Detection and Isolation of Apoptotic Bodies to High Purity
Summary
A workflow using flow cytometry or differential centrifugation is developed to detect, quantify and isolate apoptotic bodies from an apoptotic sample to high purity.
Abstract
Apoptotic bodies (ApoBDs), microvesicles and exosomes are the key members of the extracellular vesicle family, with ApoBDs being one of the largest type. It has been proposed that ApoBDs can aid cell clearance as well as intercellular communication through trafficking biomolecules. Conventional approaches used for the identification and isolation of ApoBDs are often limited by the lack of accurate quantification and low sample purity. Here, we describe a workflow to confirm the induction of apoptosis, validate ApoBD formation, and isolate ApoBDs to high purity. We will also outline and compare fluorescence-activated cell sorting (FACS) and differential centrifugation based approaches to isolate ApoBDs. Furthermore, the purity of isolated ApoBDs will be confirmed using a previously establish flow cytometry-based staining and analytical method. Taken together, using the described approach, THP-1 monocyte apoptosis and apoptotic cell disassembly was induced and validated, and ApoBD generated from THP-1 monocytes were isolated to a purity of 97-99%.
Introduction
Apoptosis, a well-studied form of programmed cell death, is required to maintain physiological homeostasis and remove potentially harmful cells within the human body1. After the induction of apoptosis, apoptotic cells (ApoCells) can undergo a series of morphological changes and disassemble into small membrane-bound vesicles termed ApoBDs. Overall, this process is known as apoptotic cell disassembly and can be divided into 3 distinct steps based on morphology2,3. Step 1 (plasma membrane blebbing) is characterized by the formation of balloon-like structures on the cell surface known as blebs4,5. Step 2 (apoptotic protrusion formation) includes the formation of long membrane protrusions such as apoptopodia, beaded-apoptopodia and microtubule spikes6,7,8. Lastly, Step 3 (ApoBD formation) includes the fragmentation of the apoptotic protrusions and/or ApoCells to generate ApoBDs6,9. Previous findings have suggested a role of ApoBDs in aiding apoptotic cell clearance and mediating intercellular communication. For example, it is proposed that the fragmentation of an ApoCell into ApoBDs may generate small 'bite-sized' pieces that could be easily removed by surrounding phagocytes2,10,11. Furthermore, ApoBDs may harbor a series of biomolecules such as DNA, RNA and proteins, which may be trafficked to surrounding cells to facilitate cell-cell communication12,13,14. To functionally investigate these processes, it is vital to confirm three key parameters including (i) validation of apoptosis induction and ApoBD formation, (ii) isolation of ApoBDs, and (iii) confirmation of ApoBD purity.
Previously, a number of methods including flow cytometry and electron microscopy have been used to study apoptosis and ApoBDs15,16,17,18. However, ApoBD detection and quantification are often difficult or overlooked. For instance, the most routinely-used flow cytometry-based apoptosis assay employs annexin V (A5, a protein that binds the externalized 'eat-me' signal phosphatidylserine (PtdSer)) and nucleic acid stain propidium iodide (PI)19. However, by using this universal stain combination, analysis assumes that there are only three types of cell subsets (viable cells, ApoCells and necrotic cells) in the sample. Furthermore, though considered as "a gold standard" by many researchers for apoptosis, flow cytometry assays and subsequent data analysis often excludes ApoBDs through an initial gating step selecting FSC/SSCintermediate-high events. Therefore, we have recently developed a novel flow cytometry assay using A5 and TO-PRO-3, another nucleic stain that can be selectively taken up by caspase 3/7-activated pannexin 1 (PANX1) channels7,20. As caspase 3-induced PANX1 activation precedes PtdSer exposure at the early stage of apoptosis, TO-PRO-3 differentially stains apoptotic and necrotic cells. In addition, this approach combined with our novel gating strategy includes all acquired events during data analysis and as a result, six cell/particle subsets are identified, including: (i) viable cells (FSC/SSCintermediate/high, A5low, TO-PRO-3low), (ii) A5– early ApoCells (FSC/SSCintermediate/high, A5low, TO-PRO-3intermediate), (iii) A5+ ApoCells (FSC/SSCintermediate/high, A5high, TO-PRO-3intermediate), (iv) necrotic cells or late ApoCells (FSC/SSCintermediate/high, A5high, TO-PRO-3high), (v) ApoBDs (FSC/SSClow, A5intermediate, TO-PRO-3low/intermediate), and (vi) debris (FSC/SSClow, A5low, TO-PRO-3low)20. Our approach emphasizes the importance of analyzing all cells/particle subsets and, more importantly, the separation of ApoBDs from cells and debris20. Thus, this approach demonstrates an efficient technique to validate the induction of apoptosis and ApoBD formation simultaneously.
Traditionally, ApoBDs have been isolated through a variety of differential centrifugation approaches whereby ApoBDs can be separated from cells or other extracellular vesicles based on density. However, such centrifugation methods are often limited by low ApoBD purity, lack of a quantification step to confirm sample purity, and/or inability to separate cell type-specific ApoBDs17,21,22. Therefore, we recently developed two approaches, a FACS-based and a new differential centrifugation-based approach which can be coupled with our previously established flow cytometry method to validate the induction of apoptosis and sample purity23. ApoBD isolation via our FACS-based approach can enrich ApoBDs to up to 99% purity, and can be coupled with a variety of cell type-specific antibodies to isolate ApoBDs from mixed cell populations, tissue samples and bodily fluids23. Furthermore, our revised differential centrifugation approach demonstrates an efficient method to isolate ApoBDs to >90% purity23.
In this paper, we describe in detail our experimental procedure to validate apoptosis induction, and to detect and quantify ApoBD formation. The ApoBD isolation workflows using FACS-based and differential centrifugation-based methods are also elaborated and compared. The representative data demonstrate that the described methodology provides an effective cutting-edge tool for future ApoBD studies.
Protocol
1. Induction of Apoptosis
- Centrifuge cell sample at 300 x g for 5 min and discard supernatant to remove any pre-existing cell debris.
NOTE: When using adherent cells, seed cells in advance and wash with 1x phosphate-buffered solution (PBS) prior to apoptosis induction. - Determine cell number and collect cells.
NOTE: Depending on the assay post-isolation, we recommend a starting cell number of at least 1 x 107 cells. - Resuspend in complete media (respective medium containing 10% (vol/vol) fetal calf serum, 50 IU/mL penicillin, 50 µg/mL streptomycin mixture) for a final concentration of 1 x 106 cells/mL.
- Aliquot ~2 x 106 cells per well of a 6 well plate.
- To induce apoptosis, remove the plate lid and irradiate cells at 150 mJ/cm2 using a UV irradiator. This should take approximately 30-60 s.
NOTE: Prior to irradiation, ensure that ~0.5 x 106 cells are retained for the 'Untreated' cell control.
NOTE: Apoptosis can also be induced via other methods such as anti-Fas or serum starvation6. - Incubate at 37°C, 5% CO2 for 2-8 hours, depending on the cell line.
- Using a bench top light microscope, visualize cells to confirm the presence of apoptotic morphologies, such as blebbing, apoptopodia formation, and ApoBD formation.
NOTE: 40X magnification is sufficient - Using a P1000 pipette, pipette and collect apoptotic samples.
- Wash the plate with 1x PBS and combine with remaining sample to ensure maximum yield.
- Collect ~1/10th for the 'Whole Apoptotic Sample' (WAS) control.
- Collect the 'Untreated' sample.
- Centrifuge both the WAS and Untreated samples at 3,000 x g for 6 min.
- Resuspend in 1 mL of 1x PBS and set aside on ice.
- For ApoBD isolation, continue to either step 2 or 3 with the remaining apoptotic sample.
2. ApoBD Isolation via FACS
- Centrifuge the entire sample at 3,000 x g for 6 min.
- Remove the majority of the supernatant without disrupting the pellet.
- Resuspend in a staining solution containing 1 mL of 1x A5 binding buffer, 75 µL of A5-FITC, and 2 µL of TO-PRO-3 per 1×107 cells.
- Incubate sample in the dark at room temperature for 10 min.
- Add 1-2 mL of 1x A5 binding buffer and centrifuge sample at 3,000 x g for 6 min to remove excess stain.
NOTE: For mixed cell populations or tissue samples, perform an antibody staining step using a combination of cell type-specific markers in 1x A5 binding buffer and incubate on ice for 20 min (or as per manufacturer's protocol) before centrifugation at 3,000 x g for 6 min. - Resuspend sample pellet in 3 mL of FACS buffer (1x PBS, 1x A5 binding buffer, 10% FSC, 2 mM EDTA) per 1×107 cells.
- Filter through a 70 µm cell strainer into a round-bottom, polypropylene (flow cytometry) tube and keep samples on ice and in the dark.
- Turn on the FACS machine and perform standard set up using a 100 µm nozzle, perform the drop delay, and ensure a stable stream.
- Load the sample and set acquisition speed to ~1000 events/s.
- Adjust FSC, SSC, APC (TO-PRO-3) and FITC (A5) voltages and position events within the FACS plots to ensure populations can be clearly separated.
- Record 20,000 events.
- Set up a gating strategy as in part 4.
- In the sort layout, add the final ApoBD gate as the desired sorting population.
- Begin acquiring the sample and perform a test sort by collecting 5,000-10,000 ApoBDs into a new tube containing ~250 µL FACS buffer.
- Perform a system back flush and load sorted ApoBDs.
- Acquire and record test-sort ApoBDs.
- Check that the ApoBD purity is ~99% by comparing FSC (y-axis) vs A5 (x-axis events).
NOTE: A5 staining may reduce slightly when re-analyzing samples due to laser bleaching. - Once high purity is achieved, load original sample and continue sorting until the desired number of ApoBDs has been obtained.
NOTE: If necessary, dual sorting can be performed to simultaneously isolate ApoCells and ApoBDs.
NOTE: When sorting over a long period of time, we recommend incubating the collection tube at 4 °C. - Once sorting is complete, collect a small portion of post-sort ApoBDs, post-sort ApoCells, Untreated, and WAS to validate apoptosis and confirm post-sort purity.
NOTE: Although the test-sort and post-sort purity should not differ significantly, this is based on the stream settings and stability.
3. ApoBD Isolation via Differential Centrifugation
- Centrifuge the remaining apoptotic sample at 300 x g for 10 min.
- Collect the supernatant, leaving ~500 µL to avoid disrupting the cell pellet, and add into a new 15 mL conical tube.
- Remove the remaining 500 µL and resuspend the cell pellet in 2 mL of 1x PBS (this represents the 'ApoCell-enriched fraction'.
- Centrifuge the collected supernatant for 20 min at 3,000 g.
- Check for a pellet and carefully remove the supernatant (the supernatant may contain small extracellular vesicles including microvesicles and exosomes).
- Resuspend the pellet in 1 mL of 1x PBS (this represents the 'ApoBD-enriched' fraction)
- Collect 100 µL of each Viable, WAS, ApoCell-enriched, and ApoBD-enriched samples in a new round-bottom, polystyrene (flow cytometry) tube.
- Add 100 µL of stain containing 2x A5 binding buffer, 1:100 A5-FITC, and 1:1,000 TO-PRO-3
- Incubate at room temperature for 10 min in the dark.
- Analyze samples by flow cytometry, using the gating strategy as described above to validate the successful induction of apoptosis and purity of the ApoBD-enriched fraction.
4. Flow Cytometry Gating Strategy
- Plot TO-PRO-3 (y-axis) against FSC (x-axis) to separate necrotic cells (TO-PRO-3high) from all other non-permeabilized events (TO-PRO-3low/intermediate).
- Select all non-permeabilized events and plot SSC against A5. Gate two populations including population 1 (P1), SSCintermediate/high, A5low/intermediate cells and population 2 (P2), all other events.
- From P2, plot TO-PRO-3 against A5 and select A5intermediate/high events to exclude all cell debris.
- Select all A5 positive events and plot FSC against A5. Separate ApoBDs (FSClow) from ApoCells (FSCintermediate/high).
NOTE: When gating ApoBDs for ApoBD isolation via the FACS-based approach, we recommend a final step by selecting ApoBDs and comparing TO-PRO-3 to A5 and selecting all events. This ensures that the final sorting gate uses fluorescence rather than FSC/SSC parameters. - For viable cell analysis, select P1 and perform one of two gating strategies. For general viable cell analysis, plot FSC against A5 and select all FSCintermediate/high cells, therefore removing remaining cell debris. Alternatively, for in-depth analysis, viable cells can be separated from A5– early ApoCells by plotting TO-PRO-3 against FSC. Select TO-PRO-3low, FSCintermediate/high viable cells and TO-PRO-3intermediate, FSCintermediate/high A5– early ApoCells.
Representative Results
Using the procedure outlined here, THP-1 monocyte apoptosis was induced and ApoBDs were detected and isolated via either a FACS-based or a differential centrifugation approach (Figure 1). Firstly, apoptosis was induced by UV irradiation and samples were collected after 2-3 h of incubation when cells exhibited apoptotic morphologies, including blebbing, apoptotic membrane protrusion formation and the generation of ApoBDs6. A TO-PRO-3 and A5-based flow cytometry method was used to confirm the induction of monocyte apoptosis and ApoBD formation by separating viable cells, necrotic cells, early ApoCells, ApoCells, ApoBDs and debris (Figure 2). Taken together, flow cytometry analysis indicated that UV treatment results in ~20% ApoCells (Figure 3).
THP-1 monocyte ApoBDs were then isolated from the WAS via two approaches. Firstly, samples were prepared for a high purity FACS-based approach where only a single centrifugation step is required to pellet the entire apoptotic sample before staining and FACS. This method is appropriate for functional assays when significantly high sample purity is required (for example, for qPCR analysis), when a specific number of ApoBDs is required, or when acquiring cell type-specific ApoBDs from a complex sample. Using this methodology, ApoBDs were isolated to ~99% purity (Figure 4).
Next, THP-1 monocyte ApoBDs were also isolated via a two-step, differential centrifugation approach. The first step includes the isolation of viable cells, ApoCells and necrotic cells. The second step includes the separation of larger ApoBDs from small extracellular vesicles such as microvesicles and exosomes, which are unable to be pelleted at 3,000 x g. Flow cytometry was then performed to confirm apoptosis induction and ApoBD sample purity and demonstrated an ApoBD-enriched sample containing ~97% ApoBDs (Figure 4). This presents a quick and effective technique to isolate ApoBD to relatively high purity and is appropriate when purifying ApoBDs from samples containing a single cell type.
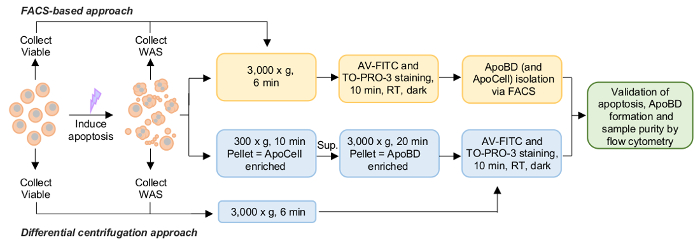
Figure 1. Schematic diagram of ApoBD isolation via either a FACS-based or differential centrifugation approach. Please click here to view a larger version of this figure.
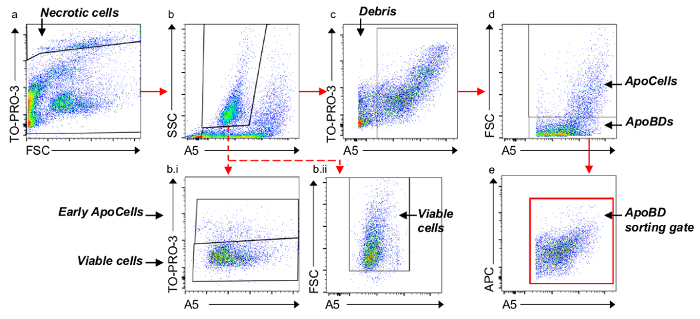
Figure 2. Flow cytometry gating strategy. Six cell/particle subsets (including viable cells, A5- early ApoCells, A5+ ApoCells, necrotic cells, ApoBDs and debris) were identified and used to select ApoBD for FACS-based isolation. (a) Membrane permeabilised necrotic cells are separated from non-permeabilised events. (b) A5low-intermediate, SSClow-high cells are separated from A5low-high events. (b.i) For in depth analysis, TO-POR-3low viable cells can be separated from TO-PRO-3intermediate A5– early ApoCells. (b.ii) Alternatively, when simply calculating ApoBD purity, viable cells can be separated from FSClow events. (c) A5low debris are excluded. (d) FSCintermediate/high ApoCells are separated from FSClow ApoBDs. (e) All A5-TO-PRO-3intermediate/high ApoBDs are selected for sorting. Please click here to view a larger version of this figure.
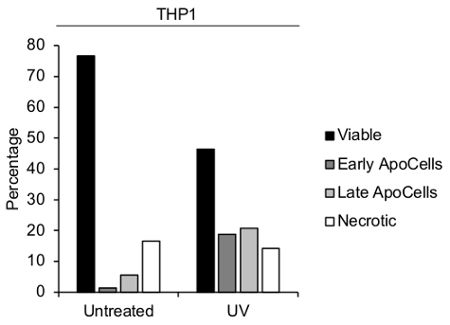
Figure 3. Validation of THP-1 monocyte apoptosis. Flow cytometry analysis of untreated or UV-irradiated THP-1 monocytes was performed to determine the levels of viable cells, A5– early ApoCells, A5+ ApoCells, and necrotic cells.
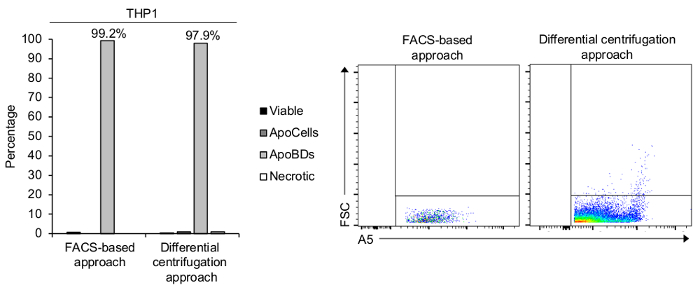
Figure 4. Purification of THP-1 monocyte-derived ApoBDs. Flow cytometry analysis was performed on isolated THP-1 monocyte-derived ApoBDs via either a FACS-based or differential centrifugation based approach, showing the enrichment of ApoBDs from viable cells, ApoCells and necrotic cells. Please click here to view a larger version of this figure.
Discussion
Since its early description in the 1950s, the field of apoptosis has advanced markedly, becoming a prominent research area. Despite the broad interest and extensive efforts, certain aspects of apoptosis, in particular the formation of ApoBDs, have not been well studied due to the lack of appropriate methodologies. These notably include the limitation in tracking apoptosis progression and ApoBD formation simultaneously using the traditional flow cytometry A5/PI analysis and the impurities of ApoBD isolation. We have recently developed approaches to address these methodological shortcomings.
Our new flow cytometry-based analytical approach allows ApoBDs, which are often ignored using traditional analytical methods, to be detected and quantified20. In addition, this modified flow cytometry method, with the use of the stain TO-PRO-3, reveals an additional A5– early apoptotic stage, hence rendering better delineation of apoptosis progression20. Conventionally, ApoBD detection have relied heavily on image-based techniques such as confocal microscopy and histology, whereas our flow cytometry method provides a high-throughput approach to quantitate ApoBD formation. Though seemingly complex, the procedure is relatively easy and only requires commercially available reagents and a basic flow cytometer. The logical detection and precise quantification of ApoBDs would further the knowledge of apoptotic cell microenvironment that dictates cell clearance and immunological responses. In fact, by coupling herein-described flow cytometry method with organelle-specific stains, we have recently reported the heterogeneous distribution of cellular contents in ApoBDs24. These findings suggest that ApoBDs can be categorized into different subsets and each ApoBD subset may exhibit different functions.
Our recently developed ApoBD isolation techniques would also contribute to advances in the field of extracellular vesicles. Traditionally, differential centrifugation methods used for ApoBD isolation may include a significant amount of smaller cells, which may affect downstream functional assays. However, our modified differential centrifugation approach can be used to isolate ApoBDs to 97% purity. Although high purity can be achieved by differential centrifugation, such method may not be suitable for isolating ApoBDs from complex samples. In contrast, our FACS-based method can enrich ApoBDs to 99% purity, and is based on the unique biological characteristics of ApoBDs including particle size, granularity and PtdSer exposure, instead of relying solely on particle density. This approach also has the potential to simultaneously identify and isolate ApoBDs of different cell origins using cell type-specific markers24. Despite a lengthy FACS procedure which could take 1-8 h depending on the quantity of ApoBDs required, accurate ApoBD isolation and subsequent downstream analysis would allow direct attribution of the molecular characteristics and functional roles of ApoBDs. For such methodologies, it is critical that ApoBD isolation approaches are coupled with techniques such as flow cytometry (as outlined here) to validate both the induction of apoptosis and purity of the ApoBD-enriched sample. Furthermore, proper FACS set up is essential for ApoBD isolation via the FACS-based approach, as an unstable stream or incorrectly performed drop delay may result in low purity.
Collectively, we have outlined cutting-edge methodologies for quantifying apoptosis induction, ApoBD detection and isolation of highly pure ApoBDs. Our approach may provide a new tool to study the apoptotic cell disassembly process and elucidate the role of this process in disease settings.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This worked was supported by grants from National Health and Medical Research Council (GNT1125033 and GNT1140187) and Australian Research Council (DP170103790) to I.K.H.P.
Materials
| Cells e.g. cultured human THP-1 monocytes (clone number: TIB-202) | ATCC | – | |
| RPMI 1640 medium | Life Technologies | 22400-089 | |
| Penicillin-streptomycin mixture | Life Technologies | 15140122 | |
| FSC | Gibco | 10099-141 | |
| 1x PBS | – | – | |
| Annexin V FITC | BD Bioscience | – | |
| TO-PRO-3 iodide | Life Technologies | T3605 | TO-PRO-3 may cause skin, eye and respiratory irration. Avoid direct contact. |
| 10x Annexin V binding buffer | BD Bioscience | 556454 | |
| EDTA | Sigma-Aldrich | 1001710526 | |
| Centrifuge tube (15 mL) | Cellstar | 188271 | |
| Microcentrifuge tube (1.5 mL) | Sarstedt | 72.690.001 | |
| Tissue culture incubator (37 °C, 5% CO2) | – | – | |
| Centrifuge | Beckman Coulter | 392932 | |
| FACS ARIA III Flow cytometer, configured with two lasers for FITC and APC detection | BD Bioscience | – | |
| FACS Canto II Flow cytometer, configured with two lasers for FITC and APC detection | BD Bioscience | – | |
| FACS Diva 6.1.1 software | BD Bioscience | – | |
| FlowJo 8.8.6 software | – | – | |
| UV Stratalinker 1800 | Stratagene | – |
References
- Poon, I. K., Lucas, C. D., Rossi, A. G., Ravichandran, K. S. Apoptotic cell clearance: basic biology and therapeutic potential. Nature Reviews. Immunology. 14, 166-180 (2014).
- Atkin-Smith, G. K., Poon, I. K. Disassembly of the Dying: Mechanisms and Functions. Trends in Cell Biology. 27, 151-162 (2017).
- Tixeira, R., et al. Defining the morphologic features and products of cell disassembly during apoptosis. Apoptosis: An International Journal on Programmed Cell Death. 22, 475-477 (2017).
- Sebbagh, M., et al. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nature Cell Biology. 3, 346-352 (2001).
- Coleman, M. L., et al. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nature Cell Biology. 3, 339-345 (2001).
- Atkin-Smith, G. K., et al. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nature Communications. 6, 7439 (2015).
- Poon, I. K., et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature. 507, 329-334 (2014).
- Moss, D. K., Betin, V. M., Malesinski, S. D., Lane, J. D. A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. Journal of Cell Science. 119, 2362-2374 (2006).
- Kerr, J. F., Wyllie, A. H., Currie, A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. British journal of cancer. 26, 239-257 (1972).
- Witasp, E., et al. Bridge over troubled water: milk fat globule epidermal growth factor 8 promotes human monocyte-derived macrophage clearance of non-blebbing phosphatidylserine-positive target cells. Cell Death and Differentiation. 14, 1063-1065 (2007).
- Orlando, K. A., Stone, N. L., Pittman, R. N. Rho kinase regulates fragmentation and phagocytosis of apoptotic cells. Experimental Cell Research. 312, 5-15 (2006).
- Holmgren, L., et al. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood. 93, 3956-3963 (1999).
- Zernecke, A., et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Science Signaling. 2, ra81 (2009).
- Schiller, M., et al. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death and Differentiation. 15, 183-191 (2008).
- Elamin, M. H., et al. Curcumin inhibits the Sonic Hedgehog signaling pathway and triggers apoptosis in medulloblastoma cells. Molecular Carcinogenesis. 49, 302-314 (2010).
- Sagulenko, V., et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death and Differentiation. 20, 1149-1160 (2013).
- Crescitelli, R., et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. Journal of Extracellular Vesicles. 2, (2013).
- Turiak, L., et al. Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice. Journal of Proteomics. 74, 2025-2033 (2011).
- Vermes, I., Haanen, C., Steffens-Nakken, H., Reutelingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. Journal of Immunological Methods. 184, 39-51 (1995).
- Jiang, L., et al. Monitoring the progression of cell death and the disassembly of dying cells by flow cytometry. Nature Protocols. 11, 655-663 (2016).
- Berda-Haddad, Y., et al. Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 108, 20684-20689 (2011).
- Lleo, A., et al. Shotgun proteomics: identification of unique protein profiles of apoptotic bodies from biliary epithelial cells. Hepatology. 60, 1314-1323 (2014).
- Atkin-Smith, G. K., et al. Isolation of cell type-specific apoptotic bodies by fluorescence-activated cell sorting. Scientific Reports. 7, 39846 (2017).
- Jiang, L., et al. Determining the contents and cell origins of apoptotic bodies by flow cytometry. Scientific Reports. 7, 14444 (2017).

