Quantification of Proliferating Human Antigen-specific CD4+ T Cells using Carboxyfluorescein Succinimidyl Ester
Summary
Presented here is a protocol for measuring proliferating CD4+ T cells in response to antigenic proteins or peptides using dye dilution. This assay is particularly sensitive to rare antigen-specific T cells and can be modified to facilitate cloning of antigen-specific cells.
Abstract
Described is a simple, in vitro, dye dilution-based method for measuring antigen-specific CD4+ T cell proliferation in human peripheral blood mononuclear cells (PBMCs). The development of stable, non-toxic, fluorescent dyes such as carboxyfluorescein succinimidyl ester (CFSE) allows for rare, antigen-specific T cells to be distinguished from bystanders by diminution in fluorescent staining, as detected by flow cytometry. This method has the following advantages over alternative approaches: (i) it is very sensitive to low-frequency T cells, (ii) no knowledge of the antigen or epitope is required, (iii) the phenotype of the responding cells can be analyzed, and (iv) viable, responding cells can be sorted and used for further analysis, such as T cell cloning.
Introduction
The ability to detect and study antigen-specific T cells is important in studies of cell-mediated immunity. However, doing so is particularly challenging for autoantigen-specific CD4+ T-cell responses, which are very weak and difficult to detect. A common method used for the detection of antigen-specific lymphocyte proliferation is [3H]-thymidine, which is a radiolabeled nucleotide incorporated into the DNA of proliferating cells. Although the [3H]-thymidine assay can detect DNA synthesis, this method is an indirect measure of cell division, because DNA synthesis can initiate independently of mitosis (i.e., during gene duplication and apoptosis1). This issue is compounded by the fact that antigen-specific proliferation of cells can result in considerable apoptosis2, leading to potential overestimation of antigen-specific proliferation. Furthermore, the [3H]-thymidine method does not provide phenotypic information for proliferating lymphocytes, such as CD4+ or CD8+ lineage proliferation in PBMCs stimulated with antigenic peptides.
In 2003, we published the first dye dilution assay using CFSE, called the CFSE-based proliferation assay3,4. CFSE is a fluorescent dye that binds stably to intracellular proteins by forming a covalent bond to intracellular lysine residues. Since CFSE-labeled proteins are divided equally among daughter cells3, cells that have divided can be distinguished from undivided cells by flow cytometry, which also allows for the quantitative phenotyping of lymphocyte populations. Indeed, the number of divisions a cell has undergone from the time of CFSE-staining can be measured to some degree5. More recently, many similar dyes such as CellTrace Violet proliferation dye (VPD) and CytoTrack dye have been developed, which work in a similar way6. This protocol focuses on CFSE, but the principles apply equally to other related dyes.
Peptide-MHC tetramer staining is a widely used method for detecting and cloning antigen-specific CD8+ T cells. This is a well-established method7,8,9,10; however, tetramer-based cloning requires existing knowledge of the epitope/MHC restriction and each epitope requires its own tetramer11, which limits the scope of discovery and cloning of novel epitope-specific T cells. The CFSE-based proliferation can be used with peptides, proteins, or cell lysates. The protocol described herein is both simple and robust, and the responding CD4+ T cells can be sorted for use in downstream functional and biochemical characterization assays12,13.
Protocol
All subjects gave informed consent prior to the collection of peripheral blood. Ethical approval for experiments using PBMC was given by St. Vincent’s Hosptial (HREC-A 135/08, and HREC-A 161/15).
1. Reagent Preparation
- Human T cell media
- Prepare RP-5 media for culturing PBMC, which consists of RPMI 1640, 1x non-essential amino acids, L-alanyl-L-glutamine dipeptide (2 mM), penicillin (100 U/mL)/streptomycin (0.1 mg/mL), and 5% pooled human serum (PHS).
- CFSE stock solutions
- Prepare a master stock by dissolving 25 mg of CFSE in ~9 mL of DMSO to achieve a final stock solution with a concentration of 5 mM.
- Prepare a working stock by diluting the master stock in PBS to achieve a working concentration of 10 µM.
2. Preparation of Human PBMCs from Whole Blood
- There are generally between 0.5–1.5 x 106 PBMCs/mL of human peripheral blood. Therefore, the amount of blood required depends upon the desired number of PBMCs. Dilute human peripheral blood with PBS at least 1:2. Separate the PBMCs by adding 15 mL of density gradient medium to a 50 mL tube, then overlay 35 mL of diluted whole blood.
- Centrifuge at 850 x g for 15 min without deceleration at room temperature (RT). This will result in three clear layers: the bottom layer containing the red blood cell pellet, middle layer of density gradient medium with white blood cells lining its top interface, and top plasma layer14.
- Remove approximately 20 mL of the top plasma layer. Collect the white blood cell layer, being careful to avoid the red blood cell pellet. Wash 3x with PBS and count viable cells using trypan blue exclusion on a hemocytometer. Dilute to 1 x 106 PBMCs/mL in PBS.
- Non-CFSE-stained cells
- These cells are used as compensation controls for flow cytometry, comprising of unstained and CD4+ single-stained cells. Add 300 µL of each control sample PBMC suspension to a 10 mL tube, top with PBS, and centrifuge at 550 x g for 5 min at RT.
- Resuspend 1 x 106 cells/mL in RP-5 media. Incubate these unlabeled cells for 7 days in a 37 °C/5% CO2 incubator with the CFSE-labeled cells (step 2.6.1).
- CFSE-stained cells
- Transfer the cells from step 2.3 into a 50 mL tube. Add 1.0 µL of CFSE working stock solution (10 µM) per 1 mL of cell suspension to the side of the tube. Mix quickly by inverting the tube several times. The final concentration of CFSE is 10 nM.
- Incubate for 5 min in a 37 °C/5% CO2 incubator. To stop the staining, add 5 mL of RP-5 media, pellet the cells by centrifuging for 5 min at 550 x g. Resuspend the PBMCs at 1 x 106/mL in RP-5 media.
- Add 1.0 mL of cell suspension to a 10 mL tube. Use one tube for each antigen to be tested.
- Antigenic stimulation of human PBMCs and cell culture
- Culture human CFSE-labelled PBMCs with antigens in RP-5 media for 7 days in a 37 °C/5% CO2 incubator. Culture 1 x 105 cells/well (100 µL) in a 96 well plate with 100 µL/well of RP-5 media containing diluted antigen.
NOTE: Antigens used, including working concentrations, are summarized in Table 1.
- Culture human CFSE-labelled PBMCs with antigens in RP-5 media for 7 days in a 37 °C/5% CO2 incubator. Culture 1 x 105 cells/well (100 µL) in a 96 well plate with 100 µL/well of RP-5 media containing diluted antigen.
3. Anti-CD4 Staining for FACS Analysis
- Pipette 200 µL of the cultured cells into FACS tubes, wash cells 1x in 1.0 mL of PBS containing 0.1% FCS, and centrifuge for 5 min at 550 x g.
- Stain with anti-human CD4 Alexa Fluor 647 (0.25 μg/mL) in 100 µL of PBS/0.1% FCS. Keep aside a sample of CFSE-labelled cells, unstained with any other fluorophores, to use for setting the FACS CFSE compensation. Incubate the cells at 4 °C protected from light for 20 min.
- Wash the cells by adding 1 mL of PBS/0.1% FCS, centrifuge at 550 x g for 5 min at RT, and resuspend in 100 µL of PBS/0.1% FCS. Immediately before FACS analysis, add 1 µL of propidium iodide (PI, 0.1 mg/mL) to all tubes to allow the dead cells to be excluded by flow cytometry.
4. Flow Cytometric Configuration and Gating Strategy
NOTE: Figure 1 shows the FACS configuration including compensation controls and gating strategy.
- Gate the forward scatter (FSC) vs. side scatter (SSC) (Figure 1A) population to include all lymphocytes.
- Gate the FSC vs. PI (Figure 1B) population on PI negative cells to exclude apoptotic cells.
- Use unstained cells to set a voltage baseline for non-fluorescent cells. Set the voltages for CD4-A647 and CFSE so that the fluorescence signal is below 1,000 for each (Figure 1C). Use the single color controls CFSE (Figure 1D) and CD4-A647 (Figure 1E) to confirm positive fluorescent signals (~10,000) for each color, using the voltages set with unstained cells.
- CFSE and PI have some spectral overlap; adjust the compensation to subtract PI fluoresence from CFSE fluoresence until the CFSE-only sample does not yield a signal in the PI channel.
NOTE: These gates were applied to all samples analyzed herein.
5. Calculation of Cell Division Index to Enumerate Antigen-specific CD4+ T Cell Proliferation
NOTE: Cell division index (CDI) refers to the number of divided cells (CD4+/CFSEdim) per 5,000 undivided cells (CD4+/ CFSEbright). When the number of undivided CD4+ cells is not exactly 5,000, the number of divided cells is corrected to express the number of divided cells per 5,000 undivided cells. For example, using the tetanus toxoid-specific proliferation (Figure 2D), there were 4,930 undivided cells (CFSEbright) and 3,268 divided cells (CFSEdim); therefore, the corrected number of divided cells is (5,000/4,930) x 3,268 = 3,304.3.
- Calculate the CDI (Table 1) by dividing the number of divided cells/5,000 undivided cells from the antigen-stimulated group by the mean number of divided cells (per 5,000 undivided cells) from the cells cultured without antigen (Table 2).
Representative Results
In vitro stimulation of human PBMCs with tetanus toxoid protein: PBMCs were stained with CFSE and stimulated for 7 days in the presence of tetanus toxoid. Almost all donors showed a strong T cell response to tetanus toxoid because they had been vaccinated, which makes tetanus toxoid a useful positive control antigen. Figure 2 demonstrates in triplicate, that the CFSE proliferation of CD4+ T cells from unstimulated PBMCs was minimal (~12 CFSEdim cells; Figure 2A,B,C), while there was a marked proliferation of CD4+ T cells in response to tetanus toxoid (>3,000 CFSEdim cells; Figure 2D,E,F).
In vitro stimulation of human PBMCs with antigenic peptides: PBMCs were stained with CFSE and stimulated for 7 days in the presence of human proinsulin C-peptide. Figure 3 demonstrates the detection of proinsulin C-peptide-specific CD4+ T cells in the peripheral blood of an individual with type 1 diabetes. Proliferation was not observed in the healthy donor’s PBMC (data not shown). In Figure 4, PBMCs stimulated with influenza A virus (H1N1 PR8) matrix protein demonstrated proliferation; however, the tetanus-specific response for this donor was relatively weak, and responses are generally variable between donors.
Taken together, these results demonstrate that the assay can be performed using full-length proteins and short synthetic peptides.

Figure 1: Compensation controls and gating strategy for CFSE-based proliferation of CD4+ T cells. Unstimulated PBMCs following 7 days of in vitro cell culture. Lymphocytes (A) were stained with propidium iodide, and propidium iodide-negative cells were gated to exclude dead cells (B). FACS compensation controls included unstained cells (C), cells stained with CFSE alone (D), and cells stained with CD4 alone (E). Please click here to view a larger version of this figure.

Figure 2: Proliferation of CFSE-stained tetanus-specific CD4+ T cells. PBMCs were CFSE-stained and cultured for 7 days in the presence of 145 ng/mL (166 LfU/mL) tetanus toxoid. Cells were stained with CD4. CFSE dilution is shown without antigen (A-C) and in response to tetanus toxoid protein (D-F). Please click here to view a larger version of this figure.

Figure 3: Proliferation of CFSE-stained human CD4+ T cells in response to proinsulin C-peptide. CFSE-stained PBMCs were cultured for 7 days in the presence of 10 µM proinsulin C-peptide. Cells were stained with CD4. CFSE dilution is shown using PBMC from a donor with type 1 diabetes. CFSE dilution in unstimulated PBMCs (A), tetanus toxoid protein stimulated (B), and human proinsulin C-peptide stimulated PBMCs (C). Please click here to view a larger version of this figure.
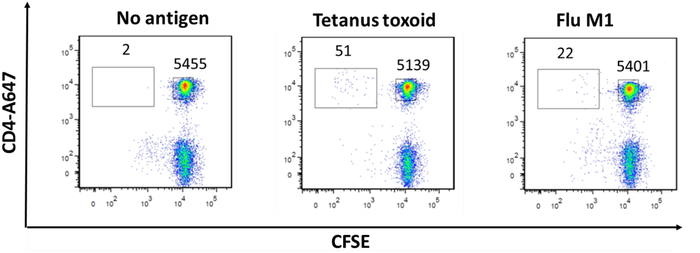
Figure 4: Proliferation of CFSE-stained human CD4+ T cells in response to influenza A virus (H1N1 PR8) matrix protein 1. PBMCs were CFSE-stained and cultured for 7 days in the presence of 0.08 µg/mL influenza A virus matrix protein 1. Cells were stained with CD4. CFSE dilution is shown for unstimulated cells (A), tetanus toxoid stimulated cells (B), and influenza A virus matrix protein-stimulated cells (C). Please click here to view a larger version of this figure.
| Antigen | Working Concentration |
| Tetanus Toxoid protein | 145 ng/mL |
| Influenza A H1N1 (PR8) Matrix protein 1 | 0.08–10 µg/mL |
| Proinsulin C-peptide PI33-63 | 10 nM |
Table 1: Enumeration of tetanus-specific CD4+ proliferation. CDI calculations using 5,000 recorded flow cytometry events from unstimulated and tetanus-stimulated human PBMC.
| Sample | Undivided cells CD4+CFSEbright | Divided cells CD4+CFSEdim | Number of divided cells CFSEdim/5,000 CFSEbright | Mean divided cells CFSEdim/5,000 CFSEbright | |
| Nil 1 | 5,004 | 11 | 11 | 12.3 | |
| Nil 2 | 4,995 | 14 | 14 | ||
| Nil 3 | 5,006 | 12 | 12 | ||
| Sample | Undivided cells CD4+CFSEbright | Divided cells CD4+CFSEdim | Number of divided cells CFSEdim/ 5000 CFSEbright | CDI Number of divided cells (Tetanus) Mean divided cells (Nil) | Mean CDI |
| Tetanus 1 | 4,930 | 3,258 | 3,304.3 | 268 | 263.7 |
| Tetanus 2 | 4,928 | 3,205 | 3,251.8 | 263.7 | |
| Tetanus 3 | 4,910 | 3,142 | 3,199.6 | 259.5 |
Table 2: Antigens used to simulate CD4+ proliferation.
Discussion
CFSE-based proliferation is a simple and robust method for detecting and enumerating antigen-specific human CD4+ T cells. It has been previously demonstrated that using the optimal concentration of CFSE is essential for optimal results4. Too much CFSE abrogates proliferation, whereas too little does not allow for divided and undivided cells to be distinguished. In contrast, relatively high concentrations (5.0 µM) of CFSE are used to label purified murine T cells3. Due to variability between batches, it is recommended that each batch be titrated to determine the optimal concentration. We use our current batch at a much lower concentration (10 nM) than in the original publication (200 nM)4.
Another important aspect of this method is optimization of the antigen dose. It has been well established that T cells cultured with less antigen can be more sensitive and exhibit enhanced function. This was originally demonstrated in the context of HIV peptides15. The amount of tetanus toxoid used in this protocol has also been optimized previously4 to elicit highly sensitive and proliferative CD4+ T cells. Interestingly, it has been shown previously that excess antigen reduces antigen-specific proliferation15. Therefore, to detect and enumerate rare T cell populations in peripheral blood, optimizing both antigen and CFSE concentration is of utmost importance.
Since we published the first human dye dilution-based assay4, many similar dyes have been developed. We found that CD4+ T cell proliferation was detected less effectively when PKH-26, a lipophilic dye, was used compared to CFSE (unpublished). We have not systematically tested other dyes on the market, so there is no comment on their relative strengths. However, the protocol for quantifying T cell proliferation is equally applicable to all dye dilution proliferation assays.
Expressing proliferation as a cell division index (CDI) incorporates the background proliferation into calculation of the magnitude of responses. In our hands, the background is usually low (e.g., <15 events/5,000 CD4+ CFSEbright). Background proliferation does vary between individuals and experiments. Using a ratio (in this case, the CDI) to express the strength of proliferation is somewhat effective in reducing its impact on the results. Background responses to proteins in the media can be problematic. It is recommended to use tissue culture media that contain human serum, not bovine serum, as bovine proteins can stimulate a weak background response that diminishes sensitivity of the assay. A high background proliferation has been found in people who have a mild infection, presumably due to proliferation that is primed in vivo in response to infection. In such cases, the assay needs to be repeated once the donor has cleared the infection.
The CFSE-based proliferation assay can be modified to efficiently clone human antigen-specific CD4+ T cells directly from PBMCs12. For example, this approach has been used to clone CD4+ T cells specific for insulin12. More recently, cloning and functional analyses of proinsulin C-peptide-specific CD4+ T cells has provided remarkable insight into the pathogenesis of T1D, particularly in patients with recent onset T1D13.
In conclusion, the CFSE-based proliferation assay is both sensitive and robust. It is quick and technically simple and can be performed using fresh or cryopreserved PBMC. The ability to incorporate CFSE into multicolor FACS analysis provides a quantitative measure of the immunophenotype of proliferating T cells from PBMC containing many cell types, and this degree of detail is not possible with assays utilizing DNA incorporation such as [3H]-thymidine.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by: The Juvenile Diabetes Research Foundation [JDRF 5-CDA-2014-210-A-N] (S. M.). The National Health and Medical Research Council (NHMRC GNT123586) (S. M.), Diabetes Australia Research Trust Millennium Award (Y17M1-MANS) (S. M.), Operational Infrastructure Support Program of the Victorian Government (S. M., A. D., E. T., M. S.), and NHMRC Postgraduate Scholarship APP1094337 and JDRF PhD Top-up Scholarship (M. S.).
Materials
| Anti-human CD4-AlexaFluor647 | Biolegend | 317422 | RRID:AB_2716180 |
| Carboxyfluorescein succinimidyl ester (CFSE) | ThermoFisher | C1157 | |
| Ficoll-Paque Plus | GE Healthcare | 71-7167-00 | |
| Glutamax (1x) | Gibco | 35050 | |
| Influenza A H1N1 (PR8) Matrix protein 1 | Sino Biological | 40010-V07E | |
| Non-Essential amino acids (1x) | Gibco | 11140 | |
| Penicillin/ Streptomycin (1x) | Gibco | 15070063 | |
| Phosphate buffered saline (PBS) | Sigma-Aldrich | D8537 | |
| Pooled human serum | Australian Red Cross | N/A | |
| Proinsulin C-peptide PI33-63 | Purar Chemicals | N/A | Custom made synthetic peptide |
| RPMI 1640 | Sigma-Aldrich | R8758 | |
| Tetanus Toxoid protein | Statens Serum Intitut | N/A |
References
- Duque, A., Rakic, P. Different effects of BrdU and 3H-Thymidine incorporation into DNA on cell proliferation, position and fate. Journal of Neuroscience. 31, 15205-15217 (2011).
- Mannering, S. I., Zhong, J. I. E., Cheers, C. T-cell activation, proliferation and apoptosis in primary Listeria monocytogenes infection. Immunology. 106, 87-95 (2002).
- Lyons, A. B., Parish, C. R. Determination of lymphocyte division by flow cytometry. Journal of Immunological Methods. 171, 131-137 (1994).
- Mannering, S. I., et al. A sensitive method for detecting proliferation of rare autoantigen-specific human T cells. Journal of Immunological Methods. 283, 173-183 (2003).
- Quah, B. J. C., Parish, C. R. The Use of Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) to Monitor Lymphocyte Proliferation. Journal of Visualized Experiments. , 4-7 (2010).
- Ten Brinke, A., et al. Monitoring T-cell responses in translational studies: Optimization of dye-based proliferation assay for evaluation of antigen-specific responses. Frontiers in Immunology. 8, 1-15 (2017).
- Gillespie, G. M. A., et al. Strong TCR Conservation and Altered T Cell Cross-Reactivity Characterize a B*57-Restricted Immune Response in HIV-1 Infection. Journal of Immunolgy. 177, 3893-3902 (2006).
- Tynan, F. E., et al. High Resolution Structures of Highly Bulged Viral Epitopes Bound to Major Histocompatibility Complex Class I: Implications for t-cell receptor engagement and t-cell immunodominance. Journal of Biological Chemistry. 280, 23900-23909 (2005).
- Blanchard, N., et al. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. Journal of Immunology. 184, 3033-3042 (2010).
- Glanville, J., et al. Identifying specificity groups in the T cell receptor repertoire repertoire. Nature. 547, 94-98 (2017).
- Wooldridge, L., et al. Tricks with tetramers: how to get the most from multimeric peptide MHC. Immunology. 126, 147-164 (2009).
- Mannering, S. I., et al. An efficient method for cloning human autoantigen-specific T cells. Journal of Immunological Methods. 298, 83-92 (2005).
- So, M., et al. Proinsulin C-peptide is an autoantigen in people with type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 115, (2018).
- Hui-Yuen, J., Mcallister, S., Koganti, S., Hill, E., Bhaduri-Mcintosh, S. Establishment of Epstein-Barr Virus Growth-transformed Lymphoblastoid Cell Lines. Journal of Visualized Experiments. , 2-7 (2011).
- Alexander-Miller, M. A., Leggatt, G. R., Berzofsky, J. A., Moss, B. Selective expansion of high or low avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proceedings of the National Academy of Sciences of the United States of America. 93, 4102-4107 (1996).

