Generation of Cancer Cell Clones to Visualize Telomeric Repeat-containing RNA TERRA Expressed from a Single Telomere in Living Cells
Summary
Here, we present a protocol to generate cancer cell clones containing a MS2 sequence tag at a single subtelomere. This approach, relying on the MS2-GFP system, enables visualization of the endogenous transcripts of telomeric repeat-containing RNA (TERRA) expressed from a single telomere in living cells.
Abstract
Telomeres are transcribed, giving rise to telomeric repeat-containing long noncoding RNAs (TERRA), which have been proposed to play important roles in telomere biology, including heterochromatin formation and telomere length homeostasis. Recent findings revealed that TERRA molecules also interact with internal chromosomal regions to regulate gene expression in mouse embryonic stem (ES) cells. In line with this evidence, RNA fluorescence in situ hybridization (RNA-FISH) analyses have shown that only a subset of TERRA transcripts localize at chromosome ends. A better understanding of the dynamics of TERRA molecules will help define their function and mechanisms of action. Here, we describe a method to label and visualize single-telomere TERRA transcripts in cancer cells using the MS2-GFP system. To this aim, we present a protocol to generate stable clones, using the AGS human stomach cancer cell line, containing MS2 sequences integrated at a single subtelomere. Transcription of TERRA from the MS2-tagged telomere results in the expression of MS2-tagged TERRA molecules that are visualized by live-cell fluorescence microscopy upon co-expression of a MS2 RNA-binding protein fused to GFP (MS2-GFP). This approach enables researchers to study the dynamics of single-telomere TERRA molecules in cancer cells, and it can be applied to other cell lines.
Introduction
The long noncoding RNA TERRA is transcribed from the subtelomeric region of chromosomes and its transcription proceeds towards the chromosome ends, terminating within the telomeric repeat tract1,2. For this reason, TERRA transcripts consist of subtelomeric-derived sequences at their 5' end and terminate with telomeric repeats (UUAGGG in vertebrates)3. Important roles have been proposed for TERRA, including heterochromatin formation at telomeres4,5, DNA replication6, promoting homologous recombination among chromosome ends7,8,9, regulating telomere structure10and telomere length homeostasis2,11,12,13. Furthermore, TERRA transcripts interact with numerous extratelomeric sites to regulate widespread gene expression in mouse embryonic stem (ES) cells14. In line with these evidence, RNA fluorescence in situ hybridization (RNA-FISH) analyses have shown that only a subset of TERRA transcripts localize at telomeres1,2,15. In addition, TERRA has been reported to form nuclear aggregates localizing at the X and Y chromosomes in mouse cells2,16. These findings indicate that TERRA transcripts undergo complex dynamics within the nucleus. Understanding the dynamics of TERRA molecules will help define their function and mechanisms of action.
The MS2-GFP system has been widely used to visualize RNA molecules in living cells from various organisms17,18. This system has been previously used to tag and visualize single-telomere TERRA molecules in S. cerevisiae12,19. Using this system, it was recently shown that yeast TERRA transcripts localize within the cytoplasm during the post-diauxic shift phase, suggesting that TERRA may exert extranuclear functions20. We have recently used the MS2-GFP system to study single-telomere TERRA transcripts in cancer cells21. To this aim, we employed the CRISPR/Cas9 genome editing tool to integrate MS2 sequences at a single telomere (telomere 15q, hereafter Tel15q) and obtained clones expressing MS2-tagged endogenous Tel15q TERRA (TERRA-MS2 clones). Co-expression of a GFP-fused MS2 RNA-binding protein (MS2-GFP) that recognizes and binds MS2 RNA sequences enables visualization of single-telomere TERRA transcripts in living cells21. The purpose of the protocol illustrated here is to describe in detail the steps required for the generation of TERRA-MS2 clones.
To generate TERRA-MS2 clones, a MS2 cassette is integrated within the subtelomeric region of telomere 15q, downstream of the TERRA promoter region and transcription start site. The MS2 cassette contains a neomycin resistance gene flanked by lox-p sites, and its integration at subtelomere 15q is performed using the CRISPR/Cas9 system22. After transfection of the MS2 cassette, single clones are selected and subtelomeric integration of the cassette is verified by PCR, DNA sequencing and Southern blot. Positive clones are infected with a Cre-expressing adenovirus in order to remove the selection marker in the cassette, leaving only MS2 sequences and a single lox-p site at the subtelomere 15q. Expression of MS2-tagged TERRA transcripts from Tel15q is verified by RT-qPCR. Finally, the MS2-GFP fusion protein is expressed in TERRA-MS2 clones via retroviral infection in order to visualize MS2-TERRA transcripts by fluorescence microscopy. TERRA transcripts can be readily detected by RNA-FISH and live-cell imaging using telomeric repeat-specific probes1,2,15,23. These approaches provide important information on the localization of the total population of TERRA molecules at single cell resolution. The generation of clones containing MS2 sequences at a single subtelomere will enable researchers to study the dynamics of single-telomere TERRA transcripts in living cells, which will help define the function and mechanisms of action of TERRA.
Protocol
1 . Selection of Neomycin Resistant Clones
- Grow AGS cells in Ham's F-12K (Kaighn's) medium supplemented with 10% Fetal Bovine Serum (FBS), 2 mM L-glutamine, penicillin (0.5 units per mL of medium), and streptomycin (0.2 µg per mL of medium) at 37 °C and 5% CO2. Transfect the cells at a 50-60% confluence with the sgRNA/Cas9 expressing vector and the MS2 cassette at a 1:10 molar ratio21.
NOTE: In a parallel experiment, verify transfection efficiency by transfecting a GFP-expressing vector (i.e., Cas9-GFP vector). At least 60-70% transfection efficiency should be achieved. - The following day, replace the culturing medium with medium containing neomycin at 0.7 µg/mL final concentration (selective medium).
NOTE: Splitting the cells and seeding them in selective medium the day after transfection will speed up the selection process. All non-transfected cells will immediately die. If transfection is performed in 6 well plates, in which case each well at a 60% confluence would contain approximately 0.7 x 106 cells, on the following day the cells can be split from a single well to a 10 cm dish containing selective medium. - Keep the cells in selective medium for 7-10 days, changing medium every one or two days, until single clones are visible.
- Picking of cell clones
- Prepare a 96 well plate containing 10 µL of 0.25% trypsin in each well.
NOTE: Prepare two 96 well plates in case more than 96 clones are expected to be picked. - With the use of a microscope, mark the position of each clone visible in the 10 cm dish by making a dot at the bottom of the dish using a marker. Each dot will correspond to a colony to be picked.
- Replace the culturing medium with just enough Phosphate Buffered Saline (PBS) to form a thin film of liquid on the clones and not let the cells dry during the clone picking.
- Pick single colonies using a 10 µL pipette. Attach the tip containing 5 µL of trypsin to the colony and slowly release the trypsin which will remain localized on the colony. Allow trypsin to detach the cells for 1 min, then scrape the colony with the tip and suck it up into the tip.
NOTE: During this procedure, flipping the dish a bit on one side so to decrease the volume of PBS around the colony being picked will help the picking process by avoiding diluting the trypsin around the colony. Using a clone ring may also help picking up single clones. - Place the cells from the colony into a well of the 96 well plate containing 10 µL of 0.25% trypsin.
- Incubate 5 min at room temperature, then fill the well with 150 µL of selective medium (Ham's F-12K (Kaighn's) medium supplemented with 10% FBS, 2 mM L-glutamine, penicillin (0.5 units per mL of medium), streptomycin (0.2 µg per mL of medium) and containing neomycin at a concentration of 0.7 µg/mL.
NOTE: During the incubation time other clones can be picked. It is recommended to pick as many clones as possible. The more clones are picked the higher the chances will be of identifying positive ones. - Once all the clones are picked and transferred in the 96 well plate, allow the cells to grow for a few days in selective medium Ham's F-12K (Kaighn's) medium supplemented with 10% FBS, 2 mM L-glutamine, penicillin (0.5 units per mL of medium), streptomycin (0.2 µg per mL of medium) and containing neomycin at a concentration of 0.7 µg/mL at 37 °C and 5% CO2, until reaching 90% confluence.
- Prepare a 96 well plate containing 10 µL of 0.25% trypsin in each well.
- Splitting of clones
- Prepare three 96 well plates coated with gelatin by adding 100 µL of gelatin per well, incubate for 30 min at room temperature, then wash two times with PBS. These plates will be used for DNA extraction (DNA plate) and for clone freezing (freezing plates).
NOTE: Gelatin will promote the attachment of the cells and of the DNA to the wells. In particular, the gelatin coating will allow the DNA to stick at the bottom of the wells during the DNA extraction and wash procedures (discussed below). During the clone-splitting procedure, it is advisable to use a multichannel pipette. - Once clones reach 90% confluence, aspirate medium from each well of the 96 well plate, wash with PBS, add 30 µL of 0.25% trypsin per well, and incubate for 5 min at 37 °C.
NOTE: Clones will grow at different rates, which will also depend on the number of cells picked per clone. Thus, this step will be performed during the several days when the different clones reach 90% confluence. - Add 70 µL of selective medium per well, disrupt cell clumps by pipetting up and down inside the wells, then transfer 30 µL of the 100 µL to the gelatinized DNA plate prefilled with 120 µL of selective medium per well and 30 µL to the gelatinized freezing plates prefilled with 50 µL medium (without selection).
- Place the DNA plate in the incubator and allow the cells to grow at 37 °C and 5% CO2 until 90% confluence.
- Add 80 µL of ice-cold freshly made 2x freezing medium (80% FBS and 20% dimethyl sulfoxide (DMSO)) to each well of the freezing plate, add parafilm (sprayed with 70% ethanol) on top of the plate so to seal each well, place the lid on top and wrap the plate with aluminum foil. Place the freezing plates at -80 °C.
- Add 90 µL of selective medium per well to the original 96 well plate containing the clones that have been split and keep it in culture until PCR screening results. This plate will be used as backup plate.
- Prepare three 96 well plates coated with gelatin by adding 100 µL of gelatin per well, incubate for 30 min at room temperature, then wash two times with PBS. These plates will be used for DNA extraction (DNA plate) and for clone freezing (freezing plates).
2. Screening of Neomycin Resistant Clones
- DNA extraction from the 96 well DNA plate.
- Once the clones cultured in the DNA plate reach 90% confluence, wash 2 times with PBS, then lyse with 50 µL lysis buffer (10 mM Tris pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% SDS, and 1 mg/mL proteinase K). Cover the plate with parafilm, sealing each well, put the lid on, cover with saran wrap, and place at 37 °C overnight.
- Add 100 µL of cold ethanol (Et-OH)/NaCl solution (0.75 M NaCl in 100% ethanol) to each well and precipitate for 6 h or overnight at room temperature.
NOTE: The protocol can be paused here. - Remove the Et-OH/NaCl solution by inverting the plate and wash 3 times with 200 µL of 70% ethanol per well.
- Add 25 µL of RNAse A solution in distilled water and incubate at 37 °C for 1 h.
- Use 3 µL of genomic DNA for PCR amplification. Perform PCR screening of the selected clones using primers annealing within the neomycin resistance gene and subtelomere 15q. PCR amplification is performed using standard polymerase enzymes and PCR protocols21.
NOTE: PCR conditions for MS2 primers (MS2-subtel15q-primer-S and MS2 primer AS) and CTR primers (CTR prime S and CTR primer AS) are the following: 98 °C for 20 s as denaturation step and then 34 cycles at 98 °C 10 s, 58°C 20 s, 72 °C 15 s, using polymerase enzyme in a 25 µL reaction mix (see Table of Materials). Primer sequences are indicated in Table 1. - Run PCR reactions on agarose gel and extract PCR bands obtained from positive clones using standard gel extraction procedures (see Table of Materials for gel extraction reagents).
- Perform DNA sequencing analyses of the gel-extracted PCR product for confirmation of the presence of the MS2 sequences21.
NOTE: The sequencing analyses can be performed using the primers used for the PCR screening. - Southern blot screening of PCR positive clones
- Grow the clones positive at PCR and sequencing screening from the original 96 well plate (the backup plate) to 6 well plates in Ham's F-12K (Kaighn's) medium supplemented with 10% FBS, 2 mM L-glutamine, penicillin (0.5 units per mL of medium), streptomycin (0.2 µg per mL of medium) and containing neomycin at a concentration of 0.7 µg/mL at 37 °C 5% CO2.
NOTE: Alternatively, if some of these clones have been lost, thaw them from one of the freezing plates (see protocol 2.6). - Once the clones are at 90% confluence in the 6 well plate, wash the cells with PBS and add 250 µL of lysis buffer containing 0.5 µg proteinase K per well.
- Scrape the cells using a cell scraper and transfer the lysate in a 1.5 mL tube.
- Incubate at 37 °C for 16 h.
- Add 1 mL of 100% ethanol, shake vigorously, and allow the DNA to precipitate at least 2 h or overnight at -20 °C.
NOTE: The protocol can be paused here. - Spin at 13,400 x g at 4 °C for 10 min, discard the supernatant, and wash the pellets with 70% ethanol. Let the pellets air dry at room temperature. Alternatively, use a vacuum concentrator.
- Resuspend the DNA pellets in 50 µL of distilled water containing RNAse A and incubate for 1 h at 37 °C.
- Digest 5-10 µg of genomic DNA using NcoI and BamHI restriction enzymes (two independent digestions) in 100 µL reaction volume by incubating the digestion reactions at 37 °C overnight.
- Run 4 µL of the digestion on agarose gel (0.8% agarose) for complete digestion confirmation.
- Add 1/10 volume of sodium acetate 3 M solution pH 5.2 and 2 volumes of 100% ethanol to the restriction digestion reactions and incubate at least 2 h or overnight at -20°C to precipitate DNA.
NOTE: The protocol can be paused here. - Centrifuge at 13,400 x g at 4 °C for 20 min, discard the supernatants, and wash the pellets with 70% ethanol. Allow the pellets to air dry at room temperature. Alternatively, use a vacuum concentrator.
- Resuspend pellets in 20 µL of distilled water and load the digested DNA on a 0.8% agarose gel.
NOTE: For a better resolution of the digested DNA, prepare a gel at least 15 cm long and run overnight at low voltage ( ̴30 volts). The electrophoresis set up should be optimized. - The following day, stain the gel with a DNA labelling agent, such as ethidium bromide at 1 µL/10 mL final concentration, for 30 min at room temperature and take a picture with a ruler close to the gel on a gel imaging instrument.
- Set the transfer of DNA to a nylon membrane and perform membrane hybridization with a MS2 sequence-specific probe using standard procedures21.
- Thaw the clones that are positive at PCR, DNA sequencing and Southern blot from one of the two freezing plates (see next step).
- Grow the clones positive at PCR and sequencing screening from the original 96 well plate (the backup plate) to 6 well plates in Ham's F-12K (Kaighn's) medium supplemented with 10% FBS, 2 mM L-glutamine, penicillin (0.5 units per mL of medium), streptomycin (0.2 µg per mL of medium) and containing neomycin at a concentration of 0.7 µg/mL at 37 °C 5% CO2.
- Thawing of clones
- Prepare one 15 mL tube containing 5 mL of pre-warmed Ham's F-12K (Kaighn's) medium supplemented with 10% FBS, 2 mM L-glutamine, penicillin (0.5 units per mL of medium), and streptomycin (0.2 µg per mL of medium) for each clone to be thawed.
- Remove one of the freezing plates from -80° and add 100 µL of pre-warmed F12K complete medium to the well containing the positive clone to be thawed.
NOTE: This procedure should be performed quickly and the 96 well plate should be placed on dry ice after each clone is thawed, in order to allow the other clones to remain frozen. This is particularly important if multiple clones need to be thawed from the same 96 well plate. - Transfer the cells to the 15 mL tube containing 5 mL of medium and centrifuge at 800 x g for 5 min at room temperature.
- Aspirate the medium, resuspend the cells in 500 µL of pre-warmed complete F12K medium, and transfer each clone to a single well of a 12 well plate.
- Elimination of the neomycin resistance gene from the MS2 cassette integrated at subtelomere 15q.
- Allow the clones to grow from a 12 well plate to a 10 cm dish in complete F12K medium.
NOTE: Neomycin should not be included in the medium unless otherwise indicated. - Add the Cre-expressing adenovirus to the cells cultured in a 10 cm dish at 70% confluence in 10 mL of complete F12K medium.
NOTE: Using a Cre-GFP expressing adenovirus will allow evaluation of the efficiency of infection, which should approach 100%. The replication-defective adenovirus will be lost after few passages in culture. - 48 hours after infection, split the cells in three 10 cm dishes. Two dishes will be used for verification of the neomycin gene removal by negative selection, growing the cells in neomycin-containing medium (first dish), and by Southern blot (second dish).
- Culture the third dish containing the clone for cell freezing and RNA extraction.
- Allow the clones to grow from a 12 well plate to a 10 cm dish in complete F12K medium.
3. Verification of TERRA-MS2 Transcript Expression by RT-qPCR
- Upon verification of neomycin gene removal, perform total RNA extraction from TERRA-MS2 clones using organic solvents (phenol and guanidine isothiocynate solution)21.
- Resuspend the RNA extracted from a 10 cm dish in 100 µL of diethyl pyrocarbonate (DEPC) water.
- Run 3 µL of RNA on a denaturating (1% formaldehyde-containing) 1x 3-(N-Morpholino) propanesulfonic acid (MOPS) gel in order to verify concentration and integrity of the RNA. Also analyse RNA concentration using a spectrophotometer.
- Treat 3 µg of RNA with DNAse I using 1 unit of DNAse I enzyme in 60 µL final reaction volume.
- Incubate the reaction for 1 h at 37 °C.
- Reverse transcription reaction and qPCR analyses
- Add the following components to a nuclease-free microcentrifuge tube: 2 µL of a 1 µM TERRA specific primer, 1 µL of dNTPs mix (10 mM each), 6 µL of DNAse I-treated RNA (corresponding to ̴300ng RNA). Adjust the volume to 13 µL with 4 µL of DEPC water.
NOTE: For each RNA to be analysed, a second tube containing the same reagents but a reference specific primer, instead of TERRA specific primer, should be prepared. - Heat the mixture to 65 °C for 5 min and incubate in ice for at least 1 min.
- Collect the content of the tubes by brief centrifugation and add 4 µL of 5X RT enzyme Buffer, 1 µL 0.1 M dithiothreitol (DTT), 1 µL (4 units) of RNAse inhibitor, and 1 µL of reverse transcriptase (see Table of Materials).
- Incubate the samples at 42 °C for 60 min, then use 2 µL of the RT reaction for qPCR analyses.
- Add the following components to a nuclease-free microcentrifuge tube: 2 µL of a 1 µM TERRA specific primer, 1 µL of dNTPs mix (10 mM each), 6 µL of DNAse I-treated RNA (corresponding to ̴300ng RNA). Adjust the volume to 13 µL with 4 µL of DEPC water.
- Prepare qPCR reaction mix in a final volume of 20 µL consisting of 10 µL of 2x qPCR master mix, 2 µL of cDNA template, 1 µL of forward primer (10 µM), 1 µL of reverse primer (10 µM), and 6 µL of water.
- Perform qPCR reaction in a thermocycler using standard protocols21.
4. Production of a MS2-GFP Expressing Retrovirus
- To generate MS2-GFP expressing retrovirus, transfect 80% confluent phoenix packaging cells with a MS2-GFP fusion protein expressing retrovirus vector (pBabe-MS2-GFP PURO) and an env gene expressing vector (such as pCMV-VSVG) using a molar ratio 4:1 of the two vectors.
- On the following day, replace the culturing medium (DMEM supplemented with 10% FBS, 2 mM L-glutamine and Pen/Strep) with fresh medium containing 10mM sodium butyrate.
- Incubate for 8 h at 37 °C, 5% CO2, then replace the medium with fresh culturing medium from which the virus will be collected.
- After 48 h, remove the retrovirus-containing medium from the phoenix cells. This medium can be directly used for infection of TERRA-MS2 clones, in which case filter the medium through a 0.45 µm filter, add polybrene (30 µg/mL final concentration) and add it to the cells (in this case the protocol continues at step 5). Alternatively, retrovirus can be precipitated in a 50 mL falcon tube by adding 1/5th volume of 50% PEG-8000/900 mM NaCl solution and incubating overnight at 4 °C on a rotator wheel.
- On the following day, pellet retrovirus particles by centrifugation at 2,000 x g for 30 min, remove supernatant, and resuspend the pellet in F12K medium without serum.
NOTE: The virus particles can be resuspended in 1/100th of the original supernatant volume. An infection test should be performed in order to verify the minimum volume of retrovirus required to efficiently infect the cells.
5. Visualization of TERRA-MS2 Transcripts in Living Cells
- Plate TERRA-MS2 clones and WT AGS cells in glass-bottomed dishes. On the day of infection, add polybrene to the medium (30 µg/mL final concentration) and the MS2-GFP expressing retrovirus.
- After 24 hours, discard the virus-containing medium and add fresh medium without phenol-red.
- Analyze the cells at an inverted microscope using the appropriate microscope setting. Image the cells with a 100X or 60X objective with large numerical aperture (1.4X) and using a sensitive camera (EMCCD). Use an environmental control system to maintain the samples at 37 °C and 5% CO2 during imaging.
Representative Results
Figure 1 represents an overview of the experimental strategy. The main steps of the protocol and an indicative timeline for the generation of TERRA-MS2 clones in AGS cells are shown (Figure 1A). At day 1, multiple wells of a 6 well plate are transfected with the MS2 cassette and sgRNA/Cas9 expressing vectors (shown in Figure 1B). Two different subtelomere 15q-specific guide RNA sequences are cloned in the Cas9 nickase-expressing pX335 vector, generating two sgRNA-pX335 vectors that are co-transfected with the MS2 cassette. One well of the plate can be transfected with a GFP expressing vector to verify the transfection efficiency. At day 2, the cells are trypsinized and transferred from a single well to a 10 cm dish containing selective medium. At day 3, medium should be changed as most untransfected cells will be dead. Cells are grown in selection until clones are visible and ready to be picked. During this time, cells should not be trypsinized and culturing medium should be changed every 1-2 days for the first week and then every 2-3 days afterwards. Once single clones are visible, they are picked and transferred in a 96 well plate where they are allowed to grow until reaching 80-90% of confluence. At this point, clones are split in 4 different 96 well plates, which are marked in Figure 1 with a color code. Once the clones cultured in the DNA plate (green) reach 80% confluence, they are lysed and genomic DNA extracted for PCR screening; the clones in the backup plate (blue) will be kept in culture until the results of the PCR screening, while the red freezing plates are frozen at -80°C. Figure 2B shows representative results of a PCR screening of neomycin-resistant clones. For this screening, two sets of primers are used: one primer pair (MS2 primers) annealing within the neomycin gene and subtelomere 15q sequence is used to verify the presence of the MS2 cassette (a representative image of the primers localization is shown in Figure 2A). A second primer pair (CTR primers) annealing at an internal chromosomal region is used to verify the presence of false negative clones (primer sequences are indicated in Table 1). Negative clones for the integration of the cassette should be negative to the MS2 primers' amplification and positive to the CTR primers' amplification. Technical problems in genomic DNA extraction resulting in the absence of genomic DNA or its contamination would preclude PCR amplification from MS2 and CTR primer reactions. In these cases, the clones involved can be re-screened by PCR upon extraction of genomic DNA from the backup plate. Bands obtained from MS2 primers amplification of positive clones can be gel-extracted and sequenced using the MS2 primers, in order to confirm the presence of ten MS2 sequences.
The clones that are positive at PCR screening are cultured from the 96 well backup plate (the blue plate in Figure 1) to a 6 well plate, then lysed for genomic DNA extraction and Southern blot analyses. Figure 2D shows representative images of a gel (left) and membrane hybridized with a radioactively labelled MS2 sequence specific probe (right) of a Southern blot screening of PCR positive clones. For confirmation of the MS2 sequences integration at subtelomere 15q, genomic DNA extracted from each clone should be digested with two different restriction enzymes (NcoI and BamHI) in two separate digestion reactions. The NcoI and BamHI restriction enzymes are suitable for verification of the MS2 cassette integration at subtelomere 15q. Other enzymes can also be used. The gel shows a complete digestion of genomic DNA using the BamHI restriction enzyme, as indicated by the presence of a smear. The results from Southern blot indicate that one clone is positive for the integration of the MS2 cassette at subtelomere 15q while several clones show multiple integration events of the cassette, as indicated by the presence of multiple bands. A positive clone should be analyzed by a second Southern blot upon NcoI digestion of genomic DNA21. A representative image of a subtelomere 15q containing the MS2 cassette and the position of BamHI and NcoI restriction enzyme sites is shown in Figure 2C.
The clones positive to PCR and Southern blot analyses are infected with a Cre-GFP expressing adenovirus in order to remove the neomycin gene present in the MS2 cassette. Figure 3A depicts a MS2-tagged subtelomere 15q before and after Cre expression. An image of a clone positive for the MS2 cassette integration at subtelomere 15q infected with a Cre-GFP expressing adenovirus is shown in Figure 3B. For a complete removal of the neomycin gene in all cells, the infection efficiency should reach approximately 100%. As shown in Figure 3C, in order to verify the elimination of the neomycin gene, Cre-GFP infected cells are split in three plates after 48 hours from the infection. One plate will be cultured in the presence of neomycin. All cells in this plate should die within 6-7-days. In a second plate, cells will be allowed to grow in complete medium without selection up to 80-90% confluence. At this point, genomic DNA is extracted and analyzed by Southern blot. The third plate is cultured in complete medium to allow the preparation of frozen stocks of the clone and for RNA extraction and RT-qPCR analyses of TERRA-MS2 transcript expression. Figure 3D shows representative images of Southern blot analyses of a positive clone before and after Cre-GFP infection. DNA agarose gel (image on the left) confirms the complete digestion of the genomic DNA using the NcoI restriction enzyme. Southern blot analysis was performed using a radioactively-labelled MS2-sequence specific probe (image on the right). This analysis confirms the removal of the neomycin gene. The presence of a smear in the Cre-GFP infected sample confirms the telomeric integration of the MS2 sequences. The position of the NcoI restriction sites within the subtelomere 15q is shown in Figure 3A.
Once the elimination of the neomycin resistance gene is confirmed, the clones can be tested for the expression of TERRA-MS2 transcripts. To this aim, total RNA is extracted from each clone and run on a denaturating MOPS gel to confirm its integrity (Figure 4A). Ribosomal RNA bands should be visible at 4,700 bases (rRNA 28S) and 1,900 bases (rRNA 18S). Retrotranscription reaction is performed using a telomeric repeat-specific primer and reference gene-specific primer (Figure 4B, top) while qPCR analyses of TERRA expression are performed using two sets of primers: one primer pair annealing within the MS2 sequence and the subtelomere 15q sequence; a second pair of primers annealing within the subtelomere 15q. Primer sequences are indicated in Table 1. The graph presented in Figure 4B shows RT-qPCR analyses of TERRA expression from AGS WT cells and two different TERRA-MS2 clones. These analyses confirm the expression of TERRA-MS2 transcripts in the two clones at levels that are comparable to TERRA transcripts expressed from subtelomere 15q in WT cells. These data indicate that the two TERRA-MS2 clones selected are suitable for the analyses of TERRA-MS2 transcripts by live cell imaging.
In order to visualize TERRA-MS2 transcripts in living cells, the selected clones are infected with a retrovirus expressing the MS2-GFP fusion protein. Figure 5A shows the procedure to generate MS2-GFP expressing retrovirus, as described in protocol step 4. AGS WT cells and TERRA-MS2 clones are infected in glass-bottomed dishes. After 24 h from the infection, cells are analyzed by fluorescence microscopy. The specificity of the signal is confirmed by the presence of TERRA-MS2-GFP foci detected in the nucleus of TERRA-MS2 clones and not in AGS WT cells (Figure 5B). In a population of MS2-GFP expressing cells, heterogeneity in terms of MS2-GFP levels among cells is expected and cells expressing low levels should be chosen for the imaging analyses. Alternatively, the MS2-GFP expressing cells can be sorted by FACS prior microscopy analyses in order to collect the subpopulation of cells expressing low levels of GFP. We have previously detected TERRA-MS2-GFP foci in 40% to 60% of cells of AGS TERRA-MS2 clones21. In these cells, one to four TERRA-MS2-GFP foci were imaged per cell. TERRA-MS2-GFP foci showing distinct sizes and dynamics can be detected21. TERRA-MS2 clones can be used to study the dynamics of single-telomere TERRA transcripts in living cells. As an example of this application, Figure 5C shows representative images from microscopy analyses of a TERRA-MS2 clone expressing the MS2-GFP fusion protein and the telomere-binding protein TRF2 fused to mCherry in order to visualize MS2-tagged TERRA transcripts and telomeres in living cells. Using this approach, we have previously observed that TERRA-MS2-GFP foci co-localize with telomeres in 44% of the cells, indicating that TERRA transcripts only transiently co-localize with chromosome ends in AGS cells21.
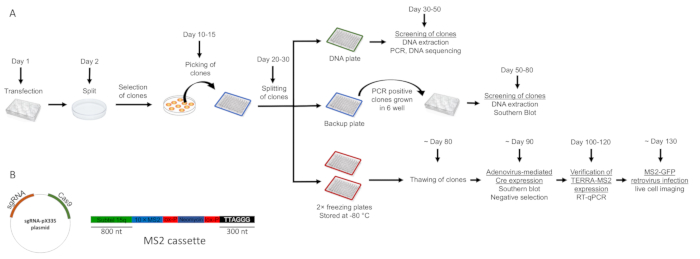
Figure 1. Overview of the experimental strategy and indicative timeline for the generation of TERRA-MS2 clones in AGS cells. A) The steps described in the protocol and an indicative time line for the selection of TERRA-MS2 clones are shown. Cells are transfected in a 6 well plate with the linearized MS2 cassette and sgRNA/Cas9 expressing vectors. A color code is used to distinguish the DNA plate, the backup plate and the freezing plates indicated in the protocol section. B) The MS2 cassette consists of an 800 nt long subtelomere 15q sequence followed by 10 repetitions of the MS2 sequences and a neomycin resistance gene flanked by lox-p sites and terminates with a 300 nt long telomeric repeat tract at its 3' end. sgRNA/Cas9 expressing vectors were generated by cloning subtelomere 15q-specific guide RNA sequences in pX335 vector using the BbsI restriction site21,22. Two different sgRNA-pX335 vectors were generated and a 1:1 mix of the two vectors was used for the transfection. The sequences of the sgRNAs are indicated in Table 1. Please click here to view a larger version of this figure.
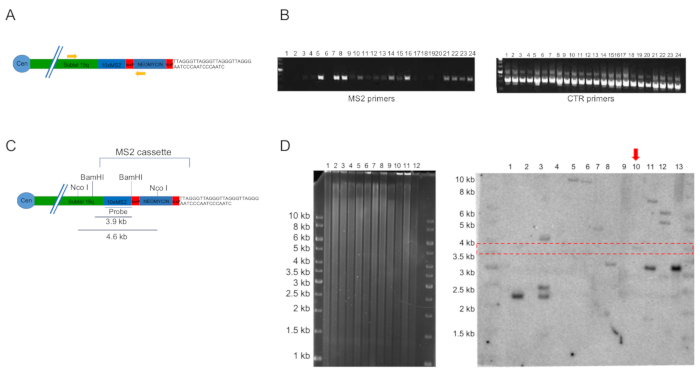
Figure 2. Representative images of PCR and Southern blot screening of neomycin resistant clones.A) Representative image of subtelomere 15q containing the MS2 cassette. The position of the MS2 primers (MS2-subtel15q-primer-S and MS2 primer AS) used for PCR screening is indicated. The loxP sites present within the cassette are shown in red. B) Representative image of PCR screening of neomycin resistant clones using two set of primers, MS2 primers and CTR primers (CTR prime S and CTR primer AS). C) Representative image of subtelomere 15q containing the MS2 cassette. BamHI and NcoI restriction sites and the MS2 probe used for the Southern blot screening are shown. D) Left: 10 µg of genomic DNA per clone were digested with BamHI and run on an agarose gel. The image was acquired after an overnight run at 30 volts. Right: genomic DNA digested with BamHI restriction enzyme was transferred to a positively charged nylon membrane and hybridized with a radioactively-labelled MS2 sequence-specific probe. The red box indicates the expected size of the positive band. The red arrow indicates one positive clone. Please click here to view a larger version of this figure.
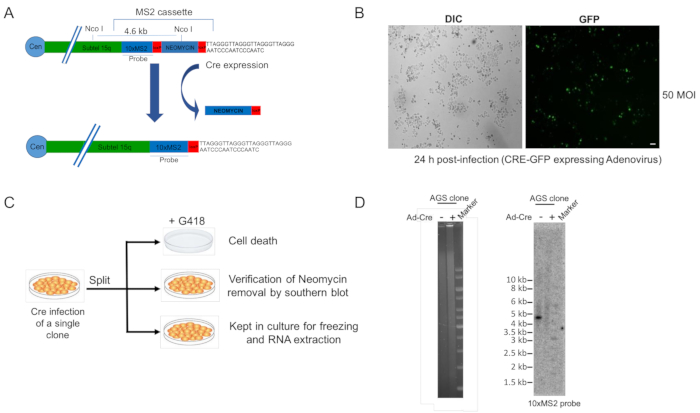
Figure 3. Elimination of the neomycin resistance gene and validation experiments. A) Representative image of subtelomere 15q containing the MS2 cassette before and after Cre expression. The MS2 specific probe used for Southern blot analyses and NcoI restriction enzyme sites are shown. The loxP sites present within the cassette are shown in red. B) Fluorescence microscopy analysis of a TERRA-MS2 clone infected with Cre-GFP expressing adenovirus. Representative image acquired at a single focal plane is shown. MOI, multiplicity of infection, is the ratio between the number of viruses used for the infection and the number of host cells. Scale bar: 30 µm C) Overview of the experimental strategy used to verify the elimination of the neomycin gene. Cre-GFP infected cells are split in three plates after 48 hours from the infection for i) negative selection, ii) Southern blot analyses and iii) preparation of frozen cell stocks and RNA extraction. D) Southern blot analyses of a TERRA-MS2 clone before and after Cre-GFP adenovirus infection. 10 µg of genomic DNA were digested with the restriction enzyme NcoI. Agarose gel confirms that complete digestion is achieved in both clones (left). The digested genomic DNA was transferred to a positively charged nylon membrane and hybridized using a MS2 sequence-specific probe. Complete elimination of the neomycin gene is confirmed by the absence of the specific band. Please click here to view a larger version of this figure.
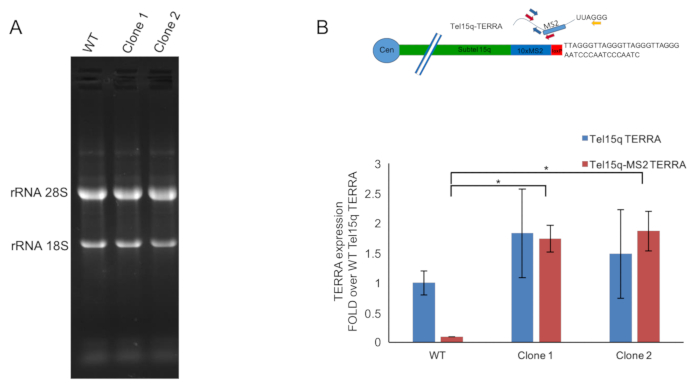
Figure 4. RT-qPCR analyses of Tel15q-TERRA and Tel15q-TERRA-MS2 transcripts expression. A) Representative image of a MOPS gel showing total RNA extracted from AGS WT cells and TERRA-MS2 clones. Bands corresponding to the ribosomal RNAs 28S and 18S are indicated. B) Top: A schematic of the MS2-tagged subtelomere 15q expressing TERRA transcripts. Primers used for TERRA retrotranscription (yellow) and qPCR analyses of Tel15q-TERRA (blue) and Tel15q-MS2 TERRA (red) are shown. The loxP site is shown in red. Bottom: RT-qPCR analyses of Tel15q-TERRA and Tel15q-MS2 TERRA transcripts expression in AGS WT cells and TERRA-MS2 clones. *p < 0.05, unpaired t-test. Please click here to view a larger version of this figure.
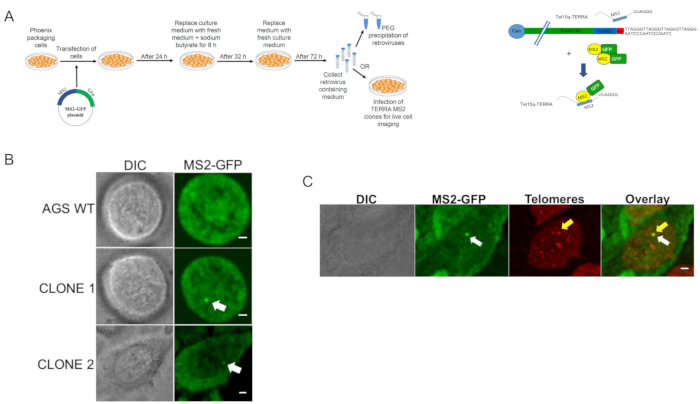
Figure 5. Live cell imaging analyses of TERRA-MS2 clones. A) An overview of the procedure to produce a MS2-GFP expressing retrovirus, as described in protocol step 4. A schematic of the MS2-tagged subtelomere 15q and TERRA transcripts recognized by the MS2-GFP fusion protein is shown on the right. B) Representative fluorescence microscopy image of AGS WT and two TERRA-MS2 clones expressing MS2-GFP. TERRA-MS2-GFP foci are indicated by arrows. Images were acquired using a spinning disc confocal microscope equipped with an EMCCD camera. The images were captured using a 100X/1.46 apochromat objective in an imaging chamber maintained at 37 °C with 5% CO2. A 488 laser was used as light source. Scale bar: 5 µm. C) Live cell imaging analyses of TERRA-MS2-GFP foci and TRF2-mCherry labeled telomeres. A co-localization event between a TERRA-MS2-GFP focus and a single telomere is shown. Images were acquired as in B, using 488 and 520 lasers as light source. A maximal projection of a z stack experiment performed at a single time point of time-lapse imaging experiment is shown. Scale bar: 5 µm. Please click here to view a larger version of this figure.
| Primer name | Primer sequence | Application |
| MS2-subtel15q-primer-S | TGCATTAAAGGGTCCAGTTG | MS2 primer forward used for PCR screening of neomycin resistant clones |
| MS2 primer AS | CCTAACTGACACACATTCCACAGA | MS2 primer reverse used for PCR screening of neomycin resistant clones |
| CTR primer S | TGT ACG CCA ACA CAG TGC TG | CTR primer forward used in PCR screening of neomycin resistant clones |
| CTR primer AS | GCT GGA AGG TGG ACA GCG A | CTR primer reverse used in PCR screening of neomycin resistant clones |
| Tel15q-S1 | GCAGCGAGATTCTCCCAAGC | Sense primer used to detect Tel15q and Tel15q-MS2 TERRA expression by qPCR |
| hTel15q-AS | TAACCACATGAGCAATGTGGGTG | Antisense primer used to detect Tel15q and Tel15q-MS2 TERRA expression by qPCR |
| Tel15q-MS2-AS | ATGTTTCTGCATCGAAGGCATTAGG | Antisense primer used to detect Tel15q-MS2 TERRA expression by qPCR |
| TERRA-RT-primer | CCCTAACCCTAACCCTAACCCTAACCCTAA | Primer used for retrotranscription of TERRA transcripts |
| sgRNA1 | TTGGGAGAATCTCGCTGGCC | Sequence of the short guide RNA 1 cloned in pX335 vector and used to direct Cas9 nickase enzymatic activity to subtelomere 15q |
| sgRNA2 | TGCATTAAAGGGTCCAGTTG | Sequence of the short guide RNA 2 cloned in pX335 vector and used to direct Cas9 nickase enzymatic activity to subtelomere 15q |
Table 1: List of the primers used in this study. A list of the primers and the sequences of the sgRNAs used in this protocol are provided. Sequences are 5' to 3'.
Discussion
In this article we present a method to generate human cancer cell clones containing MS2 sequences integrated within subtelomere 15q. Using these clones, the MS2-tagged TERRA molecules transcribed from the subtelomere 15q are detected by fluorescence microscopy by co-expression of a MS2-GFP fusion protein. This approach enables researchers to study the dynamics of TERRA expressed from a single telomere in living cells21. In this protocol, TERRA-MS2 clones are selected in the AGS cell line, which represents an interesting model system to study TERRA, since TERRA expression is upregulated in human stomach cancer samples24. However, the protocol described here can be adapted for the selection of TERRA-MS2 clones in other cell lines. The integration of the MS2 sequences can in principle be promoted at a subtelomere different from telomere 15q by using a specific MS2 cassette and CRISPR strategy. In the method presented here, a Cas9 nickase enzyme and double guide RNAs are employed in order to increase the specificity of integration22. Using this strategy, an overall 2% of positive clones are expected to be identified in AGS cells. Other versions of the Cas9 enzyme can be tested in the attempt to increase the success rate of the clones selection25. In addition, the following steps of the protocol are critical ones to take into account to maximize the efficiency of the clone selection:
I) In order to increase the chances of selecting the TERRA-MS2 clones, it is important to achieve a high transfection efficiency of the MS2 cassette and the CRISPR vectors. Poorly transfected cell lines will most likely require several rounds of transfection, clone selection and screening in order to select positive clones.
II) It is important to determine the precise concentration of the neomycin to use for the selection of the clones. Indeed, using a low concentration of the drug will result in false positive clones while a high concentration may preclude the identification of positive clones. The neomycin concentration indicated in this protocol is lethal to the AGS cells within 6-7 days from the addition to the culturing medium.
III) Clone picking is a critical step. Indeed, during this procedure it is required that only single clones are picked. For this reason, colonies that are too close to each other should be avoided in order to prevent mixing cells from different clones, resulting in the selection of a mixed population of clones which may contain MS2 sequences integrated at multiple genomic sites. These events should be identified during the Southern blot screening.
IV) The amount of genomic DNA extracted from a confluent 96 well DNA plate (protocol 2) is estimated to be around 5 µg and should suffice to screen every clone by PCR and Southern blotting with a single restriction enzyme digestion. However, in order to confirm the integration of the cassette at the expected telomere, it will be important to screen each clone by Southern blot by digesting the genomic DNA with two different restriction enzymes. To this aim, as indicated in the protocol, the clones positive at PCR screening should be cultured in 6 well plates. The amount of genomic DNA extracted from a well of a 6 well plate will be sufficient for at least two restriction digestions required for the Southern blot analyses.
In the protocol described here, the MS2 sequences are integrated within subtelomere 15q for a number of reasons: i) TERRA promoter region and TERRA transcription start sites have been identified on this subtelomere26,27; ii) TERRA expression from subtelomere 15q has been validated by using in vitro techniques4,27,28,29,30; iii) the subtelomeric region of the chromosome 15q has been sequenced and it contains a unique region adjacent to the telomeric repeat tract that can be targeted for the integration of the MS2-cassette21. PCR and Southern blot approaches were developed in order to confirm the single integration of the MS2 sequences within subtelomere 15q in the TERRA-MS2 clones. It would be interesting to also perform DNA-FISH experiments on chromosome spreads in order to visualize the chromosomal localization of the MS2 sequences in fixed metaphase chromosomes. However, the length of the 10xMS2 sequences (450 bp) imposes the use of very short probes as compared to the probes generally used in DNA-FISH experiments on chromosome spreads (Bacterial artificial chromosomes (Bac), cosmids or plasmids)31. This technical challenge has prevented us from visualizing the MS2 sequences on chromosome spreads which represents a limitation of the protocol. One further limitation of the method is the time and the effort required for selecting the TERRA-MS2 clones. In addition, specific laboratory set up and equipment for the use of viruses, radioactive material and microscopy analyses are required.
The method described here can be implemented for the generation of TERRA-MS2 clones in different cell lines in order to define the dynamics of TERRA in various biological contexts, such as during telomere dysfunction or cellular senescence, helping us to understand the function of TERRA in these processes. An interesting development of this approach will be the generation of TERRA-MS2 clones containing TetO repeats integrated at a specific subtelomere, including the MS2-tagged telomere. The expression of a TetR protein fused to a fluorescent protein (i.e., mCherry) will allow visualization of this particular telomere in living cells32. This approach will enable investigation of whether human TERRA transcripts localize and act in cis at the TERRA transcribing telomere and chromosome, or in trans by relocating to other chromosomes. This question in the biology of TERRA remains to be clarified.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We are grateful to the staff of the advanced imaging facility of CIBIO at the University of Trento and the BioOptics Light Microscopy facility at the Max F. Perutz Laboratories (MFPL) in Vienna. The research leading to these results has received funding from the Mahlke-Obermann Stiftung and the European Union's Seventh Framework Programme for research, technological development and demonstration under grant agreement no 609431 to EC. EC is supported by a Rita Levi Montalcini fellowship from the Italian Ministry of Education University and Research (MIUR).
Materials
| AGS cells | – | – | Gift from Christian Baron (Université de Montréal). |
| F12K Nut Mix 1X | GIBCO | 21127022 | Culturing medium for AGS cells |
| L-Glutamine | CORNING | MT25005CI | Component of cell culturing medium |
| Penicillin Streptomycin Solution | CORNING | 30-002-CI | Component of cell culturing medium |
| Fetal Bovine Serum | Sigma Aldrich | F2442 | Component of cell culturing medium |
| DMEM 1X | GIBCO | 21068028 | culturing medium for phoenix cell |
| CaCl2 | Sigma Aldrich | C1016 | used in phoenix cell transfection |
| HEPES | Sigma Aldrich | H3375 | used in phoenix cell transfection (HBS solution) |
| KCl | Sigma Aldrich | P9333 | used in phoenix cell transfection (HBS solution) |
| Dextrose | Sigma Aldrich | D9434 | used in phoenix cell transfection (HBS solution) |
| NaCl | Sigma Aldrich | S7653 | used in phoenix cell transfection (HBS solution) and retrovirus precipitation |
| Na2HPO4 | Sigma Aldrich | S3264 | used in phoenix cell transfection (HBS solution) |
| TRYPSIN EDTA SOLUTION 1X | CORNING | 59430C | used in cell split |
| DPBS 1X | GIBCO | 14190250 | Dulbecco's Phosphate Buffered Saline |
| DMSO | Sigma Aldrich | D8418 | Component of cell freezing medium (80% FBB and 20% DMSO) |
| G-418 Disulphate | Formedium | G4185 | selection drug for |
| Gelatin solution Bioreagent | Sigma Aldrich | G1393 | cotaing of 96 well DNA plate and freezing plate |
| Tris-base | Fisher BioReagents | 10376743 | Component of Cell lysis buffer for genomic DNA extraction |
| EDTA | Sigma Aldrich | E6758 | Component of Cell lysis buffer for genomic DNA extraction |
| SDS | Sigma Aldrich | 71729 | Component of Cell lysis buffer for genomic DNA extraction |
| Proteinase K | Thermo Fisher | AM2546 | Component of Cell lysis buffer for genomic DNA extraction |
| RNAse A | Thermo Fisher | 12091021 | RNA degradation during DNA extraction |
| Agarose | Sigma Aldrich | A5304 | DNA gel preparation |
| Atlas ClearSight | Bioatlas | BH40501 | Stain reagent used for detecting DNA and RNA samples in agarose gel |
| ethanol | Fisher BioReagents | BP28184 | DNA precipitation |
| Sodium Acetate | Sigma Aldrich | 71196 | Used for DNA precipitation at a 3M concentration pH5.2 |
| Wizard SV Gel and PCR clean-Up system | Promega | A9282 | Extraction of PCR fragments from agarose gel during PCR screening of neomycin positive clones |
| Trizol | AMBION | 15596018 | Organic solvent used for RNA extraction |
| Dnase I | THERMO SCIENTIFIC | 89836 | degradation of genomic DNA from RNA |
| dNTPs mix | Invitrogen | 10297018 | used in RT and PCR reactions |
| DTT | Invitrogen | 707265ML | used in RT reactions |
| diethyl pyrocarbonate | Sigma Aldrich | D5758 | used to inactivate RNAses in water (1:1000 dilution) |
| Ribolock | Thermo Fisher | EO0381 | RNase inhibitor |
| MOPS | Sigma Aldrich | M9381 | preparation of RNA gel |
| Paraformaldehyde | Electron Microscopy Sciences | 15710 | preparation of denaturating RNA gel (1% PFA in 1x MOPS) |
| Superscript III Reverse transcriptase | Invitrogen | 18080-093 | Retrotranscription reaction |
| Pfu DNA polymerase (recombinant) | Thermo Scientific | EP0501 | PCR reaction |
| 2X qPCRBIO SyGreen Mix Separate-ROX | PCR BIOSYSTEMS | PB 20.14 | qPCR reaction |
| Cre-GFP adenovirus | https://medicine.uiowa.edu/vectorcore | 1174-HT | used to infect TERRA-MS2 clones in order to remove the neomycn gene |
| Sodium Butyrate | Sigma Aldrich | B5887 | used to promote retrovirus particles production in phoenix cells |
| PEG8000 | Sigma Aldrich | 89510 | Precipitation of retrovirus partcles |
| 35µ-Dish Glass Bottom | Ibidi | 81158 | used in live cell imaging analyses of TERRA-MS2 clones |
References
- Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 318 (5851), 798-801 (2007).
- Schoeftner, S., Blasco, M. A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature Cell Biology. 10 (2), 228-236 (2008).
- Diman, A., Decottignies, A. Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: from Darkness to Dawn. FEBS Journal. , (2017).
- Arnoult, N., Van Beneden, A., Decottignies, A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nature Structural & Molecular Biology. 19 (9), 948-956 (2012).
- Montero, J. J., et al. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nature Communications. 9 (1), 1548 (2018).
- Beishline, K., et al. CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nature Communications. 8 (1), 2114 (2017).
- Arora, R., et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nature Communications. 5, 5220 (2014).
- Balk, B., et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nature Structural & Molecular Biology. 20 (10), 1199-1205 (2013).
- Graf, M., et al. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell. 170 (1), 72-85 (2017).
- Lee, Y. W., Arora, R., Wischnewski, H., Azzalin, C. M. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nature Structural & Molecular Biology. , (2018).
- Moravec, M., et al. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Reports. 17 (7), 999-1012 (2016).
- Cusanelli, E., Romero, C. A., Chartrand, P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Molecular Cell. 51 (6), 780-791 (2013).
- Moradi-Fard, S., et al. Smc5/6 Is a Telomere-Associated Complex that Regulates Sir4 Binding and TPE. PLoS Genetics. 12 (8), e1006268 (2016).
- Chu, H. P., et al. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell. 170 (1), 86-101 (2017).
- Lai, L. T., Lee, P. J., Zhang, L. F. Immunofluorescence protects RNA signals in simultaneous RNA-DNA FISH. Experimental Cell Research. 319 (3), 46-55 (2013).
- Zhang, L. F., et al. Telomeric RNAs mark sex chromosomes in stem cells. Génétique. 182 (3), 685-698 (2009).
- Gallardo, F., Chartrand, P. Visualizing mRNAs in fixed and living yeast cells. Methods in Molecular Biology. 714, 203-219 (2011).
- Querido, E., Chartrand, P. Using fluorescent proteins to study mRNA trafficking in living cells. Methods in Cell Biology. 85, 273-292 (2008).
- Cusanelli, E., Chartrand, P. Telomeric noncoding RNA: telomeric repeat-containing RNA in telomere biology. Wiley Interdisciplinary Reviews: RNA. 5 (3), 407-419 (2014).
- Perez-Romero, C. A., Lalonde, M., Chartrand, P., Cusanelli, E. Induction and relocalization of telomeric repeat-containing RNAs during diauxic shift in budding yeast. Current Genetics. , (2018).
- Avogaro, L., et al. Live-cell imaging reveals the dynamics and function of single-telomere TERRA molecules in cancer cells. RNA Biology. , 1-10 (2018).
- Ran, F. A., et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154 (6), 1380-1389 (2013).
- Yamada, T., et al. Spatiotemporal analysis with a genetically encoded fluorescent RNA probe reveals TERRA function around telomeres. Scientific Reports. 6, 38910 (2016).
- Deng, Z., et al. Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. Journal of Cell Science. 125 (Pt 18), 4383-4394 (2012).
- Casini, A., et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nature Biotechnology. 36 (3), 265-271 (2018).
- Nergadze, S. G., et al. CpG-island promoters drive transcription of human telomeres. RNA. 15 (12), 2186-2194 (2009).
- Porro, A., et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nature Communications. 5, 5379 (2014).
- Farnung, B. O., Brun, C. M., Arora, R., Lorenzi, L. E., Azzalin, C. M. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS One. 7 (4), e35714 (2012).
- Flynn, R. L., et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 347 (6219), 273-277 (2015).
- Scheibe, M., et al. Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Research. 23 (12), 2149-2157 (2013).
- Bolland, D. J., King, M. R., Reik, W., Corcoran, A. E., Krueger, C. Robust 3D DNA FISH using directly labeled probes. Journal of Visualized Experiments. (78), (2013).
- Masui, O., et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 145 (3), 447-458 (2011).

