaques.
Abstract
Single-cell analysis is an important tool for dissecting heterogeneous populations of cells. The identification and isolation of rare cells can be difficult. To overcome this challenge, a methodology combining indexed flow cytometry and high-throughput multiplexed quantitative polymerase chain reaction (qPCR) was developed. The objective was to identify and characterize simian immunodeficiency virus (SIV)-infected cells present within rhesus macaques. Through quantitation of surface protein by fluorescence-activated cell sorting (FACS) and mRNA by qPCR, virus-infected cells are identified by viral gene expression, which is combined with host gene and protein measurements to create a multidimensional profile. We term the approach, targeted Single-Cell Proteo-transcriptional Evaluation, or tSCEPTRE. To perform the method, viable cells are stained with fluorescent antibodies specific for surface markers used for FACS isolation of a cell subset and/or downstream phenotypic analysis. Single cells are sorted followed by immediate lysis, multiplex reverse transcription (RT), PCR pre-amplification, and high throughput qPCR of up to 96 transcripts. FACS measurements are recorded at the time of sorting and subsequently linked to the gene expression data by well position to create a combined protein and transcriptional profile. To study SIV-infected cells directly ex vivo, cells were identified by qPCR detection of multiple viral RNA species. The combination of viral transcripts and the quantity of each provide a framework for classifying cells into distinct stages of the viral life cycle (e.g., productive versus non-productive). Moreover, tSCEPTRE of SIV+ cells were compared to uninfected cells isolated from the same specimen to assess differentially expressed host genes and proteins. The analysis revealed previously unappreciated viral RNA expression heterogeneity among infected cells as well as in vivo SIV-mediated post-transcriptional gene regulation with single-cell resolution. The tSCEPTRE method is relevant for the analysis of any cell population amenable to identification by expression of surface protein marker(s), host or pathogen gene(s), or combinations thereof.
Introduction
Many intracellular pathogens rely on host cell machinery to replicate, often altering host cell biology or targeting very specific subpopulations of host cells to maximize their chances of propagation. As a result, cell biological processes are commonly disrupted, with deleterious consequences for the overall health of the host. Understanding the interactions between viruses and the host cells in which they replicate will elucidate disease mechanisms that may aid in the development of improved therapies and strategies to prevent infection. Direct analytic tools that enable the study of host-pathogen interactions are essential toward this end. Single-cell analysis provides the only means to unambiguously attribute a cellular phenotype to a particular genotype, or infection status1. For example, pathogenic infections frequently induce both direct and indirect changes in host cells. Therefore, distinguishing infected cells from their uninfected counterparts is necessary to attribute host cell changes to either direct infection or secondary effects, such as generalized inflammation. Moreover, for many pathogens, like SIV and human immunodeficiency viruses (HIV), host cell infection proceeds through multiple stages, such as early, late, or latent, each of which may be characterized by distinct gene and protein expression profiles2,3,4,5. Bulk analyses of cell mixtures will fail to capture this heterogeneity6. By contrast, highly multiplexed single-cell analyses able to quantify the expression of both viral and host genes offer a means to resolve infection-specific cellular perturbations, including variations across infection stages. Further, analyzing host-pathogen interactions in physiologically relevant settings is critical for the identification of events that occur in infected organisms. Thus, methods that can be applied directly ex vivo are likely to best capture in vivo processes.
SIV and HIV target CD4+ T cells, in which they counteract host antiviral "restriction" factors and downregulate antigen presenting molecules to establish productive infection and avoid immune surveillance7,8,9,10,11. Without treatment, the infection results in massive loss of CD4+ T cells, ultimately culminating in acquired immunodeficiency syndrome (AIDS)12. In the setting of antiretroviral therapy, latently infected cell reservoirs persist for decades, posing a formidable barrier to curative strategies. Understanding the properties of in vivo HIV/SIV-infected cells has the potential to reveal host cell features instrumental in pathogenesis and persistence. However, this has been highly challenging, primarily due to the low frequency of infected cells and lack of reagents able to readily identify them. Cells that transcribe viral RNA, are estimated to be present at 0.01–1% of CD4+ T cells in blood and lymphoid tissue13,14,15. Under suppressive therapy, latently infected cells are even less frequent at 10-3–10-7 16,17,18. Viral protein staining assays that work well for studying in vitro infections, such as for intracellular Gag, are suboptimal due to background staining of 0.01–0.1%, similar to or greater than the frequency of infected cells13,14. Surface staining for Env protein using well-characterized SIV/HIV Env-specific monoclonal antibodies has also been proven to be difficult, likely for similar reasons. Recently, novel tools aim to improve the detection of cells expressing Gag by either incorporating assays specific for gag RNA or by using alternative imaging technologies14,15,19. However, such approaches remain limited in the number of quantitative measurements performed on each cell.
Here, we describe methodology that (1) identifies single virus-infected cells directly ex vivo by sensitive and specific viral gene quantitative qPCR and (2) quantifies the expression of up to 18 surface proteins and 96 genes for each infected (and uninfected) cell. This methodology combines single-cell surface protein measurement by FACS followed by immediate cell lysis and gene expression analysis using multiplexed targeted qPCR on the Biomark system. The integrated fluidic circuit (IFC) technology allows multiplexed quantitation of 96 genes from 96 samples simultaneously, accomplished by a matrix of 9,216 chambers in which the individual qPCR reactions are performed. The live cell FACS sorting records high-content protein abundance measurements while preserving the entire transcriptome for analysis performed immediately downstream. To identify virus-infected cells, assays specific for alternatively spliced and unspliced viral RNAs (vRNA) are included in the qPCR analysis, along with a panel of user-defined assays totaling up to 96 genes, the maximum number of assays currently accommodated in the IFC. The gene expression and protein information collected for each cell are linked by well position. We previously reported results from this analysis elsewhere20. Here, we provide more detailed methodological guidelines as well as further descriptive phenotyping of SIV-infected CD4+ T cells.
This approach, which we term tSCEPTRE, can be applied to the suspensions of any viable cell population reactive to fluorescently labeled antibodies and expressing a transcriptome compatible with available qPCR assays. For example, it can be used for characterizing differential gene and protein expression in rare cells or cells not readily distinguished by surface protein markers. The sample preparation relies on a standard staining protocol using commercially available antibodies. Cytometers with single-cell sorting capability are also commercially available, but additional biosafety precautions are required for processing infectious live cells. Recording the single-cell protein expression profile for each cell by well position, referred to herein as indexed sorting, is a common feature of commercially available FACS sorting software. Computational analysis of differentially expressed host genes among cell populations of interest is not described here, but references are provided to previously published methods.
Protocol
NOTE: A schematic of the protocol workflow is shown in Figure 1. It consists of three principal steps: FACS, RT and cDNA pre-amplification, and qPCR for up to 96 genes simultaneously. Two versions of the protocol, sorting cells in limiting dilutions and sorting single cells, are described in greater detail in step 5 and step 6, respectively. These strategies address different research questions but follow similar procedures.
1. Prerequisite or Prior Analyses
- Validate all gene expression assays to be used as previously described6.
NOTE: This step is done well in advance of the experiment date. Validating all assays, commercial and custom, is required to ensure efficient and linear amplification of relevant RNA down to the single-cell level. Many commercially available and custom assays fail to meet these specifications. Processing and automated curve fitting for simultaneous qualification of expression assays of up to 96 genes are provided in Supplemental Coding Files 1–5, but individual assays can be qualified using R2 and slope of the linear fit. Representative successful and failed assay qualification plots are shown in Figure 2. - Develop a flow cytometric panel of antibodies to stain cell surface markers of interest.
- Titrate antibodies by staining a relevant sample, for example, rhesus peripheral blood mononuclear cells (PBMCs), with each antibody. Start with 20 µL of antibody per test in 100 µL staining reaction and create eight two-fold serial dilutions. Identify the optimal concentration that exhibits the maximum staining intensity while maintaining a clear separation between the negative and positive populations.
- Evaluate the combined staining on additional cell sample(s) using the mixture of all antibodies at the optimal concentration determined in step 1.2.1. Ensure that the staining is similar to that observed for individual antibody stains. If the staining is less than what is observed when any antibody was used in isolation, consider alternative fluorochrome conjugates to replace such antibodies.
2. Gene Expression Assay Preparation
- Combine 96 gene expression assays into an RNase/DNase-free 1–15 mL tube (size may vary with the number of sort plates). The panel of assays used in this study is specified in Table 1 and Table 2. The resulting material is referred to as the “Assay Mix”. Add each assay to a final concentration of 180 nM of forward and reverse primers. Add DNA Suspension Buffer to achieve the appropriate dilution of the Assay Mix.
NOTE: For practical purposes, custom (user-generated) assay stocks may be prepared at 18 µM to be consistent with the concentration of commercially available “20x” gene expression qPCR assays (Table of Materials). 18 µM mixes of custom forward and reverse primer for each gene are made from stock solutions in the DNA Suspension Buffer. Commercially available assays (Table of Materials) also include probes, but probes are not required for RT or cDNA pre-amplification and can thus be omitted for custom assays. It is recommended to include one or more housekeeping genes for use in quality control for assessing efficiency of sorting, cell recovery, and cDNA synthesis. Use of random primers to generate cDNA has not been determined, but is expected to be less efficient than gene-specific primers. - Prepare the 2x assay plate for use in step 7.1 (multiplex qPCR). For each 96 x 96 chip array anticipated, pipette 6 µL of each assay into each designated well of a 96-well PCR plate. For example, for 5 chips, 30 µL of each assay will occupy a single well in the 96-well plate. If using 96 assays, each well of the 96-well plate will contain an assay. Seal the plate with adhesive seal.
Note: Ideally, steps 2.1 and 2.2 are performed simultaneously, to avoid multiple freeze-thaw cycles for gene expression assays. All genes within the assay plate must have also been present in the Assay Mix (step 2.1). Both the assay mix and the assay plate can be stored at -20 °C or 4 °C for long- or short-term use, respectively.
3. Surface Stain Viable Cells
NOTE: Intracellular staining, permeabilization, and fixation are not compatible with this method as they compromise RNA.
- Prepare the compensation samples by adding each antibody listed in the Table of Materials to 40 µL of compensation beads at 2.5-fold higher concentration than that used for cell staining. Incubate for 20 min at 25 °C protected from light. Add 3 mL of PBS to the beads and centrifuge at 500 x g for 3 min at 25 °C. Aspirate the PBS and resuspend the beads in ~300 µL of PBS.
- Prepare the flow cytometric cell sorter for sample processing: acquire compensation tubes, create compensation matrix, and apply matrix to the acquisition files for the experimental specimens.
- Prepare the master mix of fluorescent antibody cocktail by combining the appropriate volume of each antibody as specified in the Table of Materials, in a 1.5 mL amber tube for all samples to be stained. Vortex and centrifuge the cocktail at 21,000 x g for 2 min at 25 °C to pellet the antibody aggregates.
Note: Antibodies used here are listed in the Table of Materials. - Thaw the cryopreserved cells in a 37 °C water bath for 2 min. Add 0.5–2 mL of the cell suspension to 12 mL of PBS in a 15 mL tube, centrifuge at 500 x g for 3 min at 25 °C, and aspirate the PBS. Resuspend in 3 mL of PBS and transfer to a 5 mL polystyrene tube. Centrifuge as above and aspirate the PBS, leaving ~20 µL residual PBS.
NOTE: The staining temperature may be adapted to warmer or colder temperatures as needed for specific applications by modifying the antibody titration staining conditions accordingly (see step 1.2). - Resuspend up to 2 x 107 washed cells in 80 µL of antibody cocktail and incubate for 20 min at 25 °C protected from light. For samples exceeding 2 x 107 cells, increase the staining reaction volume accordingly to maintain <2 x 107 cells/100 µL.
- Wash the cells by adding 3 mL of PBS, centrifuging at 500 x g for 3 min, and aspirating the supernatant.
- Thoroughly resuspend the cells in 300–500 µL of PBS and filter by pipetting through a 35 µm nylon cell strainer cap. Keep the cells on ice and protected from light until the sort.
4. Prepare Cell Collection Plates, Perform FACS Sort, and Generate cDNA
- Combine the RT-preamp Reaction Mix components (Table 3) by pipetting into a single RNAse/DNAse-free sterile tube.
NOTE: This step can be performed prior to or during staining in step 3. The RT enzyme may be omitted here to determine the contribution of the DNA template to qPCR signal. - Use a multichannel pipette to dispense 10 µL of RT-preamp Reaction Mix into the desired number of 96-well PCR sort collection plates. Seal the plates with adhesive film, and place the plates on pre-chilled 96-well aluminum blocks.
- Establish the cell sorting gating scheme on the flow cytometer by acquiring data from approximately 20,000 cells of the stained sample. Ensure that the compensation matrix is applied to the collected data. Draw gates and define the gating tree that identifies the cell population(s) of interest to be isolated for gene expression analysis.
NOTE: The gating tree used for the collection of potential SIV vRNA+ cells is shown in Figure 3. - Enter the appropriate instrument settings to specify the number and subset of cells to be sorted into each well. Additional detailed instruction for sorting either a limiting cell dilution series or single cells are provided in steps 5 and 6, respectively.
- FACS sort the cells into prepared 96-well PCR collection plates. Remove the adhesive seal prior to sorting and replace with a fresh seal following the sort.
NOTE: Keep the plates on pre-chilled aluminum blocks at all times, including during the sort. - Immediately after the sort, vortex and centrifuge the collection plate at 2,000 x g for 1 min at 4 °C.
- Thermocycle the plate in a PCR machine with a preheated lid using the following conditions: 50 °C for 15 min (RT), 95 °C for 2 min, followed by 18 cycles of 95 °C for 15 s and 60 °C for 4 min (pre-amplification).
- Dilute the cDNA 1:5 by transferring 5 μL of cDNA into 20 µL of DNA suspension buffer in a new 96-well PCR plate. Diluted cDNA may be stored at 4 °C or -20 °C indefinitely at this point. The cDNA is now ready to be used as a template for qPCR (steps 5.2, 7.4).
NOTE: This dilution ensures that the primers present in the RT-preamp reaction do not contribute to downstream qPCR.
5. Variation A: FACS Sort Cells into a Limiting Dilution Series to Determine the Frequency of vRNA+ Cells or Perform the Experimental Quality Control
NOTE: Before performing a single-cell sort, it may be of use to determine the frequency of cells of interest, by sorting the cells into serial dilutions in replicate. This step also provides valuable quality control for sort efficiency, cell lysis, RNA recovery, and cDNA synthesis, as described in step 5.3. Prior determination of vRNA+ cell frequency allows for more accurate estimation of the number of single cells that must be sorted to achieve sufficient sample size for appropriately powered vRNA+ cell gene expression analysis.
- FACS sort the cells into the 96-well plates prepared as in steps 4.1–4.2, and collect 1–1,000 cells per well in multiple replicates.
NOTE: The number of replicate wells at each cell dilution is typically inversely associated with the cell concentration per well. When the infected cell frequency is well below 1%, cell dilutions should focus on 100–1,000 cells per well. An example sort plate map is provided in Figure 1, top left. Exceeding 1,000 cells per well should be avoided due to resulting increases in the reaction volume and interference with downstream cDNA synthesis and quantification. - Combine the qPCR reagents in a master mix solution in Table 4. For a 25 µL reaction volume, 22.5 µL of master mix is combined with 2.5 µL of diluted cDNA template from step 3.9. Perform the qPCR using standard cycling conditions (e.g., 94 °C for 5 min, followed by 40 cycles of 94 °C for 15 s and 60 °C for 1 min).
NOTE: Singleplex qPCR reactions using a conventional real-time qPCR instrument are recommended as an economical preliminary analysis of one or a few assays to demonstrate efficient cell sorting, RNA recovery, and cDNA synthesis. It may also be used to calculate the frequency of vRNA+ cells. Multiplex qPCR reactions using the Biomark are typically more appropriate for large-scale single-cell analyses. - For quality control, plot Et values (Et = Ctmax − Ct) versus numbers of cells sorted per well on a log10 scale and apply a linear regression analysis.
NOTE: Consistent replicates, linear regression slope of 3.3 (± 0.3), and R2 >0.9 are indicative of an efficient experiment. Examples of optimal and suboptimal sort, RT-preamp experiments are shown in Figure 4. - To determine the frequency of vRNA+ cells, plot cell numbers sorted per well on the x-axis (log10 scale) and the fraction of wells positive for vRNA at each cell dilution on the y-axis. For an example, see Figure 1 (lower left, Poisson distribution). Apply a linear regression model to the data to determine the number of cells that harbor one positive cell on average, corresponding to 63.2% of wells positive (0.632 on the y-axis)21. Convert this cell dilution number (x-axis intercept) into frequency expressed as a percentage. For example, one vRNA+ cell per 48 cells is equivalent to a frequency of 2.1%.
6. Variation B: FACS Sort Cells for Single-cell Analysis
- Follow steps 4.1–4.8, and specify one cell sorted per well using the flow cytometer’s index sort feature to create individual FCS files for each cell sorted, mapped by well position.
NOTE: If the number of sort collection plates exceeds the number of available thermocyclers, cycling can be stopped after the reverse transcriptase inactivation step (95 °C for 2 min) and cDNA can be stored at 4 °C until thermocyclers are available. In this case, commence pre-amplification at the first cycle of 95 °C for 15 s. - Optional: Create cDNA “pools” consisting of cDNA from user-defined batches of single cells to screen for rare cells of interest using residual undiluted cDNA. Transfer 2 µL of undiluted single-cell pre-amplified cDNA by multichannel pipette into a new 96-well plate. Repeat by pipetting 2 µL of cDNA from all additional single cells of a designated pool into the same well. Screen cDNA pools for gene(s) of interest (e.g., vRNA) to determine those that contain positive cells using conventional qPCR. Since pooling requires only a small aliquot of cDNA from each cell, the remaining ~8 µL of cDNA is still available for single-cell analyses.
NOTE: Pooling strategies are recommended prior to performing single-cell gene expression analyses in an effort to reduce the number of single cells interrogated by resource-intensive multiplex qPCR. It is appropriate for situations in which the cells of interest (e.g., SIV mRNA+) may be identified by a preliminary single-plex qPCR assay. Straightforward high-throughput strategies to create cDNA pools include combining 2 µL of undiluted cDNA from a collection sort plate’s rows (i.e., all 12 cells in row A) or columns (i.e., all 8 cells in column 1), into a single well in a new plate. To determine the best pooling strategy, consider the expected frequency of the cells of interest from step 5.4. For example, if 10% of cells are expected to be positive, pools comprised of six single-cell cDNA samples will frequently be negative and the cells represented in that pool can thus be excluded from downstream single-cell analyses.
7. Multiplex qPCR on the Biomark Platform
NOTE: This section may follow either version A or B described above. In the study described herein, it was applied exclusively to single-cell analysis.
- Prepare the qPCR assay plate by pipetting 4 µL of each assay from the 2x assay plate (prepared in step 2.2) into a new 96-well PCR plate containing 4 µL of assay loading reagent in each well. Maintain the assay plate at 4 °C.
NOTE: The assay plate is stable at 4 °C for up to one week and at -20 °C for one month. Thus, it may be useful to prepare sufficient material for multiple chips and store appropriately. - Dispense the control line fluids from the priming syringes into the two intake valves of the chip. Remove the protective plastic from beneath the plate. Place the chip on an IFC controller with the notched side at the A1 position. From the main menu, select the "Prime" script. Run the script.
- Prepare the Real-Time Reaction Mix by mixing 50 µL of sample loading reagent with 500 µL of PCR Master Mix (Table of Materials) for each microfluidic chip. Pipette 4.4 µL into each well of a new 96-well PCR plate, henceforth designated as the “sample plate”.
- Pipette 3.6 µL of the 1:5 diluted cDNA from step 4.8 into the sample plate containing the Real-Time Reaction Mix.
NOTE: If a PCR down-selection was performed to screen for rare (e.g., vRNA+) cells for downstream analysis as discussed in step 6.2, include only the cells represented in the positive pools. - Following the completion of chip priming, load the chip inlets by dispensing 5 µL from the assay plate into the corresponding well on the notched (assay) side of the chip, and 5 µL from the sample plate into the corresponding well on the other (sample) side of the chip. Insert the chip into the IFC controller and run the "Load mix" script.
- Transfer the chip to the Biomark platform to perform the multiplexed qPCR. Proceed with the instrument setup and qPCR programing following step-by-step instruction provided by the Real-Time PCR Analysis software and using the Gene Expression (GE) 96.96 Standard V.1 protocol with 40 cycles of PCR. Save the ChipRun file in a designated folder.
NOTE: Multiple chips may be run per day and over multiple days. - Analyze the qPCR data.
- Open the Real-time PCR Analysis Software. Open the "ChipRun.bml" file from the "File | Open" menu.
- Locate "Chip Explorer" and "Chip Run Summary" in the upper left corner of the software window. Identify three components of the Chip Run Summary: Analysis Views, Sample Setup, and Detector Setup.
- Click on "Detector Setup". Under "Task", click "New" and select container type "SBS plate", and container format "SBS96". Next to "Mapping", click on the … button, and select "M96-Assay-SBS96.dsp".
- Optional: Assign each Detector (assay) a number or name in the “name” section of each well by double-clicking on the 1st well. Move to the next well by pressing "F2".
- Click on "Sample Setup". Under "Task" next to "Mapping", click the … button, and select "M96-Sample-SBS96.dsp".
- Click on "Analysis views". Under "Task" in the "qPCR" tab, select "Baseline Correction for Linear (Derivative)", and "Ct Threshold Method for User (Detectors)". In the "Ct Thresholds" tab, check the "Initialize with Auto box". Click the "Analyze" button above.
- In the upper right quadrant of "Analysis Views", click on the second tab "Results Table". From the drop-down menu, select "Heat Map View". The heat map with data will appear.
- Optional: To ensure uniform ROX fluorescence across the chip, select "Image View" instead of "Heat Map View" from the same menu. In the second from the right window above the heat map, select "ROX". In the first from the right window, select one of 1–40 cycles. Click on the fourth from the right window to switch to black-and-white display of ROX fluorescence. An image will appear that informs splatters, particles, or defects on the chip. If the ROX uniformity is grossly obscured, re-run the chip.
- Under the heat map, click "Threshold" and "Log Graph". Adjust the Ct thresholds manually for each detector by clicking on assays (columns of the heat map) and dragging the threshold as necessary to intersect the amplification curves in the exponential phase. When done, click "Analyze".
- Export the qPCR data as a .csv file. Import the data into a spreadsheet or statistical analysis software (e.g., JMP) and map the results by sample and assay positions on the chip. Organize the cells into groups based on the expression of viral genes, by creating a new column and using a conditional formula. Under "Analyze", select "Fit Y by X", and plot gene expression versus group. Apply statistical analysis.
NOTE: Representative single-cell quantitative expression of four SIV RNA species is depicted in bivariate plots in Figure 5A. Quantitative host gene expression in SIV RNA+ cells is shown in Figure 5B. - Extract quantitative protein expression values from single-cell FACS data.
- Open the .fcs files from the FACS sort (step 6.1) corresponding to 96-well plate using FlowJo version 9. With the file name highlighted, select "Platform | Event Number Gate | Create indexed sort gates". Individual cells will appear displayed by row.
- Highlight all 96 cells (not rows) and select "Workspace | Export | Select all compensated fluors". Under "Data Type", select "FCS file", click "Export", and select a designated folder.
- Drag new .fcs files for individual cells into a new FlowJo workspace. Highlight all cells, click "Add Statistics" (the "Σ" button in the upper left corner) "| Mean| All fluor parameters".
- Open "Table Editor" by clicking the fourth button from the left in the upper left corner. Highlight all fluores of the first cell and drag them into the table editor window. In the table editor window, click the same button at the top "Create and View Table". This will create a table of 96 cells and a numeric parameter for each fluor.
- Copy the output into a database software (e.g., MS Excel, JMP), by either copying/pasting or by clicking "Save and Launch Application" (fourth from the left button above the table).
NOTE: This procedure is specific for FlowJo version 9. FlowJo version 10 uses a different procedure to import indexed data. Indexed flow data can also be copied/pasted into JMP directly from .csv files created by the cell sorter.
- Merge single-cell FACS data and qPCR data by the plate number and well position. Perform graphical and statistical analyses on the combined single-cell gene (qPCR) and protein expression (FACS) data.
Note: Examples of single-cell combined qPCR and FACS data are shown in Figure 6 (host surface protein expression profiles for SIV-infected, spliced vRNA+ rhesus macaque cells), Figure 7 (CD4 gene expression versus surface CD4 protein expression in spliced vRNA+ rhesus macaque cells), and previously published20. To identify differentially expressed genes in cell population(s) of interest, single-cell analysis methods described previously are recommended20,22,23,24, which account for the proportion of cells positive for a gene as well as the continuous gene expression value.
Representative Results
The workflow for the entire protocol is depicted in Figure 1. It consists of two variations defined by the number of cells sorted: either limiting dilution or as single cells, as described in the text. Examples of primer-probe qualification analyses on 2-fold serial RNA dilutions are shown in Figure 2. The gating strategy to identify potential SIV+ cells is shown in Figure 3. A successful, suboptimal and failed quality control qPCR for the housekeeping gene GAPDH on FACS-sorted cells in limiting dilution are shown in Figure 4. Single-cell quantitative expression of four SIV RNA species and rhesus macaque genes that were differentially expressed in infected cells are shown in Figure 5. Representative bivariate, histogram, and scatterplots depict the surface protein expression profile of SIV RNA+ CD4+ T cells measured by flow cytometry in Figure 6. The trivariate bubble plot (Figure 7) displays the relationship between surface CD3 or CD4 protein, CD3 or CD4 mRNA, and quantitative viral gene (tat/rev) expression in single cells.
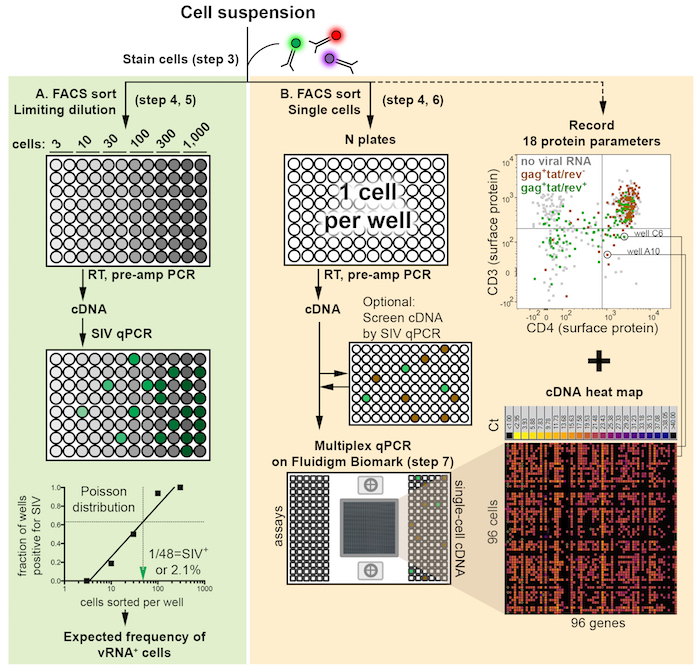
Figure 1: Schematic of experimental workflow illustrates three major components: flow cytometric sort, reverse transcription plus PCR-based pre-amplification of cDNA (RT, pre-amp), and qPCR. The sort can be performed as either a limiting dilution (A, green backdrop) or as single cells (B, orange backdrop). Immediately following the FACS sort, cells are lysed and RNA is reverse transcribed into cDNA and pre-amplified (RT, pre-amp PCR) to prepare qPCR template. Limiting dilution sorts determine the frequency of cells positive for viral RNA using Poisson distribution statistics as well as experimental efficiency and sample recovery. The green arrow head indicates the estimated number of cells sorted per well containing one cell positive for a viral gene (corresponding to a 63.2% probability of such wells being gene positive), which is converted into a cell frequency. Frequency estimates may be used to inform the number of single cells collected in a subsequent sort (B). Indexed single-cell FACS sorting deposits one cell per well and generates data files for each cell annotated by well position within the 96-well plate. Single-cell qPCR is performed in multiplex for 96 genes simultaneously. Combining the surface protein (FACS) and mRNA expression allows for profiling of individual cells (right column). An optional qPCR for a viral gene may be performed between the pre-amplification PCR and multiplexed qPCR (B, middle) to screen single cells or pools of single-cell cDNA to down-select viral RNA+ cells or pools for multiplex qPCR analysis. The heat map illustrates gene expression (Ct value) for 96 assays (columns) and 96 single cells (rows). Please click here to view a larger version of this figure.
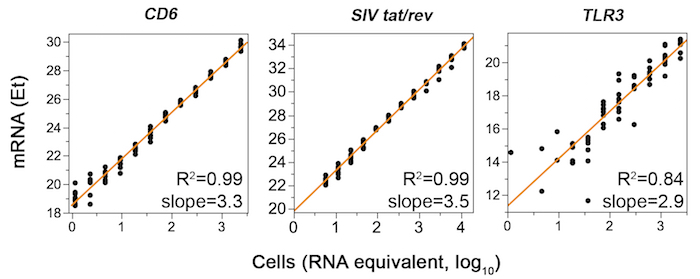
Figure 2: Representative qPCR data from primer qualification experiments are shown for successful (left and middle) and failed (right) commercially available assays (CD6, SIV tat/rev, and TLR3, respectively). For CD6 and TLR3, RNA was extracted from 106 FACS-sorted rhesus macaque PBMC CD4+ T-cells using a commercial kit. Eight replicates of a twelve-point RNA two-fold dilution series (0.023-48 ng RNA, corresponding to 1.2–2,400 cell equivalents assuming 20 pg RNA per CD4+ T-cell) were subjected to RT-preamp and qPCR. For SIV tat/rev, RNA was extracted from rhesus macaque PBMCs infected in vitro with SIVmac239. RNA dilutions were prepared spanning RNA equivalents of 6–12,000 cells. Et (40-Ct) values, which increase with gene expression, are plotted versus estimated cell numbers. Dilution series exhibiting R2 >0.97 and slope of 3.32 ±0.3 indicate successful primer qualification. Please click here to view a larger version of this figure.
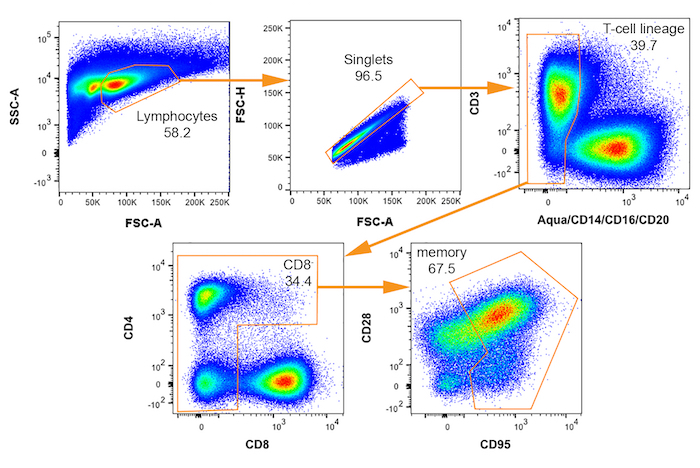
Figure 3: Bivariate FACS plots with gating scheme for isolation of rhesus macaque cells potentially infected by SIV. Sequential gates for selecting memory (CD95+) CD4+ T cells are shown from upper left to lower right with each population name indicated. Percent of parent plot that falls within each gate is indicated. Please click here to view a larger version of this figure.
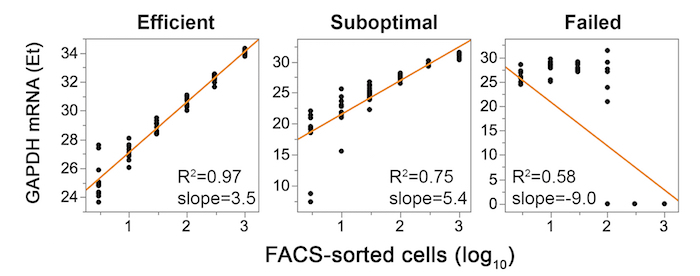
Figure 4: Representative experimental validation of qPCR performed on FACS-sorted cells in limiting dilution using GAPDH housekeeping gene for three independent experiments: successful (left), suboptimal (middle), and failed (right). Replicates of 10-100 cells per well should span no more than 2 Ets, and 300–1,000 cell wells within 1 Et. The linear regression slope should be 3.32 ± 0.3, R2 >0.95. Failure to achieve these specifications indicates technical difficulties in steps 1, 2, 3, or a combination thereof. Please click here to view a larger version of this figure.
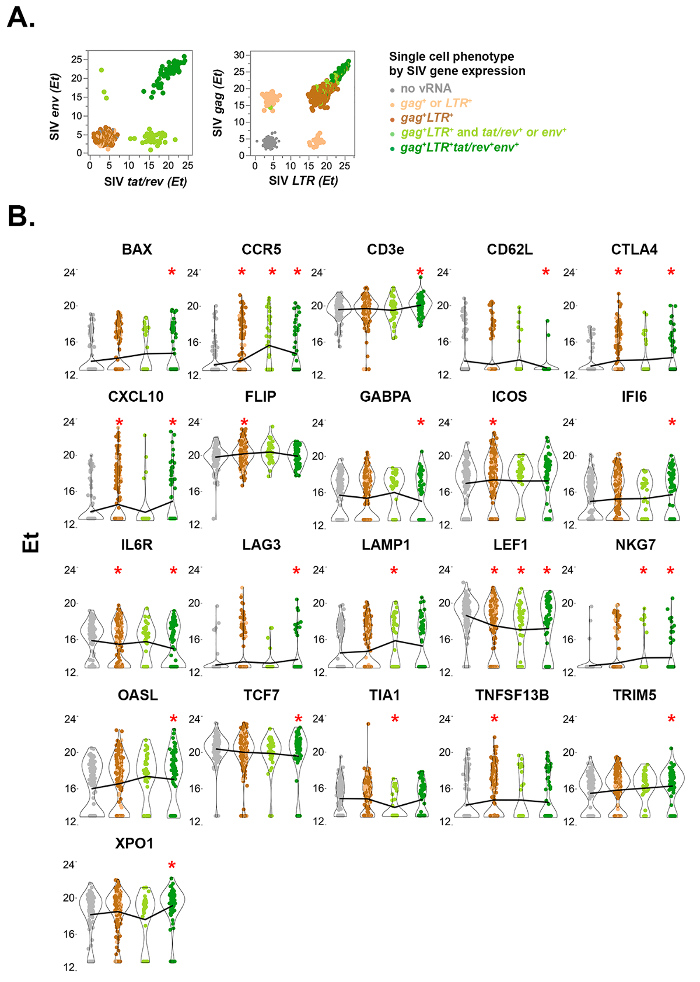
Figure 5: Single-cell quantitative viral and host gene expression in FACS-sorted rhesus macaque CD4+ T cells from mesenteric lymph node 10 days post-SIVmac251 infection. (A) Viral gene expression bivariate plots of multiply-spliced (tat/rev) and singly-spliced (env) SIV mRNA by single cells (left). Tat/rev+env– cells (light green cells along x-axis) express fewer copies of tat/rev RNA than the env+ cells (dark green), consistent with an early stage of infection and prior to Rev protein-mediated stabilization and nuclear export of partially spliced vRNAs such as env. Cells that do not express spliced viral RNA are depicted in grey (no viral RNA), brown (gag+ and LTR+), or tan (either gag+ or LTR+). Unspliced (gag+) and total (LTR+) SIV mRNA expression is shown for the same cells (right). High abundance of unspliced gag RNA in tat/rev+env+ cells is consistent with late stage productive infection during which abundant genomic RNA is expressed for packaging into budding virions. (B) Violin plots of rhesus macaque genes differentially expressed in at least one subset of SIV-infected cells compared to uninfected cells (gray). Statistical analysis was performed as described previously20,22,23,24. Asterisk indicates false discovery rate < 10% in combined likelihood ratio test comparisons relative to uninfected cells. Line connects mean values across cell groups for each gene. This figure has been modified from Bolton et al.20 Please click here to view a larger version of this figure.
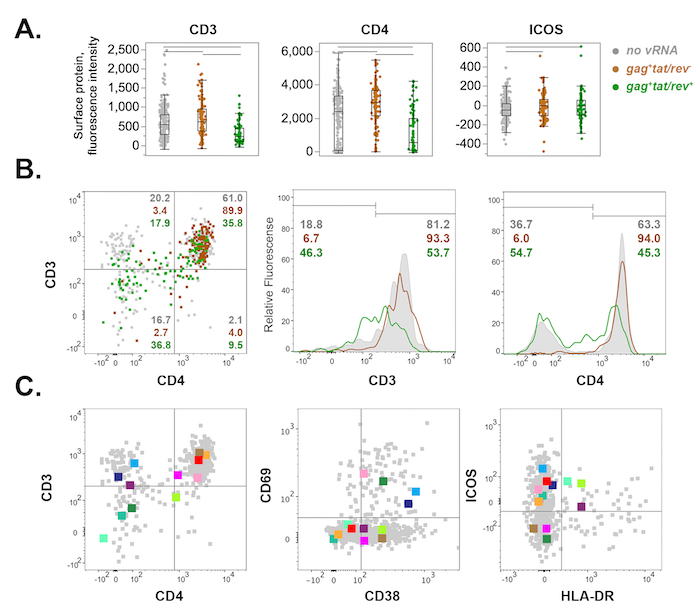
Figure 6: Representative host surface protein expression profiles of FACS-sorted cells from rhesus macaque mesenteric lymph node 10 days post-SIVmac251 infection. (A) Scatterplot display of CD3, CD4, and ICOS surface protein expression in uninfected (grey), gag+tat/rev– (brown), and gag+tat/rev+ cells (green). Fluorescence intensity is plotted for each cell (dot). Outlier box plots depict the interquartile range (IQR) and median (box), the furthest points within 1.5 x IQR from the box (whiskers), and potential outliers (disconnected points). Horizontal bars at the top indicate significant differences (p <0.05, nonparametric Wilcoxon rank test). (B) Bivariate and histogram display of surface CD3 and CD4 protein expression for cells shown in (A). Dot plot (left) indicates percentages of each cell population within a quadrant. CD3 and CD4 histograms (middle, right) depict surface protein downregulation among gag+tat/rev+ cells relative to uninfected and gag+tat/rev– cells. (C) Twelve representative tat/rev+ single cells from (A–B) are shown across three bivariate plots for surface expression of CD3/CD4 (left), CD69/CD38 (middle), and ICOS/HLA-DR (right). Please click here to view a larger version of this figure.
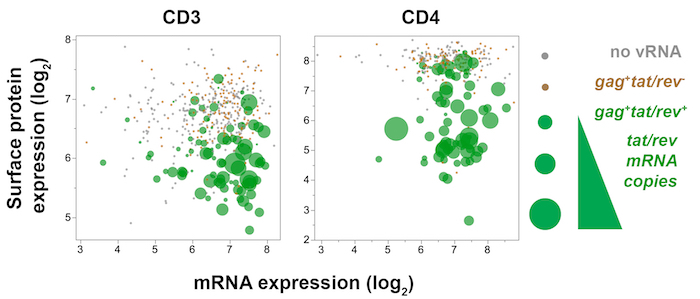
Figure 7: Trivariate plot displaying single-cell viral gene (tat/rev), host gene (CD3 or CD4), and host surface protein (CD3 or CD4) expression in SIV-infected memory CD4+ T cells from mesenteric lymph node 10 days post-SIVmac251 infection. CD4 protein expression (fluorescence) is plotted against CD4 mRNA (qPCR), while the amount of tat/rev expressed by each cell is reflected by dot size. In tat/rev+ cells (green), decreased surface CD4 and CD3 protein expression with sustained CD4 and CD3 transcripts, respectively, indicate that the surface protein expression is modulated downstream of gene expression. Please click here to view a larger version of this figure.
Table 1: Primers and probes used for the detection of SIV nucleic acids. When two sequences are indicated for a primer or probe, equimolar amounts of both sequences were used. Please click here to download this file.
Table 2: A 96-gene panel used for quantitation of amplicons on Biomark instrument. Four SIV assays are indicated with blue background. Please click here to download this file.
Table 3: Reaction mix used for reverse transcription and pre-amplification. Please click here to download this file.
Table 4: qPCR reaction mix used for real-time PCR performed on a Quant Studio 6 instrument. Please click here to download this file.
Supplemental Coding File 1. Instructions for qualifying gene expression assays. Please click here to download this file.
Supplemental Coding File 2: Sample Map template in JMP. Please click here to download this file.
Supplemental Coding File 3: Probe Map template in JMP. Please click here to download this file.
Supplemental Coding File 4: Primer Analysis script for JMP. Please click here to download this file.
Supplemental Coding File 5. Piecewise analysis script for JMP. Please click here to download this file.
Discussion
The protocol described here, termed tSCEPTRE, integrates single-cell surface protein quantitation by multiparameter flow cytometry with quantitative single-cell mRNA expression by highly multiplexed RT-qPCR. The union of these two technologies enables high-content snapshots of the combined transcriptional and protein profile of single cells in a high-throughput format. We use the method to identify heretofore elusive cells infected with SIV in vivo, and describe differentially expressed host genes and proteins. The protocol can be adapted for the study of any cell population of interest distinguishable by the expression of surface protein(s), mRNA, or a combination thereof. The described methods rely on accurate single-cell sorting and data recording by flow cytometry paired with commercially available qPCR reagents and multiplexed real-time PCR instrumentation. The output is sensitive, quantitative assessment of single-cell combined protein and gene expression data.
Other approaches linking protein and gene expression in single cells have been described1,25,26,27. Indexed FACS sorting followed by RNAseq, successfully applied to the characterization of hematopoietic stem cells, represents a particularly promising approach28. However, while RNAseq has several advantages over targeted gene expression analyses, namely unbiased full transcriptome analysis, it is subject to a higher multiple comparisons statistical cost and may be less sensitive for the quantification of low copy transcripts. Moreover, it is not currently economically feasible to perform RNAseq on thousands of cells in search of rare infected cells present at frequencies <1% (e.g., HIV, SIV). Other emerging single-cell technologies aim to generate a single readout, i.e., by FACS or PCR, for the detection of both proteins and nucleic acids in the same cell. These include the detection of proteins by fluorescent antibodies and mRNA by fluorescent oligonucleotide probes followed by FACS analysis (mRNA-Flow-FISH)14,15,29,30. Alternatively, proteins can be detected by pairs of antibody-oligonucleotide conjugates that are detected by qPCR in parallel with reversely transcribed mRNA31,32,33,34. These "hybrid" approaches provide simultaneous nucleic acid and protein measurements with single-cell resolution similar to tSCEPTRE, but they are limited where they are either not quantitative (mRNA-Flow-Fish) or may require customized antibody or mRNA probes. Our tSCEPTRE approach is quantitative for both protein and mRNA measurements and all reagents are commercially available, with the possible exception of pathogen-specific assays.
Special attention should be applied to several steps in the protocol prior to the analysis of experimental samples. First, high-parameter flow cytometry requires careful optimization of a panel of fluorescent antibodies to ensure sensitive detection and resolution35. Each antibody should be titrated individually to identify the concentration that achieves optimal separation and minimal background staining, followed by the assessment of staining when all antibody conjugates are used in combination. Second, experimental determination of the appropriate qPCR gene expression assay to measure each gene of interest is essential. Multiple commercially available assays are typically available for each gene, but in our experience, many are not quantitative at the single-cell level6. Thus, it is important to qualify all proposed assays on serially diluted RNA prior to use for quantitative gene expression. Optimization of these reagents is time consuming, but the value of reliable panels of antibody conjugates and gene expression assays that allows for sensitive detection of all molecules of interest justifies the time investment. In addition, priority should be given to commercially available assays containing probes that span exon-exon junctions (designated with suffix "m1") to improve specificity for mRNA, although alternate assays capable of detecting genomic DNA (suffix "s1" or "g1") are considered unlikely to influence gene expression results due to equivalent chromosomal copies in each cell. Preservation of RNA upstream of RT is critical and is achieved by keeping samples chilled, as noted throughout the protocol. Similarly, the duration of FACS sorting and the interval between sorting and RT-preamp should be kept to a minimum. For both limiting dilution and single-cell sorting, cDNA can be first analyzed by conventional qPCR to assess RNA recovery of highly expressed housekeeping genes. If the observed expression is not uniform among replicates of like cell numbers, or the slope of the linear fit for limiting dilutions fails validation, troubleshooting should be performed at cell sorting and downstream procedures.
Potential limitations of the approach described here include (1) detection of DNA in addition to RNA, particularly for assays not specific for mRNA splice junctions, and (2) restricted number of surface proteins and genes measured. However, DNA detection is unlikely to contribute substantially to differential host gene expression analysis for the reason stated above. Moreover, we directly measured the extent to which DNA contributes to the qPCR signal in this protocol by two approaches: a) excluding reverse transcriptase, and b) modifying the cell lysis protocol to enhance nuclear membrane lysis. In the absence of reverse transcriptase, both spliced and unspliced viral genes were still detected, but at significantly lower frequency than in the presence of reverse transcriptase (~6-fold and 2-fold reductions, respectively)20. Hence, the viral gene expression assays may overestimate the amount of viral RNA present in a cell due to the presence of cell-associated viral DNA. We attribute this finding to cytoplasmic RT products generated during host cell infection, known to occur for both spliced and unspliced SIV/HIV RNA originating from incoming virions36. Thus, it may be advisable to dedicate a portion of experiments to conditions lacking reverse transcriptase to quantitate DNA-derived template for some applications. It should also be noted that unspliced SIV/HIV RNA derived from incoming virus cannot be distinguished from de novo synthesized viral RNA. Second, we incorporated a nuclear lysis step into the RT-preamp protocol and observed an exponential increase in integrated SIV DNA (Alu-LTR) copies (Figure S3 of Bolton et al.)20. Genomic DNA is therefore unlikely to contribute substantially to the nucleic acid template extracted from the cell lysis method used here. Of note, the addition of a nuclear lysis step may be useful in future single-cell studies seeking to investigate SIV or other virus DNA-positive cells, including latently infected cells.
The number of surface proteins analyzed is dictated by the capability of the flow cytometric cell sorter. Current commercially available instruments do not exceed 30 parameters. Future studies employing more advanced flow cytometry will further broaden the protein profiling capability of this approach. The number of transcripts can also be extended beyond 96, provided supporting reagents (e.g., primers of higher concentration), equipment and instrumentation. Ultimately, emerging single-cell technologies that combine analysis of proteome (mass spectrometry), transcriptome (RNAseq), and genome (DNAseq) will supersede the targeted approaches for discovery research31,37,38,39. However, targeted qPCR will likely remain a valuable tool for validating such "omics" approaches as a gold standard for quantitative expression analyses.
Combined protein and transcription analysis by tSCEPTRE is a powerful tool for investigating rare or difficult to identify cells such as those harboring pathogens, containing oncogenes, or otherwise exhibiting an aberrant phenotype. New markers for transcriptionally active SIV/HIV-infected cells may be identified in this way, as well as the discovery of new mechanisms involved in the pathogenesis of these viral infections. Identification of latently infected cells will require further development of a protocol to asses viral DNA integrated in the host genome. Of note, the frequency of HIV/SIV infected cells is considerably lower in chronic viremic or treated infection, which will present a practical challenge in studying infected cells derived from these settings. Our approach lays the groundwork for assessing a previously intractable mechanism: that of post-transcriptional regulation at the single-cell level, and has broad applicability to host-pathogen interactions as well as more general cellular processes.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank the NIAID VRC Flow Cytometry Core and the MHRP Flow Cytometry Core facilities for maintenance and operation of FACS instruments and sorting equipment; Maria Montero, Vishakha Sharma, Kaimei Song for expert technical assistance; Michael Piatak, Jr. (deceased) for assistance with SIV qPCR assay design; and Brandon Keele and Matthew Scarlotta for SIV isolate sequences. The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense. Research was conducted under an approved animal use protocol in an AAALAC accredited facility in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
Materials
|
RNA extraction and PCR reagents and consumables |
|||
|
Genemate 96-Well Semi-Skirted PCR Plate |
BioExpress/VWR |
T-3060-1 |
|
|
Adhesive PCR Plate Seals |
ThermoFisher |
AB0558 |
|
|
Armadillo 384-well PCR Plate |
ThermoFisher |
AB2384 |
|
|
MicroAmp Optical Adhesive Film |
Applied Biosystems/ThermoFisher |
4311971 |
|
|
DEPC Water |
Quality Biological |
351-068-101 |
|
|
Glass Distilled Water |
Teknova |
W3345 |
|
|
Superscript III Platinum One-Step qRT-PCR Kit |
Invitrogen/ThermoFisher |
11732088 |
|
|
SUPERase-In Rnase Inhibitor |
Invitrogen/ThermoFisher |
AM2696 |
|
|
Platinum Taq |
Invitrogen/ThermoFisher |
10966034 |
|
|
dNTP Mix |
Invitrogen/ThermoFisher |
18427088 |
|
|
ROX Reference Dye (if separate from kit) |
Invitrogen/ThermoFisher |
12223012 |
|
|
DNA Suspension Buffer |
Teknova |
T0223 |
|
|
RNAqueous kit |
Invitrogen/ThermoFisher |
AM1931 |
|
|
TaqMan gene expression assays not listed in Table 2 |
|||
|
CD6 |
Applied Biosystems/ThermoFisher |
Hs00198752_m1 |
|
|
TLR3 |
Applied Biosystems/ThermoFisher |
Hs1551078_m1 |
|
|
Biomark reagents |
|||
|
Control Line Fluid Kit |
Fluidigm |
89000021 |
|
|
TaqMan Universal PCR Mix |
Applied Biosystems/ThermoFisher |
4304437 |
|
|
Assay Loading Reagent |
Fluidigm |
85000736 |
|
|
Sample Loading Reagent |
Fluidigm |
85000735 |
|
|
Dynamic Array 96.96 (chip) |
Fluidigm |
BMK-M-96.96 |
|
|
FACS reagents |
|||
|
SPHERO COMPtrol Goat anti-mouse (lambda) |
Spherotech Inc. |
CMIgP-30-5H |
|
|
CompBeads Anti-Mouse Ig,k |
BD Biosciences |
51-90-9001229 |
|
|
5 ml Polystyrene tube with strainer cap |
FALCON |
352235 |
|
|
Aqua Live/Dead stain |
Invitrogen/ThermoFisher |
L34976 |
dilute 1:800 |
|
Mouse Anti-Human CD3 BV650 clone SP34-2 |
BD Biosciences |
563916 |
dilute 1:40 |
|
Mouse Anti-Human CD4 BV786 clone L200 |
BD Biosciences |
563914 |
dilute 1:20 |
|
Mouse Anti-Human CD8 BUV496 clone RPA-T8 |
BD Biosciences |
564804 |
dilute 1:10 |
|
Mouse Anti-Human CD28 BV711 clone CD28.2 |
Biolegend |
302948 |
dilute 1:20 |
|
Mouse Anti-Human CD95 BUV737 clone DX2 |
BD Biosciences |
564710 |
dilute 1:10 |
|
Mouse Anti-Human CD14 BV510 clone M5E2 |
Biolegend |
301842 |
dilute 1:83 |
|
Mouse Anti-Human CD16 BV510 clone 3G8 |
Biolegend |
302048 |
dilute 1:167 |
|
Mouse Anti-Human CD20 BV510 clone 2H7 |
Biolegend |
302340 |
dilute 1:37 |
|
Anti-CD38-R PE clone OKT10 |
NHP reagent recource |
N/A |
dilute 1:100 |
|
Mouse Anti-Human CD69 BUV395 clone FN50 |
BD Biosciences |
564364 |
dilute 1:10 |
|
Mouse Anti-Human HLA-DR APC-H7 clone G46-6 |
BD Biosciences |
561358 |
dilute 1:20 |
|
Mouse Anti-Human ICOS Alexa Fluor 700 clone C398.4A |
Biolegend |
313528 |
dilute 1:80 |
|
Instruments |
|||
|
BioPrptect Containment Enclosure |
Baker |
||
|
BD FACS Aria |
BD Biosciences |
||
|
ProtoFlex Dual 96-well PCR system |
Applied Biosystems/ThermoFisher |
4484076 |
|
|
Quant Studio 6 qPCR instrument |
Applied Biosystems/ThermoFisher |
4485694 |
|
|
IFC controller HX |
Fluidigm |
IFC-HX |
|
|
Biomark HD |
Fluidigm |
BMKHD-BMKHD |
References
- Macaulay, I. C., Ponting, C. P., Voet, T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet. 33 (2), 155-168 (2017).
- Pasternak, A. O., Lukashov, V. V., Berkhout, B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 10, 41 (2013).
- Mailler, E., et al. The Life-Cycle of the HIV-1 Gag-RNA Complex. Viruses. 8 (9), (2016).
- Varmus, H. Retroviruses. Science. 240 (4858), 1427-1435 (1988).
- Frankel, A. D., Young, J. A. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 67, 1-25 (1998).
- Dominguez, M. H., et al. Highly multiplexed quantitation of gene expression on single cells. J Immunol Methods. 391 (1-2), 133-145 (2013).
- Tokarev, A., Guatelli, J. Misdirection of membrane trafficking by HIV-1 Vpu and Nef: Keys to viral virulence and persistence. Cell Logist. 1 (3), 90-102 (2011).
- Malim, M. H., Bieniasz, P. D. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2 (5), a006940 (2012).
- Sugden, S. M., Bego, M. G., Pham, T. N., Cohen, E. A. Remodeling of the Host Cell Plasma Membrane by HIV-1 Nef and Vpu: A Strategy to Ensure Viral Fitness and Persistence. Viruses. 8 (3), 67 (2016).
- Simon, V., Bloch, N., Landau, N. R. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 16 (6), 546-553 (2015).
- Kirchhoff, F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 8 (1), 55-67 (2010).
- Wigzell, H. Immunopathogenesis of HIV infection. J Acquir Immune Defic Syndr. 1 (6), 559-565 (1988).
- Reynolds, M. R., et al. Ex vivo analysis of SIV-infected cells by flow cytometry. Cytometry A. 77 (11), 1059-1066 (2010).
- Baxter, A. E., et al. Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals. Cell Host Microbe. 20 (3), 368-380 (2016).
- Grau-Exposito, J., et al. A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4+ T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients. MBio. 8 (4), (2017).
- Eriksson, S., et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9 (2), e1003174 (2013).
- Finzi, D., et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 278 (5341), 1295-1300 (1997).
- Chomont, N., et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 15 (8), 893-900 (2009).
- DeMaster, L. K., et al. A Subset of CD4/CD8 Double-Negative T Cells Expresses HIV Proteins in Patients on Antiretroviral Therapy. J Virol. 90 (5), 2165-2179 (2015).
- Bolton, D. L., et al. Combined single-cell quantitation of host and SIV genes and proteins ex vivo reveals host-pathogen interactions in individual cells. PLoS Pathog. 13 (6), e1006445 (2017).
- Quintans, J., Lefkovits, I. Precursor cells specific to sheep red cells in nude mice. Estimation of frequency in the microculture system. Eur J Immunol. 3 (7), 392-397 (1973).
- Finak, G., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
- McDavid, A., et al. Modeling bi-modality improves characterization of cell cycle on gene expression in single cells. PLoS Comput Biol. 10 (7), e1003696 (2014).
- McDavid, A., Finak, G., Gottardo, R. The contribution of cell cycle to heterogeneity in single-cell RNA-seq data. Nat Biotechnol. 34 (6), 591-593 (2016).
- Kok, Y. L., Ciuffi, A., Metzner, K. J. Unravelling HIV-1 Latency, One Cell at a Time. Trends Microbiol. , (2017).
- Rato, S., Golumbeanu, M., Telenti, A., Ciuffi, A. Exploring viral infection using single-cell sequencing. Virus Res. 239, 55-68 (2017).
- Wagner, A., Regev, A., Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 34 (11), 1145-1160 (2016).
- Nestorowa, S., et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 128 (8), e20-e31 (2016).
- Soh, K. T., et al. Simultaneous, Single-Cell Measurement of Messenger RNA, Cell Surface Proteins, and Intracellular Proteins. Curr Protoc Cytom. 75, 41-47 (2016).
- Kochan, J., Wawro, M., Kasza, A. Simultaneous detection of mRNA and protein in single cells using immunofluorescence-combined single-molecule RNA FISH. Biotechniques. 59 (4), 209-212 (2015).
- Stahlberg, A., Thomsen, C., Ruff, D., Aman, P. Quantitative PCR analysis of DNA, RNAs, and proteins in the same single cell. Clin Chem. 58 (12), 1682-1691 (2012).
- Assarsson, E., et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 9 (4), e95192 (2014).
- Fredriksson, S., et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 20 (5), 473-477 (2002).
- Genshaft, A. S., et al. targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 17 (1), 188 (2016).
- Mahnke, Y. D., Roederer, M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med. 27 (3), 469-485 (2007).
- Liang, C., Hu, J., Russell, R. S., Kameoka, M., Wainberg, M. A. Spliced human immunodeficiency virus type 1 RNA is reverse transcribed into cDNA within infected cells. AIDS Res Hum Retroviruses. 20 (2), 203-211 (2004).
- Stegle, O., Teichmann, S. A., Marioni, J. C. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 16 (3), 133-145 (2015).
- Wu, M., Singh, A. K. Single-cell protein analysis. Curr Opin Biotechnol. 23 (1), 83-88 (2012).
- Haque, A., Engel, J., Teichmann, S. A., Lonnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9 (1), 75 (2017).

