Hierarchical and Programmable One-Pot Oligosaccharide Synthesis
Summary
This protocol demonstrates how to use the Auto-CHO software for hierarchical and programmable one-pot synthesis of oligosaccharides. It also describes the general procedure for RRV determination experiments and one-pot glycosylation of SSEA-4.
Abstract
This article presents a general experimental protocol for programmable one-pot oligosaccharide synthesis and demonstrates how to use Auto-CHO software for generating potential synthetic solutions. The programmable one-pot oligosaccharide synthesis approach is designed to empower fast oligosaccharide synthesis of large amounts using thioglycoside building blocks (BBLs) with the appropriate sequential order of relative reactivity values (RRVs). Auto-CHO is a cross-platform software with a graphical user interface that provides possible synthetic solutions for programmable one-pot oligosaccharide synthesis by searching a BBL library (containing about 150 validated and >50,000 virtual BBLs) with accurately predicted RRVs by support vector regression. The algorithm for hierarchical one-pot synthesis has been implemented in Auto-CHO and uses fragments generated by one-pot reactions as new BBLs. In addition, Auto-CHO allows users to give feedback for virtual BBLs to keep valuable ones for further use. One-pot synthesis of stage-specific embryonic antigen 4 (SSEA-4), which is a pluripotent human embryonic stem cell marker, is demonstrated in this work.
Introduction
Carbohydrates are ubiquitous in nature1,2, but their presence and mode of action remain an uncharted territory, mainly due to difficult access to this class of molecules3. Unlike automated synthesis of oligopeptides and oligonucleotides, the development of automated synthesis of oligosaccharides remains a formidable task, and progress has been relatively slow.
To tackle this problem, Wong et al. developed the first automated method for the synthesis of oligosaccharides using a programmable software program called Optimer4, which guides the selection of BBLs from a library of ~50 BBLs for sequential one-pot reactions. Each BBL was designed and synthesized with well-defined reactivity tuned by various protecting groups. Using this approach, the complexities of protecting manipulation and intermediate purification can be minimized during synthesis, which have been considered the most difficult issues to overcome in the development of automated synthesis. Despite this advance, the method is still quite restricted, as the number of BBLs is too small and the Optimer program can only handle certain small oligosaccharides. For more complex oligosaccharides that require more BBLs and multiple passes of one-pot reactions and fragment condensation, an upgraded version of the software program, Auto-CHO5, has been developed.
In Auto-CHO, more than 50,000 BBLs with defined reactivity to the BBL library have been added, including 154 synthetic and 50,000 virtual ones. These BBLs were designed by machine learning based on basic properties, calculated NMR chemical shifts6, 7, and molecular descriptors8, which affect the structure and reactivity of the BBLs. With this upgraded program and new set of BBLs available, the synthesis capacity is expanded, and as demonstrated, several oligosaccharides of interest can rapidly be prepared. It is believed that this new development will facilitate the synthesis of oligosaccharides for the study of their roles in various biological processes and their impacts on the structures and functions of glycoproteins and glycolipids. It is also thought that this work will benefit the glycoscience community significantly, given that this method is available to the research community free of charge. Synthesis of the essential human embryonic stem cell marker, SSEA-45, is demonstrated in this work.
Protocol
1. Auto-CHO software manipulation
- Java Runtime Environment installation: make sure the Java Runtime Environment (JRE) has been installed in the device. If JRE has been installed, go to the next step, “software initialization”; otherwise, download and install JRE according to the user’s operating system found at: <https://www.oracle.com/technetwork/java/javase/downloads/index.html>.
- Software initialization: go to the Auto-CHO website at <https://sites.google.com/view/auto-cho/home> and download the software according to the operating system. Currently, Auto-CHO supports Windows, macOS, and Ubuntu. The latest PDF user guide is provided on the Auto-CHO website.
- For Windows users, unzip the Auto-CHO_Windows.zip and double-click on Auto-CHO.jar in the Auto-CHO_Windows folder to start the program.
NOTE: The user needs to install unzip software, such as 7-Zip, found at <https://www.7-zip.org>, for unpacking the zip file. The user may also use the java -jar Auto-CHO.jar command to start the program by the Windows Command Prompt. - For macOS users, right-click on Auto-CHO.jar and choose Open to start the program.
- For Ubuntu users:
- Install libcanberra-gtk using the following command:
$ sudo apt-get install libcanberra-gtk* - Change the access permission of Auto-CHO_Ubuntu.sh:
$ chmod 755 Auto-CHO_Ubuntu.sh - Run the Auto-CHO program:
$ ./Auto-CHO_Ubuntu.sh
- Install libcanberra-gtk using the following command:
- For Windows users, unzip the Auto-CHO_Windows.zip and double-click on Auto-CHO.jar in the Auto-CHO_Windows folder to start the program.
- Input the desired glycan structure. Choose to draw a glycan structure or read an existing structure file.
- Input by drawing:
- Click on Edit Glycan by GlycanBuilder9,10 (Figure 1, btn1; Figure 2A) or the area of Click Here to Edit the Synthetic Target to draw and edit the query structure by GlycanBuilder. Linkage and chirality information should not be ignored. Click on the Globo-H, SSEA-4, or OligoLacNAc buttons (Figure 1, examples) to display the examples.
- Select File | Export to sequence formats | Export to GlycoCT condensed to save the edited structure (optional).
- Close the GlycanBuilder dialog to complete editing.
- Input by reading a file:
- Click on Edit Glycan by GlycanBuilder (Figure 1, btn1; Figure 2A) or the area of Click Here to Edit the Synthetic Target to edit the query structure.
- Select File | Import from sequence formats to choose the query structure file with the corresponding format.
- Input by drawing:
- Search parameter settings (optional).
- Define the search parameters in the “Parameter Settings” tab (Figure 1, tab2) to get reasonable search results.
NOTE:
Threshold of High-Class RRV must be a real number and ≥0.
Threshold of Medium Class RRV must be a real number and ≥0.
Threshold of High-Class RRV should be >Threshold of Medium Class RRV.
Max Fragment Number should be an integer and ≥1.
Min BBL Number in a Fragment must be an integer and between 1 and 3.
Max BBL Number in a Fragment must be an integer and between 1 and 3.
Max BBL Number in a Fragment must be ≥Min BBL Number in a Fragment.
Min Donor/Acceptor RRV Difference must be a positive real number.
Min Donor/Acceptor RRV Ratio must be a positive real number.
Max Donor/Acceptor RRV Ratio must be a positive real number.
Max Donor/Acceptor RRV Ratio must be >Min Donor/Acceptor RRV Ratio. - Click on the OK button to enable the new settings.
- Define the search parameters in the “Parameter Settings” tab (Figure 1, tab2) to get reasonable search results.
- Select the building block library (Figure 1, tab5). The default setting is to search the experimental library only. If it is desired to search both the experimental and virtual libraries, check the following steps.
- Select the Virtual Building Block Library tab (Figure 2C, tab5). Experimental and virtual building blocks can work together to enhance the searching ability of Auto-CHO. Currently, Auto-CHO provides more than 50,000 virtual building blocks with predicted RRVs in the library.
- Select Use Experimental and Virtual Libraries and apply Filtering to display virtual building blocks with certain criteria. Click on the Show Selected Virtual BBL(s) button (Figure 2C, btn5) to show only the selected virtual building block(s).
- Click on the Show Filtered Virtual BBL(s) button (Figure 2C, btn6) to show only virtual building blocks with certain criteria defined by the user.
- Click on the Show All Virtual BBL(s) button (Figure 2C, btn7) to show all available virtual building blocks and reset the filter.
- Check one or multiple desired virtual building blocks that the user would like to use for searching.
- Select the Query Structure tab (Figure 1, tab1) and click on the Search Building Block Library button (Figure 1, btn2) to find the one-pot synthetic solutions for the query structure. Then, confirm the parameter settings.
- Search the result viewer.
NOTE: The search result is shown in the Result Visualization tab (Figure 1, tab6). The reducing end acceptors of different residue numbers are displayed in the Reducing End Acceptor column (Figure 1, viewer1).- Select a reducing-end acceptor, and solutions are displayed on the Synthetic Solution List (Figure 1, viewer2). Fragments are shown in the Fragment List (Figure 1, viewer3) to suggest how many fragments should be used in the synthesis.
NOTE: The system provides detailed information of each fragment, including the RRV of the fragment, computational yield as well as which protecting group should be deprotected for the subsequent use of the fragment in the one-pot reaction. The building blocks used to assemble the selected fragment are shown in the viewer4 of Figure 1. The viewer5 of Figure 1 also displays the fragment connection information. - View and check chemical structures and detailed information of the selected building blocks in the regions of Chemical Structure of Building Block and Building Block Browser, respectively, for experimental building blocks (Figure 1, tab4).
- Select a reducing-end acceptor, and solutions are displayed on the Synthetic Solution List (Figure 1, viewer2). Fragments are shown in the Fragment List (Figure 1, viewer3) to suggest how many fragments should be used in the synthesis.
- Output the search result to text (optional).
- Select the Result Text tab (Figure 1, tab7).
- Click on Save Result Text (Figure 2B, btn4) and choose the text file destination.
- Feedback for virtual building blocks (optional).
NOTE: Feedback can be given on virtual building blocks through the online questionnaire. Feedback can help the community to keep useful virtual building blocks and remove ineffectual ones.- Select the Virtual Building Block tab (Figure 1, tab5).
- Click on the To rate link of the virtual building block of which it is desired to rate or comment in the Feedback column.
- Fill in the feedback form after the system opens up a webpage and submit it.
NOTE: Do not change the virtual BBL ID.
2. RRV determination experiments
- In a 10 mL round-bottom flask, combine the two thioglycoside donors (0.02 mmol of each: Dr4 is the reference donor with known RRV; Dx1 is the donor molecule of unknown RRV), absolute methanol (0.10 mmol), and Drierite in dichloromethane (DCM, 1.0 mL), then stir at room temperature (RT) for 1 h.
- Take an aliquot of this mixture (30 µL) and inject the mixture into high performance liquid chromatography (HPLC) in three separate injections (10 µL for each injection). Measure the coefficient (a) between the absorption (A) and concentration of the donor molecule [D] under the baseline separation conditions (ether acetate/n-Hexane = 20/80).
- Add a solution of 0.5 M N-Iodosuccinimide (NIS) in acetonitrile (40 µL, 0.02 mmol) into the reaction mixture, followed by addition of a 0.1 M trifluoromethanesulfonic acid (TfOH) solution (20 µL, 0.002 mmol), and stir the mixture at RT for 2 h.
- Dilute the reaction mixture with DCM (4.0 mL), filter, and wash with saturated aqueous sodium thiosulfate containing 10% sodium hydrogen carbonate (2x with 5 mL volume each). Extract the aqueous layer with DCM (3x with 5 mL). Combine all organic layer, wash it with 5 mL of brine, and dry it with approximately 200 mg of anhydrous magnesium sulfate.
- Shake the mixture mildly for 30 s, filter it through a funnel with a fluted filter paper in order to remove the magnesium sulfate, then collect the filtrate in a 25 mL round-bottom flask. Remove the solvent using a rotary evaporator.
- Dissolve the residue in DCM (1.0 mL). Take an aliquot of this mixture (30 µL) and inject it into HPLC in three separate injections (10 µL for each injection). Measure the concentrations of the remaining donors ([Dx] and [Dref]) by HPLC under the same separation conditions (ether acetate/n-Hexane = 20/80) (Aref)t = 24417.0, (Ax)t = 23546.3.
- Measure the relative reactivity between Dx1 vs. Dr4, kx1/kr4 = 0.0932. Based on the relative reactivity value of Dr4, the relative reactivity value of Dx1 is 3.
NOTE: a = A/[D], (Aref)0 = 74530.1, (Ax)0 = 26143.0. kx/kref = (ln[Dx]t – ln[Dx]0)/(ln[Dref]t – ln[Dref]0) = (ln[Ax]t – ln[Ax]0)/(ln[Aref]t – ln[Aref]0) = 0.0932.
3. One-pot glycosylation of SSEA-4
- Place a 10 mL round-bottom flask under vacuum, flame-dry it, and allow the flask to cool to RT while still under vacuum. Remove the rubber septum to add a mixture of disaccharide 1 donor (38 mg, 1.1 eq., 0.057 mmol), the first acceptor 2 (40 mg, 1.0 eq., 0.053 mmol) and a Teflon-coated magnetic stir bar into the flask.
- Transfer 100 mg of powdered molecular sieves 4 Å into a 5 mL round-bottom flask. Keep this flask under vacuum, flame-dry it, and allow the flask to cool to RT while still under vacuum. Transfer the freshly dried 4 Å molecular sieves into the first flask that contains the starting material.
- Transfer 1 mL of freshly dried DCM into the flask. Stir the reaction mixture for 1 h at RT and then place it under a temperature of -40 °C. Transfer the NIS (13 mg, 1.1 eq., 0.057 mmol) into the flask.
- Inject TfOH (34 μL, 0.3 eq., 0.017 mmol, 0.5 M in ether) into the flask through the septum using a micro-volume syringe at -40 °C. Keep stirring at -40 °C for 3 h.
- After the first acceptor 2 is almost consumed, inject the solution of acceptor 3 in DCM into the flask through the septum.
- Warm the reaction mixture up to -20 °C and transfer NIS (19 mg, 1.6 eq., 0.083 mmol) into the flask. Inject TfOH (34 μL, 0.3 eq., 0.017 mmol, 0.5 M in ether) into the flask through the septum at -20 °C. Keep stirring at -20 °C for 3 h.
- After the product of the first step reaction is consumed, quench the reaction by injecting two equivalents of triethyl amine. Remove the molecular sieves through a filter funnel packed with Celite, collect the filtrate into a 25 mL round-bottom flask and further wash the filter with 10 mL of DCM.
- Transfer the filtrate into a separatory funnel and wash it with saturated aqueous sodium thiosulfate containing 10% NaHCO3 (2x with 10 mL each). Extract the aqueous layer with DCM (3x with 10 mL). Combine the organic layers and wash the mixture with brine (10 mL) and dry it by adding anhydrous MgSO4. Filter it and collect the filtrate in a 100 mL round-bottom flask.
- Remove the solvent using a rotary evaporator. Dissolve the crude mixture with approximately 1 mL of DCM and load it on top of the silica bed. Elute the product with a mixture of ethyl acetate and toluene (EtOAc/toluene, 1/4 to 1/2) and collect the fractions.
- Remove the solvent using a rotary evaporator. Dry the residue under reduced pressure to give fully protected SSEA-4 derivative 4 (74 mg, 50% based on acceptor 2) as white foam.
Representative Results
The Auto-CHO search result based on default parameter settings indicates SSEA-4 can be synthesized by a [2 + 1 + 3] one-pot reaction. Figure 3 shows the software screenshot of the SSEA-4 search result. When a trisaccharide reducing end acceptor is selected (Figure 3, label 1), the program shows four potential solutions for the query. The first solution has one fragment (Figure 3, label 2), and its calculated yield is about 94%. The fragment can be synthesized by two BBLs (Figure 3, label 3). The RRV of the first disaccharide BBL is 1462 and the RRV of the second monosaccharide is 32.0. Label 4 of Figure 3 shows the chemical structure of the first suggested BBL used in the one-pot reaction. The one-pot experiment shows that SSEA-4 can be synthesized in 43% yield by this suggestion successfully (Figure 4) and it has also been demonstrated in previous work5. The detail experimental procedures and the characterization of mentioned compounds, particularly SSEA-4 can be found in the cited reference5.
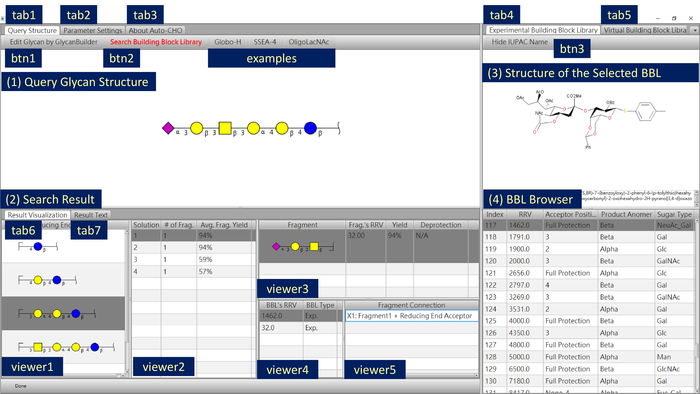
Figure 1: Auto-CHO screenshot. Users can edit query glycan structure, browse experimental and virtual building block information, and view one-pot synthetic solutions provided by the software. Please click here to view a larger version of this figure.
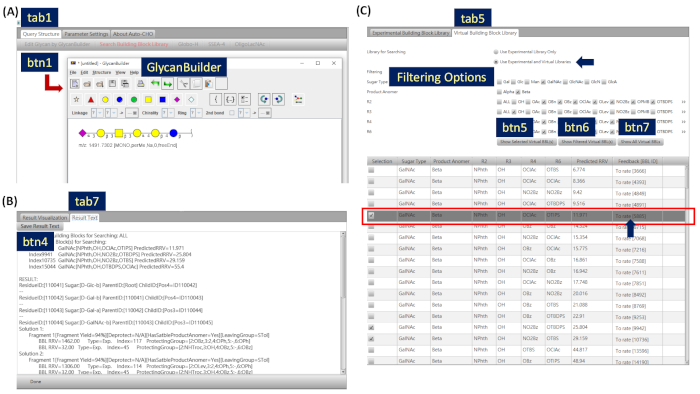
Figure 2: Partial screenshots of Auto-CHO software. (A) Click “Edit Glycan by GlycanBuilder” button (btn1) in the “Query Structure” tab (tab1) and the system pops up the GlycanBuilder dialog. (B) Select “Result Text” (tab7) and click “Save Result Text” (btn4) to save the text search results. (C) Select “Virtual Building Block Library” (tab5) and check desirable virtual building blocks for searching by filtering options. Please click here to view a larger version of this figure.
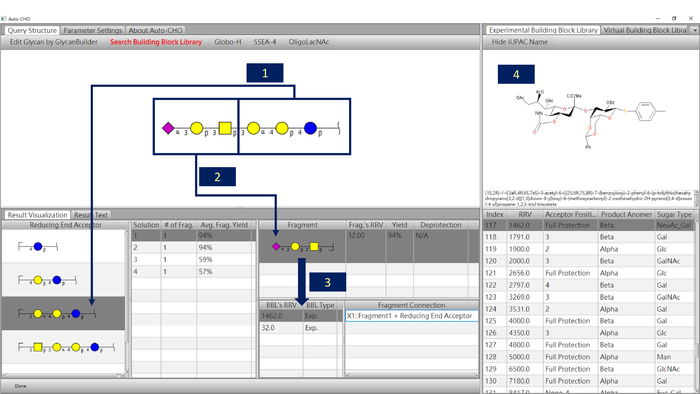
Figure 3: One-pot synthetic blueprint given by the Auto-CHO program. Label 1: Reducing end acceptor. Label 2: Fragment. Label 3: Building block of the fragment. Label 4: Chemical structure of the selected building block. Please click here to view a larger version of this figure.
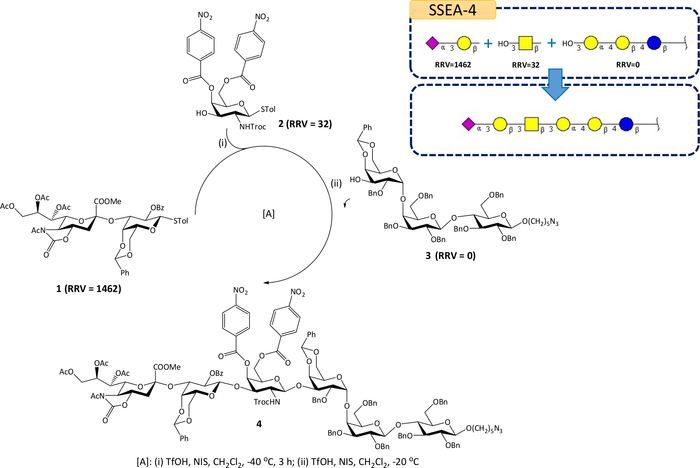
Figure 4: The [2 + 1 + 3] one-pot synthetic strategy for SSEA-4. SSEA-4 can be synthesized by three units suggested by Auto-CHO: sialyl disaccharide building block 1 (RRV=1,462), monosaccharide building block 2 (RRV=32.0), and reducing end acceptor 3 (RRV=0). This figure was modified from our previous publication5 with permission (under a Creative Commons Attribution 4.0 International License: http://creativecommons.org/licenses/by/4.0/). Please click here to view a larger version of this figure.
Discussion
The Auto-CHO software was developed for assisting chemists to proceed hierarchical and programmable one-pot synthesis of oligosaccharides5. Auto-CHO was built by Java programming language. It is a GUI software and cross-platform, which currently supports Windows, macOS, and Ubuntu. The software can be downloaded free of charge for the Auto-CHO website at <https://sites.google.com/view/auto-cho/home>, and its source code with MIT license can be accessed from the GitHub at <https://github.com/CW-Wayne/Auto-CHO>.
The BBL library of Auto-CHO contains 154 experimental BBLs and more than 50,000 virtual BBLs with accurately predicted RRVs. Currently, the sugar types of virtual BBLs include Gal, Glc, Man, GalNAc, GlcNAc, GlcN, and GlcA. All library searches are processed in the local machine and we do not collect any query structure from users. Since Auto-CHO cannot guarantee the success of high-yield synthesis among virtual BBLs given by the program (due to many structural constraints or unknown factors in the chemical reactions), Auto-CHO provides online feedback questionnaire for virtual BBLs. It is believed that user feedback from the research community can help keep valuable virtual BBLs and eliminate unsuitable ones. An email address is provided for technical assistance in the software user guide. Users can contact this address if they encounter technical questions or problems.
Two search strategies are provided here. For parameter settings (section 1.4), it is suggested to set parameters with stricter criteria at the beginning. If Auto-CHO does not return satisfied synthetic solutions, it is advised to use more flexible parameters in the next search run. For the selection of BBL library (section 1.5), it is suggested to search the experimental library only at first. If the software does not return any suitable solution, it is advised to search experimental and virtual libraries in the following iterations.
In summary, this protocol demonstrates the operation of Auto-CHO software and use of Auto-CHO for one-pot synthesis of the SSEA-4 molecule. In addition, the programmable one-pot protocol is described. Auto-CHO is the GUI and open-source software with library includes validated and virtual BBLs, and it supports hierarchical one-pot synthesis of oligosaccharides. It is believed that this software can benefit the research community and more essential oligosaccharides can be synthesized by one-pot reactions through Auto-CHO for further research.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by Academia Sinica including the Summit Program, Ministry of Science and Technology [MOST 104-0210-01-09-02, MOST 105-0210-01-13-01, MOST 106-0210-01-15-02], and NSF (1664283).
Materials
| Acetonitrile | Sigma-Aldrich | 75-05-8 | |
| Anhydrous magnesium sulfate | Sigma-Aldrich | 7487-88-9 | |
| Cerium ammonium molybdate | TCI | C1794 | |
| Dichloromethane | Sigma-Aldrich | 75-09-2 | |
| Drierite | Sigma-Aldrich | 7778-18-9 | |
| Ethyl acetate | Sigma-Aldrich | 141-78-6 | |
| Methanol | Sigma-Aldrich | 67-56-1 | |
| Molecular sieves 4 Å | Sigma-Aldrich | ||
| n-Hexane | Sigma-Aldrich | 110-54-3 | |
| N-Iodosuccinimide | Sigma-Aldrich | 516-12-1 | |
| Sodium bicarbonate | Sigma-Aldrich | 144-55-8 | |
| Sodium thiosulfate | Sigma-Aldrich | 10102-17-7 | |
| Toluene | Sigma-Aldrich | 108-88-3 | |
| Trifluoromethanesulfonic acid | Sigma-Aldrich | 1493-13-6 |
References
- Apweiler, R., Hermjakob, H., Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica Et Biophysica Acta. 1473 (1), 4-8 (1999).
- Sears, P., Wong, C. -. H. Toward Automated Synthesis of Oligosaccharides and Glycoproteins. Science. 291 (5512), 2344-2350 (2001).
- Kulkarni, S. S., et al. “One-Pot” Protection, Glycosylation, and Protection-Glycosylation Strategies of Carbohydrates. Chemical Reviews. 118 (17), 8025-8104 (2018).
- Zhang, Z., et al. Programmable One-Pot Oligosaccharide Synthesis. Journal of the American Chemical Society. 121 (4), 734-753 (1999).
- Cheng, C. -. W., et al. Hierarchical and programmable one-pot synthesis of oligosaccharides. Nature Communications. 9 (1), 5202 (2018).
- . . ChemDraw. , (2019).
- Cheeseman, J. R., Frisch, &. #. 1. 9. 8. ;. . Predicting magnetic properties with chemdraw and gaussian. , (2000).
- Yap, C. W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. Journal of Computational Chemistry. 32 (7), 1466-1474 (2011).
- Ceroni, A., Dell, A., Haslam, S. M. The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code for Biology and Medicine. 2, 3 (2007).
- Damerell, D., et al. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biological Chemistry. 393 (11), 1357-1362 (2012).

