Fabrication of Anisotropic Polymeric Artificial Antigen Presenting Cells for CD8+ T Cell Activation
Summary
Here, we present a protocol to quickly and reproducibly generate biologically inspired, biodegradable articifical antigen presenting cells (aAPC) with tunable size, shape, and surface protein presentation for T cell expansion ex vivo or in vivo.
Abstract
Artificial antigen presenting cells (aAPC) are a promising platform for immune modulation due to their potent ability to stimulate T cells. Acellular substrates offer key advantages over cell-based aAPC, including precise control of signal presentation parameters and physical properties of the aAPC surface to modulate its interactions with T cells. aAPC constructed from anisotropic particles, particularly ellipsoidal particles, have been shown to be more effective than their spherical counterparts at stimulating T cells due to increased binding and larger surface area available for T cell contact, as well as reduced nonspecific uptake and enhanced pharmacokinetic properties. Despite increased interest in anisotropic particles, even widely accepted methods of generating anisotropic particles such as thin-film stretching can be challenging to implement and use reproducibly.
To this end, we describe a protocol for the rapid, standardized fabrication of biodegradable anisotropic particle-based aAPC with tunable size, shape, and signal presentation for T cell expansion ex vivo or in vivo, along with methods to characterize their size, morphology, and surface protein content, and to assess their functionality. This approach to fabricating anisotropic aAPC is scalable and reproducible, making it ideal for generating aAPC for "off-the-shelf" immunotherapies.
Introduction
Artificial antigen presenting cells (aAPC) have shown promise as immunomodulatory agents because they can generate a robust antigen-specific T cell response. Essential to these platforms are their ability to efficiently present crucial signals for T cell activation. Acellular aAPC are an attractive alternative to cell-based aAPC because they are easier and less costly to fabricate, face fewer challenges during scale-up and translation, and alleviate risks associated with cell-based therapies. Acellular aAPC also allow for a high degree of control over signal presentation parameters and physical properties of the surface that will interface with T cells1.
aAPC must recapitulate a minimum of two signals essential for T cell activation. Signal 1 provides antigen recognition and occurs when the T cell receptor (TCR) recognizes and engages with an MHC class I or II bearing its cognate antigen, culminating in signaling through the TCR complex. To bypass the antigen specificity requirement, aAPC systems often bear an agonistic monoclonal antibody against the CD3 receptor, which nonspecifically stimulates the TCR complex. Recombinant forms of MHC, particularly MHC multimers, have also been used on the surface of aAPC to provide antigen specificity2,3. Signal 2 is a costimulatory signal that directs T cell activity. To provide the costimulation necessary for T cell activation, the CD28 receptor is generally stimulated with an agonistic antibody presented on the aAPC surface, although other costimulatory receptors such as 4-1BB have been successfully targeted4. Signal 1 and 2 proteins are typically immobilized on the surface of rigid particles to synthesize aAPC. Historically, aAPC have been fabricated from a variety of materials, including polystyrene4,5 and iron dextran6. Newer systems utilize biodegradable polymers like poly(lactic-co-glycolic acid) (PLGA) to generate aAPC that can be easily coupled to signal proteins, are suitable for direct administration in vivo, and can facilitate the sustained release of encapsulated cytokines or soluble factors to augment T cell activation7,8.
In addition to the presence of necessary signal proteins, receptor engagement over a sufficiently large surface area during the aAPC/T cell interaction is essential for T cell activation. Thus, physical parameters of the aAPC such as size and shape drastically alter their available contact area and affect their ability to stimulate T cells. Micron-sized aAPC have been shown to be more effective at stimulating T cells than their nanoscale counterparts9,10. However, nano-aAPC can have superior biodistribution and better drainage to the lymph nodes that may enhance their performance in vivo over micro-aAPC11. Shape is another variable of interest in particle-based aAPC systems. Anisotropic aAPC have recently been shown to be more effective than isotropic particles at stimulating T cells, mainly due to enhanced interaction with target cells coupled with reduced non-specific cell uptake. Cells preferentially bind to the long axis of ellipsoidal particles, and the larger radius of curvature and flatter surface allow for more contact between the aAPC and T cell12. The long axis of ellipsoidal particles also discourages phagocytosis, resulting in increased circulation time compared to spherical particles following in vivo administration12,13. Because of these advantages, ellipsoidal particles mediate greater expansion of antigen-specific T cells in vitro and in vivo compared to spherical particles, an effect observed at both the micro and nanoscales12,13. There are various strategies to fabricate anisotropic particles, but thin-film stretching is a simple, widely accepted method used to generate a range of diverse particle shapes14. Following synthesis, particles are cast into films and stretched in one or two dimensions at a temperature above the glass transition temperature of the particle material. The film is then dissolved to retrieve the particles. Despite growing interest in anisotropic particles, current approaches for fabricating particle-based aAPC are mostly limited to isotropic systems, and methods of altering particle shape can be difficult to implement, incompatible with certain aAPC synthesis strategies, and lack precision and reproducibility15. Our thin-film stretching technique can be performed manually or in an automated fashion to rapidly generate anisotropic particles synthesized from a variety of biodegradable polymers, stretched to a desired aspect ratio in one or two dimensions15.
Based on our previous work, we developed a biodegradable particle-based approach combined with scalable thin-film stretching technology to rapidly generate aAPC with tunable size and shape in a standardized fashion for T cell expansion ex vivo or in vivo. Our protein conjugation strategy can be used to couple any protein(s) of interest to carboxyl groups on the particle surface at a desired density, giving this aAPC system a high degree of flexibility. We also describe methods to characterize the size, morphology, and surface protein content of aAPC, and to evaluate their functionality in vitro. This protocol can be easily adapted to expand immune cells ex vivo or in vivo for a variety of immunotherapeutic applications.
Protocol
All methods described here have been approved by the Institutional Animal Care and Use Committee (IACUC) of Johns Hopkins University.
1. Fabrication of Spherical PLGA Particles of Tunable Size
- Preparation of materials for particle synthesis
- Prepare 5% w/w polyvinyl alcohol (PVA) solution.
- Add 500 mL of deionized (DI) water to an Erlenmeyer flask with a magnetic stir bar and place on hot plate stirrer at 500 rpm and monitor temperature with thermometer. Cover flask with tinfoil to prevent evaporation.
- When water temperature reaches approximately 70 °C, add 25 g total of PVA in small batches over time, waiting for PVA to dissolve before adding more.
- Once all PVA is dissolved (typically 30-60 min), let solution cool and sterile filter. Store at 4 °C for future use.
- Prepare film casting solution of 10% w/w PVA and 2% w/w glycerol.
- Add 500 mL of DI water to an Erlenmeyer flask with a magnetic stir bar. Add 8 mL of glycerol at room temperature and mix by trituration.
- Place flask on hot plate stirrer at 500 rpm and monitor temperature with thermometer. Cover flask with tinfoil to prevent evaporation.
- When solution temperature reaches approximately 70 °C, add 50 g total of PVA in small batches over time, waiting for PVA to dissolve before adding more.
- Once all PVA is dissolved (typically 60 min), let solution cool and sterile filter using a bottle-top vacuum filter system with a pore size of 0.22 μm. Store at room temperature for future use.
- Prepare a 50 mL 1% w/w PVA solution. Add 40 mL of DI water and 10 mL of 5% PVA solution (made in 1.1.1) to a 100-150 mL beaker.
- Prepare a 100 mL 0.5% w/w PVA solution. Add 90 mL of DI water and 10 mL of 5% PVA solution to a 150-250 mL beaker.
- Add a magnetic stir bar to the 0.5% PVA solution and place in a chemical hood on a stir plate at room temperature at 500 rpm.
- Prepare 5% w/w polyvinyl alcohol (PVA) solution.
- Microparticle synthesis
- Weigh out 100 mg of poly(lactic-co-glycolic acid) (PLGA) into a scintillation vial and dissolve in 5 mL of dichloromethane (DCM). Vortex to dissolve the PLGA.
- Place homogenizer in the 50 mL 1% PVA solution so that the homogenizer is as close to the bottom of the beaker as possible without touching it. Turn on homogenizer and adjust to desired speed—3,200 rpm for 5 μm diameter particles, 5,000 rpm for 3 μm, 15,000 rpm for 1 μm (increasing homogenization speed decreases particle size). Once at the desired speed, add the PLGA solution to the beaker and homogenize for 1 minute.
- After homogenization, pour the 1% PVA, PLGA microparticle solution into the 100 mL 0.5% PVA solution on a stir plate and stir for at least 4 hours for solvent evaporation in a chemical hood.
- Wash the particles 3 times in DI water.
- Pour the particle solution into 50 mL conical tubes and centrifuge at 3000 x g for 5 minutes. Pour out supernatant and add approximately 20 mL of DI water.
- Resuspend particles by vortexing. Once resuspended, fill up conical tubes to 50 mL with DI water.
- Wash again the same way two more times.
- Nanoparticle synthesis
- Weigh out 200 mg of PLGA into a scintillation vial and dissolve in 5 mL of DCM. Vortex to dissolve the PLGA.
- Place a beaker with 50 mL 1% PVA solution into container filled with ice. Place the sonicator probe in the beaker as close to the bottom as possible without touching. Begin sonication and immediately add PLGA solution into the beaker. Sonicate with a power of 12 W for 2 minutes to generate nanoparticles with an approximate diameter of 200 nm.
- After sonication, pour the 1% PVA, PLGA nanoparticle solution into a 100 mL 0.5% PVA solution on a stir plate and stir for at least 4 hours for solvent evaporation in a chemical hood.
- Pour the particle solution into 50 mL conical tubes and centrifuge at 3,000 x g for 5 minutes to remove microparticles. Remove supernatant and pour into high speed centrifuge tubes.
- Wash the particles 3 times with DI water. Centrifuge at 40,000 x g for 15 minutes. Pour out supernatant and resuspend in DI water by vortexing. Wash again the same way two more times.
2. Fabrication of Polymeric Particles of Tunable Shape
- After washing PLGA particles three times, resuspend the particles in approximately 1 mL of DI water. Add the film casting solution to particles for final particle concentration of 2.5 mg/mL particles.
- Pipette the particle suspension in 10 mL aliquots into 75 x 50 mm rectangular Petri dishes for one-dimensional stretching or in 15 mL aliquots into 100 x 100 mm square Petri dishes for two-dimensional stretching. Remove bubbles by either pipette or pushing bubbles to the side and let films dry overnight in a chemical hood.
- Once films have dried, remove films from plastic dishes with tweezers and cut off edges of films with scissors. Save edges in a 50 mL conical tube to be used as spherical particles.
- Load a film onto the automated thin film stretching device by mounting a film onto aluminum blocks. For 1D stretching, mount film onto aluminum blocks of one axis of the stretcher (Figure 2) by placing two short edges of film in between two pieces of neoprene rubber. Using an Allen wrench, screw the metal grips on top of rubber to hold the film in place. For 2D stretching, mount all four edges onto four aluminum blocks to stretch on both axes (Figure 2).
- Measure and record the length of film in between aluminum blocks on one axis for 1D stretching or both axes for 2D stretching. Calculate the distance required to stretch the film in one or two directions based on desired fold-stretch.
- Place the film-loaded stretching device in an oven at 90 °C and bring the film to temperature over 10 minutes. Place a large beaker in the oven with a small amount of water.
- Stretch film.
- When stretching is completed, remove the stretching device from oven and let the film cool to room temperature for 20 minutes.
- For 1D stretching, cut the film out of the stretching device at the edges. For 2D stretching, cut out and save the center square of film that is uniformly stretched on both axes. Place films in conical tubes with 2 films per tube and discard the rest of the film.
- Add approximately 25 mL of DI water to each conical tube and vortex until the films are dissolved.
- Once the films are dissolved, wash particles 3 times with DI water.
- For microparticles, fill up conical tubes to 50 mL and centrifuge at 3000 x g for 5 minutes, pour out supernatant, add approximately 20 mL of DI water, and vortex to resuspend particles.
- For nanoparticles, transfer particles to high speed centrifuge tubes and centrifuge at 40,000 x g for 15 minutes, pour out supernatant, add approximately 20 mL of DI water, and vortex to resuspend particles.
- After the third wash, resuspend particles in approximately 1 mL of DI water. Record the weight of the microcentrifuge tube and add particles to the tube. Freeze particles in a -80 °C freezer for 1 hour or flash freeze in liquid nitrogen. Once frozen, lyophilize particles overnight.
- Once lyophilized, weigh particles in the microcentrifuge tube and subtract recorded weight of the empty tube to determine the weight of lyophilized particles.
3. Surface Protein Conjugation to Create Artificial Antigen Presenting Cells
- Prepare 2-(N-Morpholino)ethanesulfonic acid (MES) buffer. Make a 0.1 M solution of MES in water and adjust the pH to 6.0 by titration with 1M sodium hydroxide (NaOH).
- Resuspend lyophilized PLGA/PBAE micro/nanoparticles at 20 μg/mL in MES buffer by vortexing. Fill a polypropylene microcentrifuge tube with 900 μL of MES buffer and add 100 μL of the particle solution to the tube.
- Prepare EDC/NHS solution. Dissolve 40 mg 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and 48 mg N-hydroxysulfoxuccinimide (NHS) in 1 mL MES buffer.
- Add 100 μL EDC/NHS solution to the particles and vortex to mix.
- Incubate the tube on an inverter at room temperature for 30 minutes.
- Spin down microparticles at 5,000 x g for 5 minutes, or nanoparticles at 17,000 x g for 5 minutes. Discard the supernatant and resuspend in 1 mL PBS by vortexing (microparticles) or sonicating at 2-3 W for 5 seconds (nanoparticles).
- For microparticles, add 8 μg of the desired signal 1 protein, and 10 μg anti-mouse CD28 (clone 37.51) to the particles. For nanoparticles, add 16 μg signal 1 and 20 μg anti-mouse CD28. Add PBS to bring the volume in the tube to 1.1 mL.
- Incubate the tube on an inverter at 4 °C overnight.
- The next day, wash the particles. Spin down microparticles at 5,000 x g for 5 minutes, or nanoparticles at 17,000 x g for 5 minutes. Discard the supernatant, and resuspend particles in 1 mL of sterile PBS by vortexing (microparticles) or sonication at 2-3 W for 5 seconds (nanoparticles). Repeat twice.
- Resuspend the particles at the desired concentration in culture medium for immediate use. For long-term storage, resuspend particles at 10 mg/mL in a 100 mM sucrose solution. Freeze, lyophilize, and store the particles at -80 °C.
4. Characterization and Evaluation of aAPC
- Characterization of aAPC size and shape (Figure 3)
- Characterize microparticle size and shape by imaging particles using scanning electron microscopy (SEM). For SEM imaging, spread lyophilized particles onto carbon tape adhered to an aluminum tack and sputter coat with gold-palladium.
- Analyze size and aspect ratio of particles using image analysis software.
- To determine particle size, set scale using scale bar and measure particle diameter. Repeat for approximately 100 particles to determine the average particle diameter and generate a histogram of particle sizes.
- To determine aspect ratio, measure the distance across the long axis and the short axis of particles and divide long axis by short axis. Repeat for approximately 50 particles of each shape to determine the average particle aspect ratio for each shape and generate histograms.
- Characterize nanoparticle size and shape by imaging particles using transmission electron microscopy (TEM). Analyze size and aspect ratio of nanoparticles using image analysis software as described in 5.1.2. Alternatively, measure spherical nanoparticle size using dynamic light scattering (DLS) or nanoparticle tracking analysis (NTA).
- Protein Conjugation Efficiency
- Prepare aAPC according to Methods 1-3, but in Step 3.7 use fluorescently labeled signal 1 protein and anti-mouse CD28. After conjugation and washing, resuspend the aAPC in 1 mL PBS for a final concentration of 2 mg/mL aAPC.
- Prepare protein standards in a black polystyrene 96-well microplate. Add 5 μg of fluorescently labeled signal 1 protein to PBS in the first well of the plate and make 10 1:2 dilutions in PBS across the row of the plate. Leave the last well blank. Repeat this step to generate another set of standards with fluorescently labeled anti-CD28.
- Pipette 100 μL of the aAPC solution into replicate wells of the black polystyrene microplate in triplicate.
- Read the fluorescence on a fluorescence plate reader at the appropriate wavelengths. Use the fluorescence readings from the protein standards to generate standard curves for each fluorescent antibody. Using the standard curve, calculate the concentrations of signal 1 and anti-CD28 in each sample well, and then calculate the amount of surface protein and conjugation efficiency (Figure 4A, Figure 5A).
- Evaluation of aAPC for in vitro stimulation of CD8+ T cells
- Prepare B’ media (RPMI medium supplemented with L glutamine, 10% FBS, 1% Non-essential amino acid solution, 1% Sodium pyruvate, 1% MEM Vitamin solution, 92 μM β-mercaptoethanol, 10 ng/mL ciprofloxacin, and 30 U/mL IL-2.
- Sacrifice a Black 6 mouse via carbon dioxide exposure.
- Harvest the spleen from the mouse according to a previously established protocol16. Collect the spleen in a 50 mL conical tube with 10- 15 mL PBS. Using a pestle, mash the spleen through a 70 µm cell strainer into a 50 mL conical tube. During mashing, wash the strainer with 40 mL PBS.
- Spin the splenocytes at 300 x g for 5 minutes. Pour off the supernatant and resuspend the splenocytes in 4 mL of Ack Lysing Buffer to lyse the red blood cells. Allow the tube to sit undisturbed for 1 minute, then add PBS to bring the volume in the tube to 20 mL.
- Spin the cells at 300 x g for 5 minutes. Pour off the supernatant and resuspend the cells in 1 mL of PBS. Count the cells using a hemocytometer.
- Spin the cells at 300 x g for 5 minutes and resuspend in the desired volume of cell separation buffer. Isolate CD8+ T cells from the single cell suspension using a CD8+ negative selection T cell isolation kit, following the manufacturer’s protocol.
- Following magnetic separation, spin CD8+ T cells at 300 x g for five minutes and remove the cell separation buffer. Resuspend cells in 1 mL of PBS and count the cells. Label cells with carboxyfluorescein succinyl ester (CFSE) according to the manufacturer’s protocol.
- Incubate 8,333 CD8+ T cells and 0.0833 mg (or desired dose) of aAPC in 150 μL B’ media in each well of a 96 well U-bottom tissue culture-treated plate.
- Incubate at 37 °C for 7 days. After 3-4 days, refresh the culture medium by adding 75 μL fresh medium to each well.
- After 3 days of incubation, analyze CFSE labelled cells on a flow cytometer to assess proliferation. Each peak on the flow cytometry CFSE histogram represents a generation of cells due to the CFSE dilution with each successive cell division.
- After 7 days, use a hemocytometer to count the number of cells in each well. Prior to counting, stain dead cells with a Trypan Blue solution. Exclude dead cells from final cell counts. Normalize the final cell concentration to the initial concentration to calculate the fold-expansion (Figure 4 and Figure 5).
Representative Results
A schematic for the automated 2D thin film stretching device is given in Figure 1. A schematic and description for a 1D thin film stretching device is given in Ho et al.17 The stretcher is constructed from aluminum parts using standard milling and machining techniques. Similar to the 1D stretcher, the 2D stretcher consists of metallic grips and guide rails. Bidirectional lead screws are used to translate linear to rotational motion. The lead screws are the attached via mechanical taps to identical stepper motors with sufficient torque. The 8 stepper motor control wires can be soldered onto 8-pin heat-resistant amphenol connectors for easy attachment to the control console in an oven for thin film stretching. Polytetrafluoroethylene (PTFE) coated wire of sufficient length must be used to connect the stepper motors to the drivers in the control console. The recommended computer control scheme is given in Figure 1A. The two motors must be connected through heat resistant wiring to 2 independent drivers. The two drivers must then be connected to a microcontroller to interface with a computer. The drivers should be connected to the X-Axis and Y-Axis outputs on the microcontroller. The drivers and microcontroller both require an external power supply. Prior to connecting the power supply to these three components it is recommended that a 4 A fuse be inserted in between each of the powered connections to protect the components from current overload. Finally, the microcontroller can be linked via a Parallel Port Input to a computer using a DB25 Male to Male cable. The electronics used to control the stepper motors are heat sensitive and therefore must be placed outside of any heat source (such as an oven) used during operation to heat the thin films to sufficient temperature to enable stretching. Although the recommended motors are heat resistant up to the temperatures specified in this protocol for stretching particles, the motors and drivers will build up additional heat while they are attached to the main power supply. Therefore, it is recommended that the device only be turned on during the period of actual film stretching to minimize potential heat build-up.
PLGA nano- and microparticles were synthesized using the single emulsion techniques described in this protocol and imaged using TEM (Figure 3A) and SEM (Figure 3B), respectively. Spherical nanoparticles had a diameter of 237.3 ± 4.0 nm as measured by DLS and 224 nm as measured by NTA (Figure 3C). Microparticles were synthesized by homogenization at 5000 rpm to generate spherical particles with an average diameter of 3 ± 1 μm (Figure 3D). The particles were stretched using the automated film stretching device at 90 °C in one dimension to generate prolate ellipsoidal nano- and microparticles and stretched at 70 °C in two dimensions to generate oblate ellipsoidal particles. The aspect ratios of the microparticles of all three shapes were analyzed by measuring the long axis and short axis distance of particles and dividing the two. Spherical microparticles had an aspect ratio of 1.05 ± 0.04, while 1D stretched prolate ellipsoidal particles had a larger aspect ratio of 3.6 ± 0.8 (Figure 3E). 2D stretched oblate ellipsoidal particles had an aspect ratio of 1.2 ± 0.2, roughly maintaining an aspect ratio of one.
EDC/NHS reaction chemistry was used to conjugate a fluorescently labelled peptide-loaded MHC IgG dimer and anti-CD28 antibody to the surface of stretched and spherical PLGA particles. Conjugation efficiency results demonstrate similar amounts of protein on the surface of spherical and ellipsoidal micro-aAPC (Figure 4A) and nano-aAPC (Figure 5A) and demonstrate that protein coupling during aAPC synthesis occurs in a concentration-dependent manner. To evaluate the effect of shape on aAPC functionality, spherical and prolate ellipsoidal aAPC conjugated with gp100-loaded MHC IgG dimer and anti-CD28 were used to stimulate PMEL transgenic CD8+ T cells. T cells were labelled with CFSE and evaluated by flow cytometry after 3 days to assess proliferation (Figure 4B, 5B). Prolate ellipsoidal aAPC were found to induce higher levels of T cell proliferation at sub-saturating doses than spherical aAPC, with the best separation achieved at a 0.01 mg dose. After 7 days, the T cells were manually counted. Prolate ellipsoidal aAPC more effectively stimulated T cells compared to their spherical counterparts at the microscale (Figure 4C) and nanoscale (Figure 5C), and dose-dependent T cell expansion was observed.
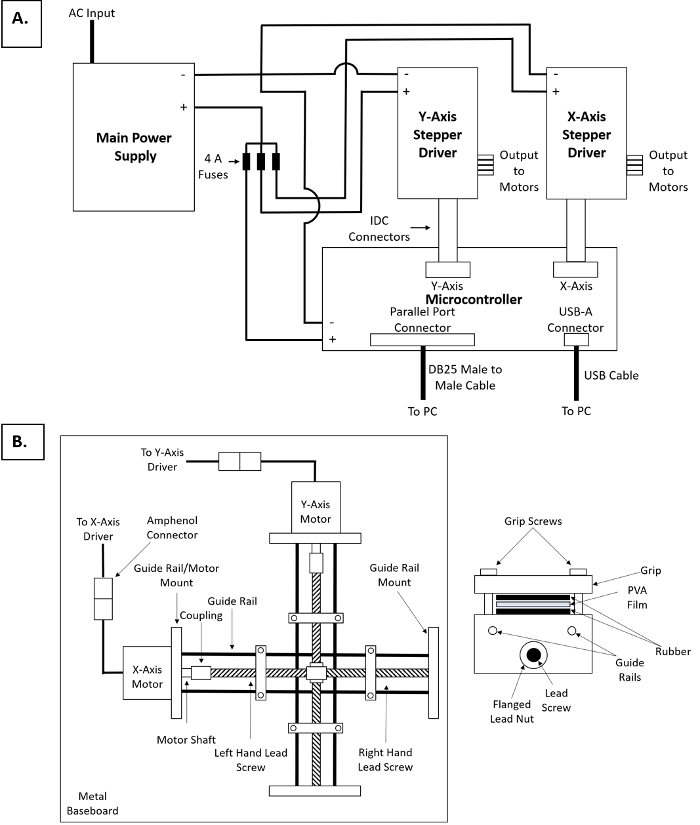
Figure 1: Schematic representation of automated thin film stretcher. (A) Schematic of control console for thin film stretcher. (B) Schematic of mechanical hardware for thin film stretcher. (Left) Overhead view of mechanical hardware. (Right) Cross-section of gripping mechanism for thin films. Please click here to view a larger version of this figure.
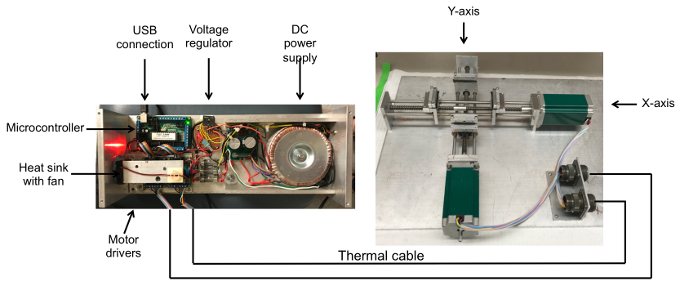
Figure 2: Photographs of assembled automated thin film stretcher to stretch polymeric particles into anisotropic shapes. The 2D thin film stretching device is composed of two axes with aluminum mounts that grip the film. The two axes contain lead screws in opposing directions so that they move apart from each other. To automate the stretching procedure, a USB linked microcontroller is connected to two stepper motor drivers that relay signals to unipolar stepper motors through a thermal cable. Please click here to view a larger version of this figure.
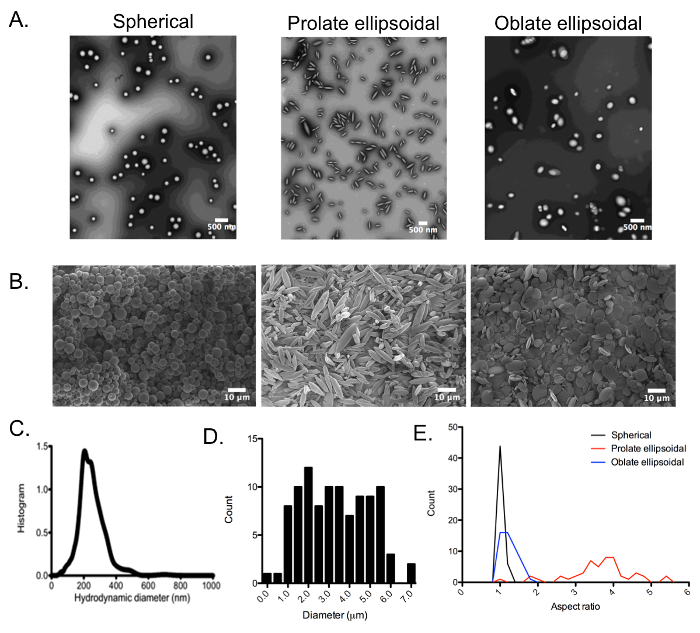
Figure 3: Size and aspect ratio analysis of spherical and ellipsoidal PLGA particles. (A) TEM and (B) SEM images of spherical, 1D stretched prolate ellipsoidal, and 2D stretched oblate ellipsoidal PLGA (A) nanoparticles and (B) microparticles. (C) Spherical nanoparticles were sized by NTA and determined to be 224 nm in diameter. SEM images of PLGA microparticles were analyzed for (D) size distribution of spherical particles and (E) aspect ratios of all particle shapes. (C) Reproduced and adapted with permission from Small13, Copyright Wiley-VCH 2015. Please click here to view a larger version of this figure.
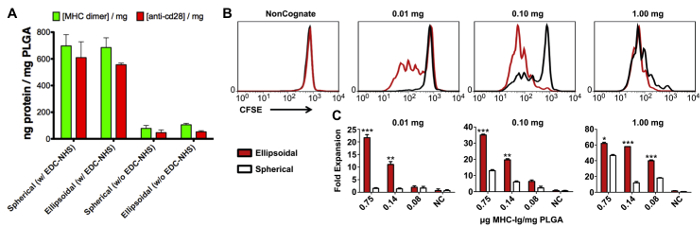
Figure 4: Characterization and functional assessment of spherical and prolate ellipsoidal micro-aAPC. (A) Conjugation efficiency of fluorescently-labelled peptide-loaded MHC IgG dimer and anti-CD28 antibody to the surface of spherical and prolate ellipsoidal microparticles. (B) CD8+ T cells were labelled with CFSE and incubated with spherical and 1D-stretched micro-aAPC at 0.01, 0.1, and 1 mg doses, or non-cognate controls. After 3 days, cells were evaluated by flow cytometry to assess proliferation. (C) T cells were also evaluated after 7 days by manual counting. Cell counts were normalized to the initial count to calculate fold-expansion. For comparison between prolate ellipsoidal and spherical fold expansion, * = p < 0.05, ** = p < 0.01, and *** = p < 0.001. Error bars represent standard error of the mean (SEM) for 3 replicates. Reproduced and adapted with permission from Biomaterials12, Copyright Elsevier 2014. Please click here to view a larger version of this figure.
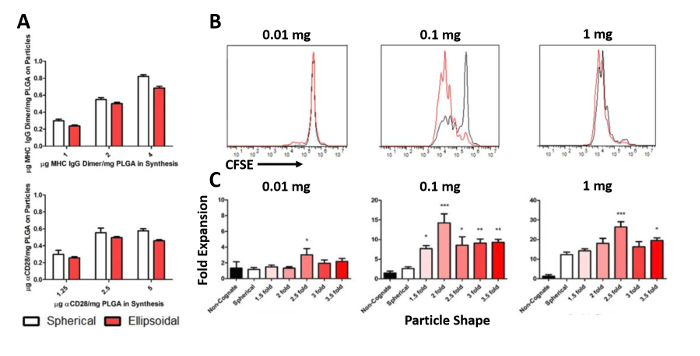
Figure 5: Characterization and functional assessment of spherical and prolate ellipsoidal nano-aAPC. (A) Conjugation efficiency of fluorescently-labelled peptide-loaded MHC IgG dimer and anti-CD28 antibody to the surface of spherical and prolate ellipsoidal nanoparticles. (B) CD8+ T cells were labelled with CFSE and incubated with spherical and prolate ellipsoidal nano-aAPC of varying fold-stretch at 0.01, 0.1, and 1 mg doses. After 3 days, cells were evaluated by flow cytometry to assess proliferation. (C) T cells incubated with prolate ellipsoidal particles of varying fold stretch (ranging from 1.5 to 3.5) were also evaluated after 7 days by manual counting. Cell counts were normalized to an untreated condition to calculate fold-expansion. * = p < 0.05, ** = p < 0.01, and *** = p < 0.001 compared to spherical. Error bars represent standard error of the mean (SEM) for 3 replicates. Reproduced and adapted with permission from Small13, Copyright Wiley-VCH 2015. Please click here to view a larger version of this figure.
Discussion
This protocol details a versatile method for the precise generation of anisotropic polymeric particles. The thin film stretching technique described here is scalable, highly reproducible and inexpensive. Alternative techniques for generating anisotropic particles suffer from many limitations, including high cost, low throughput, and limited particle size. The thin film stretching approach is also advantageous because the particles are modified to be anisotropic after synthesis, and, as a result, is compatible with a wide range of particle sizes and synthesis techniques. Figure 1 details the setup of the automated two-dimensional stretching device. This device can also be used without the electronic components by manually turning the screws until the film has reached the desired degree of stretching. However, we have found that the automated process is much more consistent and rapid than manual operation15. Various techniques have been developed to synthesize anisotropic particles, such as microfluidic approaches17,18,19, layer-by-layer coating21, and other bottom-up synthesis approaches21,22. However, these approaches do not enable strong control over particle geometry and are not as versatile in terms of shapes that can be generated and particle materials that can be used. A popular top-down method for fabricating nonspherical particles is Particle Replication in Non-Wetting Templates (PRINT)24. Although PRINT enables precise control over particle shape, it requires expensive machinery and is not as accessible and simple to implement as the thin film stretching method.
The single emulsion technique can be used to fabricate PLGA particles of various sizes, ranging from the nano to microscale12,13. By varying homogenization speed or sonication amplitude, microparticle and nanoparticle size, respectively, can be modulated. Once spherical particles have been generated, the thin film stretching method described here can be used to deform the particles into various shapes15. In this protocol, we describe the generation of prolate and oblate ellipsoidal particles by stretching in one or two dimensions, respectively. Spherical particles are cast into a thin plastic film, which is heated above the glass transition temperature of PLGA and stretched in one or two dimensions to deform the particles. The aspect ratio of the particles is highly controllable. By tuning the degree of film stretch, the aspect ratio of the particles can be modulated, and we have found that measured particle aspect ratio is highly correlated with the predicted value12,13. Various other shapes can be generated by modifying the temperature during stretching or the degree of stretching. For example, biconcave discoidal particles resembling the shape of red blood cells can be generated by stretching microparticles 1.5-fold in two dimensions at 90 °C.15. This film stretching technique has also been used to transform spherical polystyrene particles into many anisotropic shapes, including worms, barrels, and rectangular discs21. The film stretching device can be used by manually controlling the screws or the device can be automated as shown in Figure 1 to make the process more efficient and consistent15. This simple technique reliably produces anisotropic particles that retain their shape under physiologic conditions24. Furthermore, this method has been applied to other polymeric materials, in addition to PLGA, such as polycaprolactone (PCL) and hybrid particles made of PLGA and poly(beta-amino ester) (PBAE).
This protocol also describes how PLGA particles of varied size and shape can be conjugated with the surface proteins required for CD8+ T cell activation to act as aAPC. Proteins can be covalently conjugated to anisotropic and spherical PLGA micro- and nanoparticles by EDC/NHS mediated coupling of primary amines on proteins to carboxyl groups on the particle surface. The efficiency of protein conjugation can be measured by coupling fluorescently labeled protein to the surface of particles as described in this protocol, and we have found that this technique couples protein to particles at 15-20% efficiency12,13. Prolate ellipsoidal micro- and nanoparticle aAPC are more effective than their spherical counterparts at activating CD8+ T cell proliferation and expansion in vitro12,13. Ellipsoidal aAPC have enhanced binding to and interaction with T cells due to their larger surface area for contact12. Anisotropic particles also have superior properties over spherical particles in vivo due to their enhanced biodistribution and resistance to phagocytosis13. This platform is highly modular and has the potential to be adapted to many other drug delivery applications. Using this procedure, polymeric particles of tunable shape and size can be generated and the particle surface can be conjugated with any protein of interest.
Divulgations
The authors have nothing to disclose.
Acknowledgements
EBA (DGE-1746891) and KRR (DGE-1232825) thank the NSF Graduate Research Fellowship program for support. RAM thanks the National Research Service Award NIH NCI F31 (F31CA214147) and the Achievement Rewards for College Scientists Fellowship for support. The authors thank the NIH (R01EB016721 and R01CA195503), the Research to Prevent Blindness James and Carole Free Catalyst Award, and the JHU Bloomberg-Kimmel Institute for Cancer Immunotherapy for support.
Materials
| Poly(vinyl alcohol), MW 25000, 88% hydrolyzed | Polysciences, Inc. | 02975-500 | |
| Glycerol | Sigma-Aldrich | G9012 | |
| Digital Thermometer | Fluke | N/A | Model name: Fluke 52 II |
| Immersion Temperature Probe | Fluke | N/A | Model name: Fluke 80PK 22 |
| Digital Hotplate & Stirrer | Benchmark Scientific | H3760-HS | |
| Multipoint stirrer | Thermo Fisher Scientific | 50093538 | |
| Resomer RG 504 H, Poly(D,L-lactide-co-glycolide) | Sigma-Aldrich | 719900 | |
| Dichloromethane | Sigma-Aldrich | D65100 | |
| Homogenizer | IKA | 0003725001 | |
| Sonicator | Sonics & Materials, Inc. | N/A | Model number: VC 505 |
| Sonicator sound abating enclosure | Sonics & Materials, Inc. | N/A | Part number: 630-0427 |
| Sonicator probe | Sonics & Materials, Inc. | N/A | Part number: 630-0220 |
| Sonicator microtip | Sonics & Materials, Inc. | N/A | Part number: 630-0423 |
| High speed centrifuge | Beckman Coulter | N/A | Model number: J-20XP (discontinued), alternative model: J-26XP |
| High speed centrifuge rotor | Beckman Coulter | 369691 | Model number: JA-17 |
| High speed polycarbonate centrifuge tubes | Thermo Fisher Scientific | 3118-0050 | 50 mL, screw cap |
| Rectangular disposable petri dish | VWR International | 25384-322 | 75 x 50 x 10 mm |
| Square disposable petri dish | VWR International | 10799-140 | 100 mm x 100 mm |
| LEAF Purified anti-mouse CD3ε Antibody | Biolegend | 100314 | |
| InVivoMab anti-mouse CD28, clone 37.51 | Bio X Cell | BE0015-1 | |
| N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride | Sigma-Aldrich | E6383 | |
| N-Hydroxysulfosuccinimide sodium salt | Sigma-Aldrich | 56485 | |
| MES | Sigma-Aldrich | M3671 | |
| Alexa Fluor 488 anti-mouse CD3 Antibody | Biolegend | 100212 | |
| APC anti-mouse CD28 Antibody | Biolegend | 102109 | |
| Corning 96 Well Solid Polystyrene Microplate | Sigma-Aldrich | CLS3915 | flat bottom, black polystyrene |
| Protein LoBind Tubes, 1.5 mL | Eppendorf | 22431081 | |
| RPMI 1640 Medium (+ L-Glutamine) | ThermoFisher Scientific | 11875093 | |
| Fetal Bovine Serum | Sigma-Aldrich | F4135 | Heat Inactivated, sterile-filtered |
| Ciprofloxacin | Sigma-Aldrich | 17850 | |
| 2-Mercaptoethanol | Sigma-Aldrich | M6250 | |
| Recombinant Human IL-2 (carrier-free) | Biolegend | 589102 | |
| Sodium Pyruvate (100 mM) | ThermoFisher Scientific | 11360070 | |
| MEM Non-Essential Amino Acids Solution (100X) | ThermoFisher Scientific | 11140050 | |
| MEM Vitamin Solution (100X) | ThermoFisher Scientific | 11120052 | |
| CD8a+ T Cell Isolation Kit, mouse | Miltenyi Biotech | 130-104-075 | |
| CellTrace CFSE Cell Proliferation Kit | ThermoFisher Scientific | C34554 | |
| LS Columns | Miltenyi Biotech | 130-042-401 | |
| MidiMACS Separator | Miltenyi Biotech | 130-042-302 | |
| MACS Multistand | Miltenyi Biotech | 130-042-303 | |
| Flow Cytometer | Accuri C6 | ||
| Synergy 2 Multi-Detection Microplate Reader | BioTek | ||
| autoMACS Running Buffer | Miltenyi BIotech | 130-091-221 | |
| Cell Strainer | ThermoFisher Scientific | 22363548 | Sterile, 70 µm nylon mesh |
| ACK Lysing Buffer | ThermoFisher Scientific | A1049201 | |
| C57BL/6J (Black 6) Mouse | The Jackson Laboratory | 000664 | Male, at least 7 weeks old |
| U-Bottom Tissue Culture Plates | VWR | 353227 | Sterile, 96-well tissue culture treated polystyrene plates |
| 40 V DC Power Supply | Probotix | LPSK-4010 | |
| PTFE Coated Wire | Mouser | 602-5858-100-01 | This is for a 100 ft. spool but an equivalent wire will work |
| Stepper Motor Driver | Probotix | MondoStep5.6 | |
| IDC Connector Kit | Probotix | IDCM-10-12 | |
| Microcontroller | Probotix | PBX-RF | |
| 4A Fuses | Radio Shack | 2701026 | Equivalent fuses will work as well |
| DB25 Male to Male Cable | Probotix | DB25-6 | |
| USB-A to USB-B Cable | Staples | 2094915 | Equivalent cable will work as well |
| 8-Pin Amphenol Connectors Male and Female | Mouser | 654-97-3100A-20-7P and 654-97-3106A20-7S | |
| Stepper Motor | Probotix | HT23-420-8 | |
| Right Hand Lead Screw | Roton | 60722 | |
| Left Hand Lead Screw | Roton | 60723 | |
| Screws | McMaster Carr | 92196A151 | |
| Neoprene Rubber | McMaster Carr | 8698K51 | |
| Right Handed Flanged Lead Nut | Roton | 91962 | |
| Left Handed Flanged Lead Nut | Roton | 91963 | |
| Linux Control Computer | Probotix | LCNC-PC | Any computer with matching specification and Linux operating system will work |
| Corning bottle-top vacuum filter system | Sigma-Aldrich | CLS431097 | |
| Trypan Blue Solution, 0.4 % | ThermoFisher Scientific | 15250061 |
References
- Eggermont, L. J., Paulis, L. E., Tel, J., Figdor, C. G. Towards efficient cancer immunotherapy: Advances in developing artificial antigen-presenting cells. Trends in Biotechnology. 32 (9), 456-465 (2014).
- Maus, M. V., Riley, J. L., Kwok, W. W., Nepom, G. T., June, C. H. HLA tetramer-based artificial antigen-presenting cells for stimulation of CD4+ T cells. Clinical Immunology. 106 (1), 16-22 (2003).
- Oelke, M., et al. Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-Ig-coated artificial antigen-presenting cells. Nature Medicine. 9 (5), 619-624 (2003).
- Rudolf, D., et al. Potent costimulation of human CD8 T cells by anti-4-1BB and anti-CD28 on synthetic artificial antigen presenting cells. Cancer Immunology, Immunotherapy. 57 (2), 175-183 (2008).
- Tham, E. L., Jensen, P. L., Mescher, M. F. Activation of antigen-specific T cells by artificial cell constructs having immobilized multimeric peptide-class I complexes and recombinant B7-Fc proteins. Journal of Immunological Methods. 249 (1-2), 111-119 (2001).
- Perica, K., et al. Magnetic field-induced T cell receptor clustering by nanoparticles enhances T cell activation and stimulates antitumor activity. ACS Nano. 8 (3), 2252-2260 (2014).
- Steenblock, E. R., Fadel, T., Labowsky, M., Pober, J. S., Fahmy, T. M. An artificial antigen-presenting cell with paracrine delivery of IL-2 impacts the magnitude and direction of the T cell response. The Journal of Biological Chemistry. 286 (40), 34883-34892 (2011).
- Zhang, L., et al. Paracrine release of IL-2 and anti-CTLA-4 enhances the ability of artificial polymer antigen-presenting cells to expand antigen-specific T cells and inhibit tumor growth in a mouse model. Cancer Immunology, Immunotherapy. 66 (9), 1229-1241 (2017).
- Mescher, M. F. Surface contact requirements for activation of cytotoxic T lymphocytes. The Journal of Immunology. 149 (7), 2402-2405 (1992).
- Steenblock, E. R., Fahmy, T. M. A comprehensive platform for ex vivo T-cell expansion based on biodegradable polymeric artificial antigen-presenting cells. Molecular Therapy. 16 (4), 765-772 (2008).
- Fifis, T., et al. Size-dependent immunogenicity: therapeutic and protective properties of nano-vaccines against tumors. The Journal of Immunology. 173 (5), 3148-3154 (2004).
- Sunshine, J. C., Perica, K., Schneck, J. P., Green, J. J. Particle shape dependence of CD8+ T cell activation by artificial antigen presenting cells. Biomaterials. 35 (1), 269-277 (2014).
- Meyer, R. A., et al. Biodegradable nanoellipsoidal artificial antigen presenting cells for antigen specific T-cell activation. Small. 11 (13), 1519-1525 (2015).
- Champion, J. A., Katare, Y. K., Mitragotri, S. Particle shape: a new design parameter for micro- and nanoscale drug delivery carriers. Journal of Controlled Release. 121 (1-2), 3-9 (2007).
- Meyer, R. A., Meyer, R. S., Green, J. J. An automated multidimensional thin film stretching device for the generation of anisotropic polymeric micro- and nanoparticles. Journal of Biomedical Materials Research Part A. 103 (8), 2747-2757 (2015).
- Ho, C. C., Keller, A., Odell, J. A., Ottewill, R. H. Preparation of monodisperse ellipsoidal polystyrene particles. Colloid and Polymer Science. 271 (5), 469-479 (1993).
- Shum, H. C., et al. Droplet microfluidics for fabrication of non-spherical particles. Macromolecular Rapid Communications. 31 (2), 108-118 (2010).
- Lan, W., Li, S., Xu, J., Luo, G. Controllable preparation of nanoparticle-coated chitosan microspheres in a co-axial microfluidic device. Lab on a Chip. 11 (4), 652-657 (2011).
- Yang, S., et al. Microfluidic synthesis of multifunctional Janus particles for biomedical applications. Lab on a Chip. 12 (12), 2097-2102 (2012).
- Zhou, Z., Anselmo, A. C., Mitragotri, S. Synthesis of protein-based, rod-shaped particles from spherical templates using layer-by-layer assembly. Advanced Materials. 25 (19), 2723-2727 (2013).
- Jang, S. G., et al. Striped, ellipsoidal particles by controlled assembly of diblock copolymers. Journal of the American Chemical Society. 135 (17), 6649-6657 (2013).
- Petzetakis, N., Dove, A. P., O’Reilly, R. K. Cylindrical micelles from the living crystallization-driven self-assembly of poly(lactide)-containing block copolymers. Chemical Science. 2 (5), 955-960 (2011).
- Rolland, J. P., et al. Direct fabrication and harvesting of monodisperse, shape-specific nanobiomaterials. Journal of the American Chemical Society. 127 (28), 10096-10100 (2005).
- Meyer, R. A., et al. Anisotropic biodegradable lipid coated particles for spatially dynamic protein presentation. Acta Biomaterialia. 72, 228-238 (2018).

