Identifying Amino Acid Overproducers Using Rare-Codon-Rich Markers
Summary
This study presents an alternative strategy to the conventional toxic analog-based method in identifying amino acid overproducers by using rare-codon-rich markers to achieve accuracy, sensitivity, and high-throughput simultaneously.
Abstract
To satisfy the ever-growing market for amino acids, high-performance production strains are needed. The amino acid overproducers are conventionally identified by harnessing the competitions between amino acids and their analogs. However, this analog-based method is of low accuracy, and proper analogs for specific amino acids are limited. Here, we present an alternative strategy that enables an accurate, sensitive, and high-throughput screening of amino acid overproducers using rare-codon-rich markers. This strategy is inspired by the phenomenon of codon usage bias in protein translation, for which codons are categorized into common or rare ones based on their frequencies of occurrence in the coding DNA. The translation of rare codons depends on their corresponding rare transfer RNAs (tRNAs), which cannot be fully charged by the cognate amino acids under starvation. Theoretically, the rare tRNAs can be charged if there is a surplus of the amino acids after charging the synonymous common isoacceptors. Therefore, retarded translations caused by rare codons could be restored by feeding or intracellular overproductions of the corresponding amino acids. Under this assumption, a selection or screening system for identifying amino acid overproducers is established by replacing the common codons of the targeted amino acids with their synonymous rare alternatives in the antibiotic resistance genes or the genes encoding fluorescent or chromogenic proteins. We show that the protein expressions can be greatly hindered by the incorporation of rare codons and that the levels of proteins correlate positively with the amino acid concentrations. Using this system, overproducers of multiple amino acids can be readily screened out from mutation libraries. This rare-codon-based strategy only requires a single modified gene, and the host is less likely to escape the selection than in other methods. It offers an alternative approach for obtaining amino acid overproducers.
Introduction
The current production of amino acids relies heavily on fermentation. However, the titers and yields for most amino acid production strains are below the rising demands of the global amino acid market that is worth billions of dollars1,2. Obtaining high-performance amino acid overproducers are critical for the upgrade of the amino acid industry.
Traditional strategy to identify amino acid overproducers exploits the competitions between amino acids and their analogs in protein synthesis3,4. These analogs are able to charge the tRNAs that recognize the corresponding amino acids and thus inhibit the elongations of the peptide chains, leading to arrested growth or cell death5. One way to resist the analog stresses is to increase the concentrations of intracellular amino acids. The enriched amino acids will outcompete the analogs for the finite tRNAs and ensure the correct synthesis of functional proteins. Therefore, strains that survive the analogs can be selected and are likely the overproducers of the corresponding amino acids.
Although proved successful in selecting overproducers for amino acids such as L-leucine6, the analog-based strategy suffers from severe drawbacks. One major concern is the analog resistance originated from the process of mutagenesis or through spontaneous mutations. Strains with resistance can escape the selection by blocking, exporting, or degrading the analogs5. Another concern is the toxic side effects of the analogs on other cellular processes7. As a consequence, strains that survive the analog selection may not be the amino acid overproducers, while the desired overproducers could be falsely exterminated due to the negative side effects.
Here, a novel strategy based on the law of codon bias is presented in order to achieve accurate and rapid identifications of amino acid overproducers. Most amino acids are encoded by more than one nucleotide triplet that is favored differently by the host organisms8,9. Some codons are rarely used in the coding sequences and are referred to as the rare codons. Their translations into amino acids rely on the cognate tRNAs that carry the corresponding amino acids. However, the tRNAs that recognize rare codons usually have much lower abundances than the tRNAs of the common codons10,11. Consequently, these rare tRNAs are less likely to capture the free amino acids in the competitions with other isoacceptors, and translations of the rare-codon-rich sequences begin to decelerate or even are terminated when the amounts of amino acids are limited10. The translations could, theoretically, be restored if there is an amino acid surplus after charging the synonymous common tRNAs due to overproductions or extra feedings of the corresponding amino acids12. If the rare-codon-rich gene encodes a selection or screening marker, strains exhibiting the corresponding phenotypes can then be readily identified and are likely the overproducers of the targeted amino acids.
The above strategy is applied to establish a selection and a screening system for the identification of amino acid overproducers. The selection system uses antibiotic resistance genes (e.g., kanR) as markers while the screening system uses the genes encoding fluorescent (e.g., green fluorescent protein [GFP]) or chromogenic (e.g., PrancerPurple) proteins. The marker genes in both systems are modified by replacing defined numbers of the common codons for the targeted amino acid with its synonymous rare alternative. Strains in the mutation library that harbor the rare-codon-rich marker gene are selected or screened under proper conditions, and the overproducers of the targeted amino acids can be readily identified. The workflow begins with the construction of the rare-codon-rich marker gene system, followed by the optimization of the working conditions, and then the identification and verification of the amino acid overproducers. This analog-independent strategy is based on the dogma in protein translation and has been practically verified to enable accurate and rapid identifications of amino acid overproducers. Theoretically, it could be directly employed to amino acids with rare codons and to all microorganisms. In all, the rare-codon-based strategy will serve as an efficient alternative to the conventional analog-based approach when proper analogs for specific amino acids are unavailable, or when a high false positive rate is the major concern. The protocol below uses leucine rare codon to demonstrate this strategy in identifying Escherichia coli L-leucine overproducers.
Protocol
1. Construction of the plasmids expressing the rare-codon-rich marker genes
- Select a marker gene that contains an appropriate number of the common codons for the targeted amino acid.
NOTE: For L-leucine, the kanamycin resistance gene kanR, which contains 29 leucine codons, of which 27 are common codons, is used for the construction of the selection system13. The gfp gene, which contains 17 common codons out of 19 leucine codons, or the purple protein-encoding gene prancerpurple (ppg), which harbors 14 leucine common codons, is used for the screening system (Supplementary Table 1). - Replace the common codons in the marker genes with the synonymous rare codon. For L-leucine, replace its codons in kanR, gfp, or ppg with the rare codon CTA, generating kanR-RCs, gfp-RC, or ppg-RC, respectively13 (Supplementary Table 1).
NOTE: The frequency of the rare codon in the marker genes will affect the stringency of the selection or screening system. In general, increasing the number of rare codons will increase the stringency of the selection or screening system. To achieve the appropriate selection or screening strength, design a series of marker genes that harbor different numbers of rare codons and compare their effects. - Generate building blocks of the rare-codon-rich marker genes using tools such as GeneDesign14 (http://54.235.254.95/gd/) for gene synthesis. Alternatively, order the marker genes from commercial gene synthesis services.
- On the GeneDesign page, choose Building block design (constant length overlap).
- Paste the sequences of the rare-codon-rich marker genes in the Sequence box.
- Define the overlap length between the assembly oligos; keep in mind that the default 40 bp works fine for most sequences.
NOTE: See the online manual for more instructions on the settings of the other parameters. - Click the Design building blocks button and order the oligonucleotides listed on the page.
- Synthesize the rare-codon-modified genes by polymerase chain reaction (PCR)-based accurate synthesis15.
- Ligate the rare-codon-rich kanR-RC to vector pET-28a, gfp-RC to pSB1C3, and ppg-RC to CPB-37-44116.
NOTE: The pET-28a and pSB1C3 plasmid maps are available on the SnapGene online plasmid database (Supplementary Table 1); the CPB-37-441 plasmid map is available on the ATUM chromogenic protein website.- On ice, add the vector and the marker fragments in a molar ratio of 1:1 to 7.5 μL of assembly mix (see Table of Materials) to a total volume of 10 μL. Incubate the sample at 50 °C for 1 h.
- Transform 5 μL of the assembly product into 50 μL of competent cells (see the Table of Materials) at 42 °C for 30 s.
- Recover the cells in SOC (Super optimal broth with catabolite repression) medium at 37 °C for 1 h, plate them on LB (lysogeny broth) agar medium, and incubate them at 37 °C overnight.
- Inoculate the colony into LB medium and incubate at 37 °C for 8 h.
- Isolate the plasmid using a preferred commercial kit.
2. Optimizing the selection conditions
- Make the parent strain used for mutagenesis into competent cells17.
- Transform 50 μL of the competent cells with 1 μL of plasmid that carries the wild-type kanR, and transform another set of the competent cells with the plasmid containing kanR-RC29 with all leucine codon replaced by the rare codon CTA.
- Add 950 μL of SOC medium and incubate the sample in a shaker at 250 rpm at 37 °C for 1 h.
- Plate 100 μL of the cell culture onto the LB agar medium containing 50 μg·mL-1 kanamycin, and incubate it at 37 °C for approximately 8 h until colonies appear.
- Pick the colonies that harbor the wild-type kanR and the leucine rare-codon-rich kanR-RC29 and use each to inoculate 5 mL of fivefold diluted LB medium (0.2x LB) containing 50 μg·mL-1 kanamycin. Incubate the samples in a shaker at 250 rpm at 37 °C.
NOTE: The medium is crucial for the selection system. It should contain just enough carbon and nitrogen to allow expression of the antibiotic resistance protein from the wild-type gene rather than from the rare-codon-modified derivatives. In this case, the L-leucine concentration in 1x LB medium is too high to completely inhibit the protein expression from the rare-codon-rich kanR. Thus, a diluted LB medium with a dilution factor such as 5 is used to generate a clear difference in protein expressions from the wild-type and the rare-codon-rich genes. - Transfer 200 μL of each of the cell cultures to a 96-well plate in triplicate at defined time points (e.g., 8 h, 16 h, and 24 h). Measure the OD600 (optical density at 600 nm) using a plate reader.
NOTE: If a decrease in the cell OD600 cannot be detected for strains harboring the rare-codon-rich marker genes in comparison to the OD600 of the strain harboring the wild-type marker genes, try to increase the amount of rare codon in the marker genes or use a more diluted medium. - Perform the amino acid feeding assay to test if the expressions of the rare-codon-rich marker genes (e.g., kanR-RC29) can be restored by increasing the concentration of the targeted amino acids.
NOTE: For the selection of L-leucine overproducers, L-leucine is used for feeding.- Inoculate the strains harboring the kanR-RC29 marker gene into 5 mL of the 0.2x LB (containing 50 μg·mL-1 kanamycin) with or without the supply of 1 g·L-1 L-leucine. Inoculate another 5 mL of the 0.2x LB with strains that harbor the wild-type kanR as control. Incubate the samples in a shaker at 250 rpm at 37 °C.
- Measure the OD600 for each culture at defined time points (e.g., 15 h, 17 h, 19 h, and 22 h).
3. Optimizing the screening conditions
- Transform 50 μL of the parent strain used for mutagenesis with 1 μL of plasmid (~50 ng·μL-1) that carries the wild-type gfp or the wild-type ppg. Also, transform the parent strain with the rare-codon-rich derivatives of the marker genes.
- Add 950 μL of SOC medium and incubate the sample in a shaker at 250 rpm at 37 °C for 1 h.
- Plate 100 μL of the cell culture onto the LB agar medium containing the appropriate antibiotics (25 μg·mL-1 chloramphenicol for the gfp plasmid carrying a cmR marker, or 50 μg·mL-1 kanamycin for the ppg plasmid carrying a kanR marker) and incubate overnight at 37 °C.
- Pick one colony that harbors the wild-type gfp or ppg and one colony that harbors the corresponding rare-codon-rich derivative, and transfer them to 5 mL of the properly diluted LB medium individually. Incubate the samples in a shaker at 250 rpm at 37 °C.
NOTE: For E. coli strains harboring the leucine rare-codon-rich gfp-RC or ppg-RC, the undiluted LB medium (1x LB) can be used to create significant differences in the expressions of the wild-type and the rare-codon-rich marker genes. Also, inducible promoters are used to drive the expressions of the screening marker genes. Begin the induction when the cells enter the exponential phase to achieve better discriminations. - For fluorescence markers, transfer 200 μL of each of the cell cultures into a 96-well clear-bottomed black plate in triplicate at defined time points (e.g., 2 h, 4 h, and 6 h). Measure the OD600 and the fluorescence and calculate the fluorescence intensity (the ratio of fluorescence to OD600). For chromogenic markers, measure the color development of the cell cultures.
NOTE: If lower fluorescence intensities cannot be detected for strains harboring the gfp-RC in comparison to that of the strains harboring the wild-type gfp, or if a lighter color cannot be detected for cells expressing the purple protein from the ppg-RC than from the wild-type gene, try to increase the number of rare codon on the marker genes or use a more diluted medium. - Perform the feeding assay (see steps 2.7.1 and 2.7.2). Measure the fluorescence intensity or the color development at defined time points (e.g., 12 h, 18 h, and 24 h).
4. Identification of the amino acid overproducers
- Inoculate 100 μL of the culture of the mutants into 5 mL of LB medium (2% v/v) and incubate it in a shaker at 250 rpm at 37 °C until the values of OD600 reach 0.4.
- Make the mutants into competent cells17.
- Transform 50 μL of the mutant cells with 1 μL of the plasmid (~50 ng·μL-1) carrying the selection marker kanR-RC29 or the screening markers gfp-RC or ppg-RC. Add 950 μL of SOC medium and rotate the sample in a 37 °C shaker for 1 h.
- Select amino acid overproducers.
- Centrifuge the cell culture at 4,000 x g for 5 min, discard the supernatant and add 5 mL of 0.2x LB medium containing 50 μg·mL-1 kanamycin. Incubate the sample in a 37 °C shaker overnight.
- Plate the overnight culture (e.g., 100 μL, depends on the cell density of the culture) onto 0.2x LB agar medium containing 50 μg·mL-1 kanamycin and incubate at 37 °C for 12 h.
NOTE: The colonies developed are the candidates of the targeted amino acid overproducers and, in this case, the L-leucine overproducers.
- Screen the amino acid overproducers.
- Plate the appropriate number of cells (e.g., 100 μL) harboring the screening marker (as described in step 4.3) onto the LB agar medium containing the appropriate antibiotic (25 μg·mL-1 chloramphenicol for screening with gfp-RC and 50 μg·mL-1 kanamycin for screening with ppg-RC) and incubate at 37 °C for 8 h.
- Inoculate the LB medium containing the appropriate antibiotic (see step 4.5.1) with each single colony from the plate. Incubate the samples in a shaker at 250 rpm at 37 °C.
- Measure the OD600 and the fluorescence and calculate the fluorescence intensity if gfp-RC is used. Measure the color development of the cell cultures if ppg-RC is used. Note that the strains exhibiting a higher fluorescence intensity or a deeper color than that of the parent strain are the candidate amino acid overproducers and, in this case, the L-leucine overproducers.
NOTE: The fluorescence-activated cell sorting is also suitable for identifying single-cell high-producers when the rare-codon-rich fluorescent protein genes are used for screening.
- Verify the amino acid productivities of the candidate strains.
- Inoculate 5 mL of LB medium with each of the candidate strains and let the cells grow overnight in a shaker at 250 rpm at 37 °C.
- Harvest the cells from 1 mL of the cell culture by centrifugation at 4,000 x g for 2 min. Discard the supernatant and resuspend the pellet with 1 mL of sterile water.
- Inoculate 20 mL of M9 medium containing 4% glucose with 200 μL of the cell suspension and incubate in a 250 mL shaker at 250 rpm at 37 °C for 24 h.
- Centrifuge 1 mL of the culture medium at 4,000 x g for 5 min. Transfer 200 μL of the supernatant to a clean 1.5 mL tube. Prepare L-leucine solutions (HPLC (high-performance liquid chromatography) grade) of 0.01, 0.05, 0.1, 0.5, and 1 g·L-1 as standards.
- Add 100 μL of 1 mM triethylamine and 100 μL of 1 M phenyl isothiocyanate to the supernatant and the standards, mix them gently, and incubate them at room temperature for 1 h18.
CAUTION: Triethylamine and phenyl isothiocyanate can cause severe skin burns and eye damage and is harmful if inhaled. Wear gloves and a mask and, if possible, perform this step in a fume hood. - Add 400 μL of n-hexane to the same tube and vortex it for 10 s. Filter the lower phase containing the amino acid derivatives through a 0.2 μm polytetrafluoroethylene membrane.
CAUTION: n-hexane can cause skin irritation. Wear gloves and protective clothing. If it comes in contact with skin, rinse the skin with plenty of water. - Prepare mobile phase A by mixing 0.1 M sodium acetate (pH 6.5) and acetonitrile in a 99.3:0.7 volumetric ratio. Prepare acetonitrile (80% v/v) as mobile phase B. Filter all mobile phases through 0.2 μm polytetrafluoroethylene membranes.
CAUTION: Acetonitrile is harmful if inhaled and can cause skin and eye irritations. Wear gloves and protective clothing and perform this step in a fume hood. - Run 1 μL of the sample on an ultra-HPLC equipped with a C18 column according to the elution program in Table 1 with a flow rate of 0.42 mL·min-1 and a column temperature of 40 °C. Detect the targeted amino acids at 254 nm with a diode array detector and calculate their concentrations by mapping the peak areas to the standard curve.
| Time (min) | Mobile phase A (%) | Mobile phase B (%) |
| 0 | 98 | 2 |
| 3.5 | 70 | 30 |
| 7 | 43 | 57 |
| 7.1 | 0 | 100 |
| 11 | 98 | 2 |
Table 1: Elution program for the quantification of amino acids.
Representative Results
For the selection system, a sharp decrease in OD600 for strains harboring the rare-codon-rich antibiotic resistance gene should be observed in comparison to the strain harboring the wild-type antibiotic resistance gene when cultured in a suitable medium (Figure 1a). Under the same conditions, the decrease in cell OD600 becomes more obvious as the number of rare codons in the antibiotic resistance gene increases (Figure 1a). It should be noted that the inhibition of rare codon on protein expressions mostly takes place under starved conditions. Therefore, if the LB medium is not properly diluted, no significant decrease in cell OD600 will be observed for the strain harboring the rare-codon-rich marker gene in comparison to the strain harboring the wild-type gene (Figure 1b). After extra feeding of the corresponding amino acid, the OD600 for the strain harboring the rare-codon-rich antibiotic resistance gene will increase significantly and approach that of the strain harboring the wild-type gene (Figure 1c).
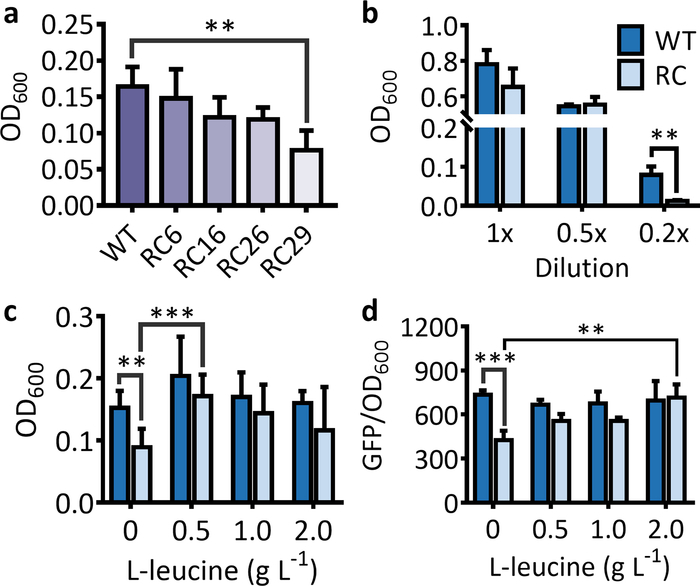
Figure 1: Effects of rare codon on the expressions of marker genes used for the selection and the screening systems. (a) The cell OD600 for an E. coli strains harboring the antibiotic resistance gene (kanR) with 6, 16, 26, and 29 leucine rare-codon (RC6, RC16, RC26, and RC29) replacement after 5 h of incubation. (b) The cell OD600 for an E. coli strain harboring the wild-type (WT) and the rare-codon-rich kanR (RC) in 1x, 0.5x, and 0.2x LB media after 5 h of incubation. (c) Effects of feeding L-leucine on the cell growth for E. coli strains harboring the leucine rare-codon-rich kanR gene after 5 h of incubation. The values and error bars represent the mean and the SD (n = 6). The feeding of L-leucine significantly increased the OD600 for cells harboring the rare-codon-rich kanR. The only exception was for the feeding of 2 g·L-1 L-leucine due to a high SD in OD600 for the feeding treatment. (d) Effects of rare-codon and L-leucine feeding on GFP expressions from the wild-type (WT) and the leucine rare-codon-rich (RC) genes after 16 h of incubation. The feeding of 0.5–2 L-1 L-leucine significantly increased the fluorescence intensity for cells harboring the rare-codon-rich gfp. The values and error bars represent the mean and the SD (n = 3). **P < 0.01, ***P < 0.001 as determined by two-tailed t-test, and only the most significant results were shown. Please click here to view a larger version of this figure.
For the screening system, the fluorescence intensity and the number of fluorescent cells will be significantly lower for the strain that expresses the fluorescent protein from the rare-codon-rich gene than from the wild-type gene (Figure 1d and Figure 2). When using the purple protein, the color developed from the rare-codon-rich ppg should be lighter than that from the wild-type gene when expressed under the same conditions for the same incubation period (Figure 3). Feeding of the corresponding amino acid will restore protein expressions from the rare-codon-rich genes. For strains harboring the rare-codon-rich gfp, the fluorescence intensity (Figure 1d) and the number of fluorescent cells (Figure 2) should increase significantly and approach that of the strains containing the wild-type gfp. When undiluted LB is used, the amino acids in the medium would be sufficient to allow slow expression of the rare-codon-rich ppg even without extra L-leucine feeding, and the expressed purple protein would become visible once the cells are pelleted (Figure 3). However, this does not conceal the fact that gene expression from the rare-codon-rich ppg was dramatically enhanced by feeding of the L-leucine to 2 g·L-1, especially when observed in liquid culture (Figure 3). Therefore, the liquid culture is a better choice for screening based on chromogenic proteins, and the use of diluted LB medium would bring a more significant difference between the phenotypes induced by the wild-type and the rare-codon-rich genes.
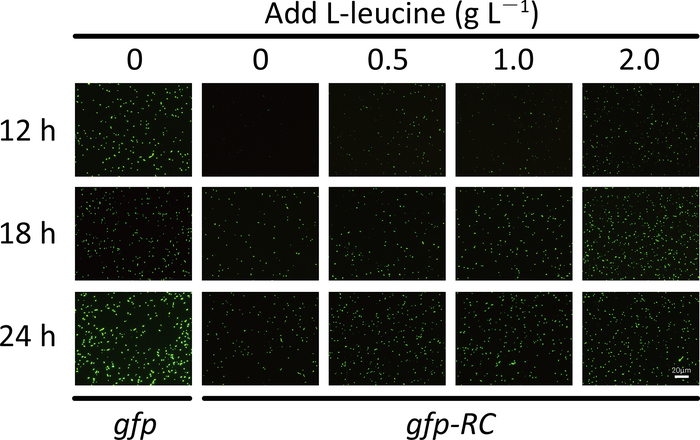
Figure 2: The number of fluorescent E. coli cells that harbor the wild-type gfp or the leucine rare codon-rich gfp (gfp-RC) after the addition of L-leucine. Cells were cultured in 1x LB medium. Scale bar = 20 μm. Please click here to view a larger version of this figure.
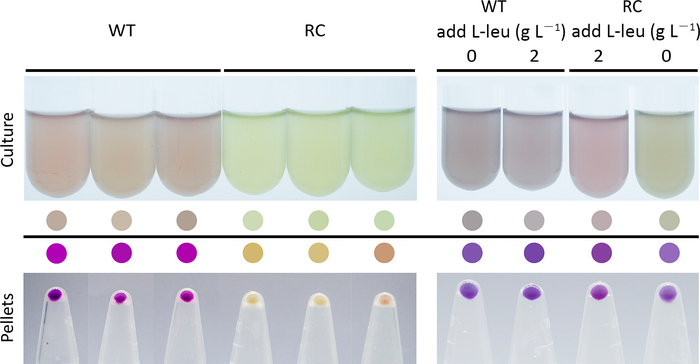
Figure 3: Color development for cells harboring the wild-type (WT) and the leucine rare-codon-rich (RC) ppg genes that encode a purple protein in 1x LB medium (left panel) and the effect of L-leucine feeding on cell culture color development (right panel). The ppg genes were induced when the cells entered the exponential phase and the images were captured 3 h after the induction. The L-leucine was added to the medium together with the inducer in the feeding assay. The colored circles were generated by picking the colors of the cell cultures and the cell pellets. Please click here to view a larger version of this figure.
The rare-codon-based strategy is able to identify overproducers of the targeted amino acids from the mutation library, and these mutants should produce higher amounts of the targeted amino acids than the parent strains (Figure 4).
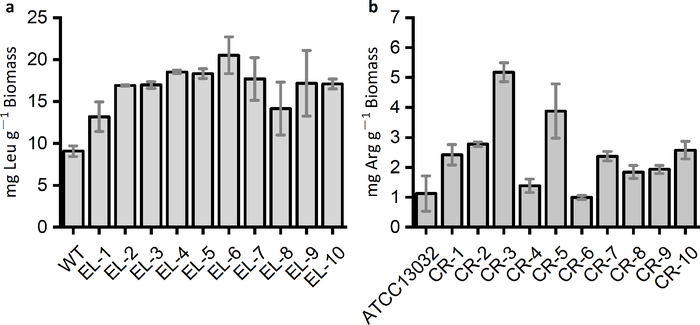
Figure 4: Amino acids produced by the wild-type and the mutated strains identified by the rare-codon-based strategy. (a) L-leucine productions of E. coli strains identified from mutation libraries by the kanR-RC29 (EL-1 to EL-5) and the gfp-RC that harbors 29 and 19 leucine rare codons (EL-6 to EL-10), respectively. (b) L-arginine productions of Corynebacterium glutamicum strains selected by the rare-codon-rich kanR, which contained eight arginine rare codons (AGG). The marker gene was introduced into the C. glutamicum mutation libraries derived from the wild-type strain ATCC13032. The selection medium was 0.3x CGIII supplied with 25 μg·mL-1 kanamycin. Please click here to view a larger version of this figure.
Discussion
The number of rare codons in the marker genes and the selection or screening medium are critical to inhibit protein expressions from the rare-codon-modified marker genes. If no significant difference can be detected between protein expressions from the wild-type marker genes and their derivatives, increasing the number of rare codons or using a nutrient-limited medium may amplify the differences. However, if the inhibition effect is too strong, the protein expressions may not be recovered even by extra feeding of the corresponding amino acids. In this case, the number of rare codons in the marker genes should be reduced to relieve part of the stress. Another way to fine-tune the selection or screening stringency is to adjust the copy numbers and the expression levels of the rare-codon-rich marker genes. Decreasing the copy number and the expression levels of the marker genes usually leads to stronger differentiations between the amino acid overproducers and the initial strains. Therefore, vectors containing the low copy number replication origins such as p15A or pSC101, as well as weak promoters, should be used. If the marker gene is driven by an inducible promoter, low induction is recommended.
The rare-codon-based strategy for the selection or screening for amino acid overproducers is a reverse adaptation of the commonly used strategy of “codon optimization”, which aims at facilitating the expressions of exogenous proteins. In codon optimization, the rare codons on the targeted genes are replaced by the synonymous common ones with respect to the host; thus, the genes from other organisms could be translated much more rapidly into proteins than those exogenous genes with high proportions of rare codons19. Therefore, it is reasonable to assume that the “reverse optimization”, which switches the common codons to their synonymous rare ones, should inhibit gene expressions. However, the gene expressions should be restored by enhanced charging of the corresponding rare tRNAs when the targeted amino acids accumulate intracellularly. The incorporation of rare codons increases the threshold of the amino acid concentration in protein expressions, which offers a potential strategy to select or screen for amino acid overproducers when combined with the proper marker genes.
Besides the antibiotic resistance genes, the fluorescent protein genes, and the chromogenic protein genes used in the protocol, various marker genes could be employed to establish the rare-codon-based selection or screening system. For instance, lethal genes such as tolC20 and sacB21 could be used to select amino acid overproducers. In this case, common codons on the genes that belong to the antidote system should be replaced by the synonymous rare codons of the targeted amino acids. Strains that overproduce the targeted amino acids are able to launch the antidote system and, thus, survive the toxic effects induced by the lethal genes.
It should be noted that side effects may occur when using high amounts of amino acids in the feeding assay. This is because some amino acids are toxic to the microorganisms. For instance, a concentration of around 100 mg·L-1 for L-serine is able to inhibit the growth of E. coli22. However, although lower than that of the wild-type gene, we found that feeding up to 2 g·L-1 L-serine could still restore the expressions of antibiotic resistance genes that rich in serine rare codon13. Therefore, the amino acid toxicity, at least for L-serine, would not jeopardize the reliability of the feeding assay. To overcome the potential negative effects of amino acid toxicity on the productivities of the targeted strains, strategies such as random mutagenesis and the enhancement of amino acid exportations23 could be applied. In fact, the rare-codon-based method is suitable for identifying tolerant strains capable of withstanding or overproducing amino acids above the toxic levels. The key mutations that confer amino acid tolerance could be identified and introduced into the targeted strains, which would be the ideal hosts for the constructions of amino acid overproducers.
The rare-codon-based selection or screening system ensures high fidelity. In other words, strains identified by the system are supposed to be the overproducers of the targeted amino acids. However, in some cases, the candidates that survive the antibiotic selection cannot produce higher amounts of the targeted amino acids than the parent strain. This could be attributed to the antibiotic resistance acquired by the strains through mutagenesis and then a loss of the selection plasmid24. As a consequence, strains without enhanced amino acid productivities could survive the antibiotic stress and escape the selection. These false positive strains could be eliminated by inserting another selection marker into the selection plasmid, such as a wild-type gene that confers resistance to another antibiotic. Strains that lost the selection plasmid are less likely to obtain dual resistance to the two antibiotics and will be eliminated during selection.
Mutants identified by the rare-codon-based system should be able to overproduce the targeted amino acids in comparison to the initial strains. However, the amino acid productivities for the selected strains may still be lower than the industrial requirements. This does not suggest a failure of the rare-codon-based strategy as the strain performances are independent of the selection or screening process but depend on factors such as the characteristics of the initial strain, the approach of mutagenesis, the size of the mutation library, and the fermentation conditions. In order to obtain high-production strains, attention should be paid to the strategies of strain engineering, such as by random mutagenesis or through rational design of the amino acid biosynthetic pathways. Combining adaptive laboratory evolution and the rare-codon-based strategy would facilitate obtaining amino acid overproducers.
The methionine and the tryptophan do not have alternative codons among the 20 proteinogenic amino acids. Therefore, this strategy may not be employed directly to these amino acids. One possible solution is to use engineered tRNAs that are able to recognize the stop codons to carry these amino acids. Thus, the corresponding stop codons could be adopted as the artificial rare codons of these amino acids25,26.
One of the biggest shortcomings concerning the conventional analog-based strategy for the selection of amino acid overproducers is the high false positive rate5,27. Strains that go through mutagenesis could easily acquire resistance toward the toxic amino acid analogs, and the tolerance may even be acquired without the aid of mutagens27. These strains could easily escape the selection pressures from the amino acid analogs and, consequently, the selected strains are usually not the true amino acid overproducers which greatly sacrifices the efficiency of the selection process.
In contrast, the rare-codon-based strategy outcompetes the traditional analog-based method by enabling accurate and rapid identifications of amino acid overproducers. To our knowledge, this is the first strategy that adopts the natural law of codon bias. It only relies on a single rare-codon-rich marker gene and, thus, eliminates the use of toxic analogs. The marker genes are generally nontoxic to the host strains, and the protein expressions from rare-codon-rich genes depend primarily on the intracellular concentrations of the corresponding amino acids because of the universal and stringent law of codon bias across all species. This would prevent the strains from escaping the selection pressures. Besides, due to the great diversity of marker genes, the rare-codon-based strategy could offer various choices for both the selection and the screening of amino acid overproducers.
Due to the universal phenomenon of codon bias in all living organisms28, the rare-codon-based selection or screening strategy could theoretically be employed to other microorganisms besides E. coli, especially those with industrial potentials. When changing to a different host, the choice of rare codons used for designing the marker genes should be based on the codon usage frequencies and the abundances of the corresponding tRNAs for the specific host. The medium used for selection or screening should also be optimized accordingly. One example is the commonly used C. glutamicum in amino acid fermentations. A rare-codon-modified kanR gene containing eight arginine rare codons (AGG) has been shown effective in selecting C. glutamicum L-arginine overproducers by a previous study13 (Figure 4b). Explorations of the rare-codon-based strategy should facilitate the constructions and understandings of amino acid overproducers. Besides amino acids, the rare-codon-based strategy could also be employed with isobutanol, 3-methyl-1-butanol, 2-methyl-1-butanol, and other products that share the same biosynthetic pathways with certain amino acids29. Strains identified by marker genes that harbor the rare codons of these amino acids are capable of overproducing the precursor compounds, which could be channeled to the synthesis of the amino acid derivatives. Therefore, the rare-codon-based strategy could serve as an indirect yet rapid method to reflect the potentials of the strains in accumulating these chemicals either intra- or extracellularly. Key mutations that confer increased amino acid productivities from various overproducers could be identified by deep sequencing and be introduced individually or simultaneously into industrial strains to further improve the amino acid productions.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The work was jointly supported by the National Natural Science Foundation of China (grant no. 21676026), the National Key R&D Program of China (grant no. 2017YFD0201400), and the China Postdoctoral Science Foundation (grant no. 2017M620643). Works in the UCLA Institute of Advancement (Suzhou) were supported by the internal grants from Jiangsu Province and Suzhou Industrial Park.
Materials
| Acetonitrile | Thermo | 51101 | |
| EasyPure HiPure Plasmid MiniPrep Kit | Transgen | EM111-01 | |
| EasyPure Quick Gel Extraction Kit | Transgen | EG101-01 | |
| Gibson assembly master mix | NEB | E2611S | |
| Isopropyl β-D-1-thiogalactopyranoside | Solarbio | I8070 | |
| L-leucine | Sigma | L8000 | |
| Microplate reader | Biotek | Synergy 2 | |
| n-hexane | Thermo | H3061 | |
| Phenyl isothiocyanate | Sigma | P1034 | |
| PrancerPurple CPB-37-441 | ATUM | CPB-37-441 | |
| TransStar FastPfu Fly DNA polymerase | Transgen | AP231-01 | |
| Triethylamine | Sigma | T0886 | |
| Ultra-high performance liquid chromatography | Agilent | 1290 Infinity II | |
| Wild type C. glutamicum | ATCC | 13032 | |
| XL10-Gold E. coli competent cell | Agilent | 200314 | |
| ZORBAX RRHD Eclipse Plus C18 column | Agilent | 959759-902K |
References
- Tatsumi, N., Inui, M. . Corynebacterium glutamicum: biology and biotechnology. , (2012).
- Tonouchi, N., Ito, H., Yokota, A., Ikeda, M. Present global situation of amino acids in industry. Amino Acid Fermentation. , 3-14 (2017).
- Gusyatiner, M., Lunts, M., Kozlov, Y., Ivanovskaya, L., Voroshilova, E. . DNA coding for mutant isopropylmalate synthase, L-leucine-producing microorganism and method for producing L-leucine. , (2005).
- Park, J. H., Lee, S. Y. Towards systems metabolic engineering of microorganisms for amino acid production. Current Opinion in Biotechnology. 19 (5), 454-460 (2008).
- Norris, R., Lea, P. The use of amino acid analogues in biological studies. Science Progress. , 65-85 (1976).
- Park, J. H., Lee, S. Y. Fermentative production of branched chain amino acids: a focus on metabolic engineering. Applied Microbiology and Biotechnology. 85 (3), 491-506 (2010).
- Bach, T. M., Takagi, H. Properties, metabolisms, and applications of L-proline analogues. Applied Microbiology and Biotechnology. 97 (15), 6623-6634 (2013).
- Crick, F. H. C. On the genetic code. Science. 139 (3554), 461-464 (1963).
- Plotkin, J. B., Kudla, G. Synonymous but not the same: the causes and consequences of codon bias. Nature Reviews Genetics. 12 (1), 32-42 (2011).
- Dittmar, K. A., Sørensen, M. A., Elf, J., Ehrenberg, M., Pan, T. Selective charging of tRNA isoacceptors induced by amino‐acid starvation. EMBO Reports. 6 (2), 151-157 (2005).
- Elf, J., Nilsson, D., Tenson, T., Ehrenberg, M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 300 (5626), 1718-1722 (2003).
- Sørensen, M. A. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. Journal of Molecular Biology. 307 (3), 785-798 (2001).
- Zheng, B., et al. Utilization of rare codon-rich markers for screening amino acid overproducers. Nature Communications. 9 (1), 3616 (2018).
- Richardson, S. M., Wheelan, S. J., Yarrington, R. M., Boeke, J. D. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Research. 16 (4), 550-556 (2006).
- Xiong, A. -. S., et al. PCR-based accurate synthesis of long DNA sequences. Nature Protocols. 1 (2), 791-797 (2006).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 343-345 (2009).
- Green, M. R., Sambrook, J. . Molecular cloning: a laboratory manual. , (2012).
- Cohen, S. A., Bidlingmeyer, B. A., Tarvin, T. L. PITC derivatives in amino acid analysis. Nature. 320 (6064), 769-770 (1986).
- Zhou, J., Liu, W. J., Peng, S. W., Sun, X. Y., Frazer, I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. Journal of Virology. 73 (6), 4972-4982 (1999).
- Gregg, C. J., et al. Rational optimization of tolC as a powerful dual selectable marker for genome engineering. Nucleic Acids Research. 42 (7), 4779-4790 (2014).
- Pelicic, V., Reyrat, J. M., Gicquel, B. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. Journal of Bacteriology. 178 (4), 1197-1199 (1996).
- Avcilar-Kucukgoze, I., et al. Discharging tRNAs: a tug of war between translation and detoxification in Escherichia coli. Nucleic Acids Research. 44 (17), 8324-8334 (2016).
- Mundhada, H., Schneider, K., Christensen, H. B., Nielsen, A. T. Engineering of high yield production of L-serine in Escherichia coli. Biotechnology and Bioengineering. 113 (4), 807-816 (2016).
- Makosky, P. C., Dahlberg, A. E. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie. 69 (8), 885-889 (1987).
- Feng, L., Tumbula-Hansen, D., Toogood, H., Söll, D. Expanding tRNA recognition of a tRNA synthetase by a single amino acid change. Proceedings of the National Academy of Sciences of the United States of America. 100 (10), 5676-5681 (2003).
- Naganuma, M., et al. The selective tRNA aminoacylation mechanism based on a single G• U pair. Nature. 510 (7506), 507 (2014).
- Hoesl, M. G., et al. Chemical evolution of a bacterial proteome. Angewandte Chemie International Edition. 54 (34), 10030-10034 (2015).
- Hershberg, R., Petrov, D. A. Selection on codon bias. Annual Review of Genetics. 42, 287-299 (2008).
- Huo, Y. -. X., et al. Conversion of proteins into biofuels by engineering nitrogen flux. Nature Biotechnology. 29, 346 (2011).

