Production of E. coli-expressed Self-Assembling Protein Nanoparticles for Vaccines Requiring Trimeric Epitope Presentation
Summary
A detailed method is provided here describing the purification, refolding, and characterization of self-assembling protein nanoparticles (SAPNs) for use in vaccine development.
Abstract
Self-assembling protein nanoparticles (SAPNs) function as repetitive antigen displays and can be used to develop a wide range of vaccines for different infectious diseases. In this article we demonstrate a method to produce a SAPN core containing a six-helix bundle (SHB) assembly that is capable of presenting antigens in a trimeric conformation. We describe the expression of the SHB-SAPN in an E. coli system, as well as the necessary protein purification steps. We included an isopropanol wash step to reduce the residual bacterial lipopolysaccharide. As an indication of the protein identity and purity, the protein reacted with known monoclonal antibodies in Western blot analyses. After refolding, the size of the particles fell in the expected range (20 to 100 nm), which was confirmed by dynamic light scattering, nanoparticle tracking analysis, and transmission electron microscopy. The methodology described here is optimized for the SHB-SAPN, however, with only slight modifications it can be applied to other SAPN constructs. This method is also easily transferable to large scale production for GMP manufacturing for human vaccines.
Introduction
While traditional vaccine development has focused on the inactivated or attenuated pathogens, the focus of modern vaccines has shifted toward subunit vaccines1. This approach can lead to a more targeted response, and potentially more efficacious vaccine candidates. However, one of the main drawbacks is that subunit vaccines are not particulates like whole organisms which can result in reduced immunogenicity2. A nanoparticle as a repetitive antigen display system can have the benefits of both the targeted subunit vaccine approach as well as the particulate nature of the whole organism1,3.
Among the existing types of nanovaccines, rationally designed protein assemblies allow for the design and development of vaccine candidates that can present multiple copies of the antigen potentially in a native-like conformation1,4,5,6. One example of these protein assemblies are the self-assembling protein nanoparticles (SAPNs)7. SAPNs are based on coiled-coil domains and are traditionally expressed in Escherichia coli8. SAPN vaccine candidates have been developed for a variety of diseases such as malaria, SARS, influenza, toxoplasmosis, and HIV-19,10,11,12,13,14,15,16,17,18,19. The design of each SAPN candidate is specific to the pathogen of interest, however, the production, purification, and refolding techniques are generally broadly applicable.
One of our current interests is an effective HIV-1 vaccine. In RV144—the only Phase III clinical trial of an HIV-1 vaccine that demonstrated modest efficacy—the reduced risk of infection was correlated with IgG antibodies to the V1V2 loop of the envelope protein20,21. The native-like trimeric presentation of this region is thought to be important for protective immunogenicity22. To present the V1V2 loop in as close to native-like conformation as possible, we developed a proof of principle SAPN vaccine candidate that contained the HIV-1 envelope post-fusion six-helix bundle (SHB) to present the V1V2 loop into the correct conformation9. This candidate was recognized by known monoclonal antibodies to HIV-1 envelope protein. Mice immunized with V1V2-SHB-SAPN raised V1V2 specific antibodies, that, most importantly, bound to gp70 V1V2, the correct conformational epitopes9. The SHB-SAPN core could have other functions beyond the role as a carrier for the HIV-1 V1V2 loop. Here we describe a detailed methodology for the expression, purification, refolding, and validation of the SHB-SAPN core. The sequence selection, nanoparticle design, the molecular cloning, and transformation of E. coli have been previously described9.
Protocol
1. Expression of the SHB-SAPN Protein in E. coli BL21(DE3)
- Mix 95 mL of component A and 5 mL of component B of the media in a 2 L sterile glass Erlenmeyer flask as per the manufacturer’s instructions (see the Table of Materials). Add ampicillin to a final concentration of 100 μg/mL.
- Inoculate the media with E. coli from a previously established glycerol stock culture. Incubate culture at 30 °C with shaking at 200 rotations per minute (rpm) for 48 h.
NOTE: The used E. coli BL21 (DE3) stock contained the ampicillin resistant expression vector23 with the SHB-SAPN gene. Although the general protocol of the media recommends 24 h of incubation at 37 °C, 48 h of incubation at 30 °C gave higher yield for SHB-SAPN. - Transfer the culture to two 50 mL conical tubes. Centrifuge the tubes at 4,000 x g for 10 min with a fixed angle rotor at 4 °C. Remove the supernatant and save the pellet to harvest cells.
NOTE: The cell pellet can be either processed immediately or frozen at -80 °C until use.
2. Lysis of E. coli BL21(DE3) by sonication
NOTE: Use nonpyrogenic plasticware and glassware baked at 250 °C for at least 30 min. Tris(2-carboxyethyl)phosphine (TCEP) as a reducing agent breaks the disulfide bonds within and between proteins. TCEP is necessary in the buffers during this protocol if the displayed antigen contains S-S bonds. For SHB-SAPN core only, the presence of TCEP in the buffers is not essential.
- Prepare imidazole-free buffer (8 M Urea, 50 mM sodium phosphate monobasic, 20 mM Tris base, 5 mM TCEP) pH 8.0 (adjusted with 5 N NaOH) and filter it using a 0.22 µm vacuum bottle filtration unit.
- Resuspend the pelleted cells (from step 1.3) with 40 mL of imidazole-free buffer in one 50 mL conical tube. Sonicate the resuspended cells with a probe on ice for 5 min (4 s of sonication, 6 s of rest) with a sonication output of 150 W.
- Centrifuge the cellular lysate (40 mL) at 29,000 x g at 4 °C for 25 min in a fixed angle rotor to generate clarified supernatant. Transfer the supernatant to a 150 mL sterile flask and discard the pellet. Dilute the supernatant to 100 mL using the imidazole-free buffer (later in the protocol referred to as “sample”).
NOTE: This dilution step is needed to prevent the FPLC system pressure from becoming too high during lysate loading on the column.
3. Protein purification using a His-column
NOTE: This protocol was performed using an FPLC instrument, but it can be adapted to gravity flow.
- Prepare the following buffers and filter them using a 0.22 µm vacuum bottle filtration unit: (i) imidazole-free buffer “Buffer A” (8 M Urea, 50 mM sodium phosphate monobasic, 20 mM Tris base, 5 mM TCEP) pH 8.0; (ii) 500 mM imidazole buffer “Buffer B” (8 M Urea, 50 mM sodium phosphate monobasic, 20 mM Tris base, 5 mM TCEP, 500 mM Imidazole) pH 8.0; and (iii) isopropanol wash (20 mM Tris, 60% isopropanol) pH 8.0.
NOTE: pH for each buffer was adjusted with 5 N NaOH. - Equilibrate the His-column.
NOTE: For lab scale production, this protocol uses a 5 mL prepacked His-column, but any larger sized column can be used.- Open the FPLC software and click on the New method option. It will immediately open to the Method settings menu. Under the drop-down menu for column position choose C1 port 3.
- On the Shown by technique drop down menu choose affinity. On the Column type drop-down menu choose others, Histrap HP, 5 mL. The column volume and the pressure boxes will be automatically set to the appropriate values.
- Click on the Method outline button. Drag the following buttons from the Phase library popup menu: equilibration, sample application, column wash, and elution next to the arrow in that exact order. Close the Phase library menu.
- Click on the equilibration button. The values listed in the table should be “initial buffer B” (4%), “final buffer B” (4%), and “volume (CV)” 5.
- Click on the Sample application box. In the sample loading box click the radio button for Inject sample on column with sample pump. Make sure the box next to the Use flow rate from method settings is checked in the sample injection with system pump box. Next to the volume box on the right side of the screen change the value to 20 mL.
- Click on the Column wash button. The values listed in the table should be “initial buffer B” (4%), “final buffer B” (4%), and “volume (CV)” 5. Next to the fraction collection scheme unclick the Enable box.
- Click on the elution button. The values listed in the table should be “initial buffer B” 0%, “final buffer B” 100%, and “volume (CV)” 5. Next to the fraction collection scheme click on the Enable box. Unclick the Use fraction size from method settings and adjust fraction size to 4 mL in the fill-in box below.
- Click the save as button on the top of the of the software. Name the file “equilibration”.
- Connect a 5 mL prepacked His-column to the corresponding column port 3 on the FPLC. Both pump A and pump B tubing as well as the sample pump tubing should be placed into 0.22 µm filtered deionized water. Run the equilibration program.
- Place both pump A and pump B as well as the sample pump tubing into the imidazole-free buffer (Buffer A) and run the equilibration protocol again.
- Bind the sample to the column and purify the protein.
- Open the FPLC software and click on the New method option. It will immediately open to the Method settings menu. Under the drop-down menu for Column position choose C1 port 3. On the Shown by technique drop down menu choose affinity. On the column type drop-down menu choose others, Histrap HP, 5 mL. The column volume and the pressure boxes will automatically be set to the appropriate values.
- Click on the Method outline button. Drag the buttons from the Phase library popup menu: equilibration, sample application, column wash (Wash 1), column wash (Wash 2), column wash (Wash 3), and Elution next to the arrow in that exact order. Close the Phase library menu.
- Click on the Equilibration button. The values listed in the table should be “initial buffer B” 4%, “final buffer B” 4%, and “volume (CV)” 5.
- Click on the Sample application box. In the sample loading box click the radio button for Inject sample on column with sample pump. Make sure the box next to the Use flow rate from method settings is checked in the sample injection with system pump box.
- Next to the volume box on the right side of the screen change the value to 100 mL. Next to the fraction collection scheme click the Enable button. Unclick the Use fraction size from method settings box and then change the fraction size to 4 mL.
- Click on the first column wash button (Wash 1). The values listed in the table should be “initial buffer B” 4%, “final buffer B” 4%, and “volume (CV)” 10. Next to the fraction collection scheme click the Enable box. Unclick the Use fraction size from method settings and then change the fraction size to 4 mL.
- Click on the second column wash button (Wash 2). The values listed in the table should be “initial buffer B” 0%, “final buffer B” 0%, and “volume (CV)” 5. Next to the fraction collection scheme click the Enable box. Unclick the Use fraction size from method settings and then change the fraction size to 4 mL.
- Click on the third column wash button (Wash 3). The values listed in the table should be “initial buffer B” 0%, “final buffer B” 0%, and “volume (CV)” 5. Next to the fraction collection scheme click the Enable. Unclick the Use fraction size from method settings box and then change the fraction size to 4 mL.
- Click on the Elution button. In the table right click the information listed and, on the menu that comes up, click Delete step. Drag the isocratic gradient button onto the table twice, so that there are two entries.
- The value for the first entry should read “initial buffer B” 30%, “final buffer B” 30%, and “Volume (CV)” 10. The value for the second entry should read “initial buffer B” 100%, “final buffer B” 100%, and “volume (CV)” 10. Next to the fraction collection scheme click the Enable button. Click the box next to Use fraction size from method settings.
- Click the Save as button on the top of the of the software. Name the file “purification”. Pump A tubing of the FPLC should be placed into the imidazole-free wash buffer while pump B tubing should be placed into the 500 mM imidazole buffer. The sample pump tubing should be placed into the 100 mL sample.
- Run the “purification” program and wait for the time when the 60% isopropanol is needed (Wash 2). Pause the program, move pump A tubing from the imidazole-free wash into the 60% isopropanol wash. Restart the program.
- Once the isopropanol step is completed, pause the program again and move pump A tubing back into the imidazole-free wash buffer. Restart the purification program (the rest of the run is automated).
4. Purity assessment and protein identification by SDS-PAGE
- Combine all the fractions corresponding to (i) flow-through (the cell lysate that did not bind to the His column), (ii) Wash 1, (iii) Wash 3 (60% isopropanol wash), and (iv) Wash 3 in separate 50 mL conical tubes. Do not combine the 2 mL fractions from the elution steps.
- Mix 15 µL from each of the pooled fractions and all fractions from elution steps with 2x Laemmli sample buffer in a 0.5 mL microcentrifuge tube and denature them at 95 °C for 10 min.
- While the protein denatures, set up the gel running apparatus with 3 stain-free 4–20% precast polyacrylamide gels in 1x Tris-glycine SDS-PAGE running buffer.
- Load 8 µL of molecular weight marker to the first well and 30 µL of denatured sample to the other wells of the gel. Run the gels at 200 V until the dye front hits the bottom of the gel (about 30 min). Remove the gels from the apparatus and briefly rinse with deionized water. Image the gel immediately using the stain-free imaging system.
- Identify the fractions that contain protein bands with the correct size (18.07 kDa). Pool all these fractions.
5. Protein identification by western blot
- Run a western blot using an His-specific antibody (anti-6x HisTag) and a SHB-specific antibody (167-D-IV) to identity the purified full-length protein. The anti-6x HisTag antibody recognizes the N terminus of the protein and the 167-D-IV antibody recognizes the C terminus demonstrating the presence of the full-length protein.
- Determine the protein concentration of the (i) flow-through, (ii) Wash 1, (iii) Wash 2 (60% isopropanol wash), (iv) Wash 3, and all fractions from the elution steps with the spectrometer instrument at an absorbance of 280 nm. Generate dilutions that contain 100 ng of protein in 15 µL of imidazole-free buffer for each of these groups.
- Add 15 µL of 2x Laemmli sample buffer to each 15 µL of the samples and denature them as in step 4.1. Once the denaturing is completed spin down the tubes to ensure all the protein can be transferred.
- Load samples and the pre-stained marker into a stain-free 4–20% precast polyacrylamide gel. Run the electrophoresis at 200 V until the dye front reaches the bottom of the gel.
- While the gel runs, make 1 L of TBS-T (20 mM Tris, 150 mM NaCl, and 0.1% Tween 20) and 200 mL of 5% non-fat milk in TBS-T.
- Use a western blot transfer system to transfer protein onto a nitrocellulose membrane. Use a pre-assembled transfer stack and place the gel on it. Set up the system to run at 25 V for 7 min. Check for the presence of the pre-stained marker on the nitrocellulose membrane indicating a complete transfer.
NOTE: All the subsequent steps are performed on an orbital shaker set at 100 rpm at room temperature (RT). - Once the transfer is completed, wash blots two times with TBS-T for 10 min each.
- Block the nitrocellulose membranes (blot) with 5% non-fat milk in TBS-T for at least 1 h. Wash blots two times with TBS-T for 10 min each.
- Dilute the primary 167-D-IV and anti-6x HisTag antibodies to 1 mg/mL in TBS-T (stock Ab). Dilute the stock Abs of the 167-D-IV 10,000-fold and the anti-6x HisTag 5,000-fold by adding 2 µL of the stock 167-D-IV and 4 µL of the stock anti-6x HisTag antibodies to two different tubes containing 20 mL of TBS-T. Add the total 20 mL volume of primary antibodies, one to each blot, and incubate blots for 1 h. Wash blots two times with TBS-T for 10 min.
- Dilute 4 µL of 1 mg/mL of the mouse anti-human secondary antibody conjugated with alkaline phosphatase in 20 mL of TBS-T (1:5,000 dilution). Dilute 4 µL of 1 mg/mL of the goat anti-mouse secondary antibody conjugated with alkaline phosphatase in 20 mL of TBS-T (1:5,000 dilution). Add secondary antibodies to corresponding blots.
NOTE: The anti-human secondary antibody binds to 167-D and the anti-mouse binds to the anti-6x HisTag. Wash blots two times with TBS-T for 10 min. - Add enough BCIP/NBT alkaline phosphatase substrate to cover the blots. Develop the blots for about 10 min until bands appear. Rinse blots with cold tap water and let them dry before scanning with a flatbed scanner.
6. Refolding the SHB-SAPN
- Add the pooled protein (10‒20 mL total) to a 10 kDa molecular weight cut off dialysis cassette and dialyze it into 8 M urea, 20 mM Tris, 5% glycerol, 5 mM TCEP pH 8.5 at RT (18‒26 °C) overnight.
- Slowly dialyze the urea off the sample by decreasing the urea concentration in the dialysis buffer stepwise by 2 M every 2 h. At a urea concentration of 2 M, move the dialysis apparatus to 4 °C (do not use TCEP in the dialysis buffer from this step). Finish the refolding by dialyzing the sample into 120 mM urea, 20 mM Tris, 5% glycerol, pH 8.5 at 4 °C overnight.
- Remove the refolded protein (SHB-SAPN) from the dialysis cassette. Filter the SHB-SAPN using a 0.22 µm polyvinylidene fluoride (PVDF) syringe filer. Aliquot SHB-SAPN into sterile tubes and freeze them at -80 °C, leaving at least 100 µL at RT for subsequent analyses.
7. Validation of particles by size and appearance
- Dynamic light scattering (DLS)
- Measure the mean particle size of the SHB-SAPN by the following parameters: select protein as the material, create a complex buffer for 120 mM urea, 20 mM Tris, 5% glycerol 25 °C for temperature, select disposable cuvettes for the analysis, select automatic measurement, set for 5 runs.
- Add 45 µL of SHB-SAPN to a disposable cuvette and run the software by clicking on the green arrow. Select the percentage volume for the readout.
- Nanoparticle tracking analysis (NTA)
- Dilute the sample by 1:20 in the refolding buffer. Make 10 mL of diluted sample.
- Using 3 mL syringes, flush the NTA instrument with the refolding buffer and load about 1.5 mL of sample to equilibrate the instrument. Use rest of the sample for the analysis.
- Create a new SOP in the NTA software by clicking on the SOP tab. Under the tab change the number of captures to 3 and change the capture time to 30 s. Press the autofocus button on the left side of the screen to bring the sample into focus. Use the manual focus knob on the side of the machine to fine tune the focus.
- Run the created SOP, when the system prompts load a small volume of sample with the syringe. After the system has taken all the captures it will automatically bring up the analysis screen. Slide the detection limit bar so that all the real particles are marked with red crosses. Press the run analysis button and the analysis will automatically begin.
- Transmission electron microscopy (TEM)
- Glow discharge formvar/carbon 400 mesh copper TEM support films.
- Add 3 µL of sample at a 0.075 mg/mL concentration to the grid for 30 s. Wick off the liquid using a filter paper.
- Wash the grid with 3 µL of deionized water three times, each time wicking off the water with filter paper.
- Add 3 µL of 0.5% uranyl acetate to the support film and allow it to sit for 30 s. Wick off most of the uranyl acetate but leave a thin film on the surface. Allow the samples to dry before imaging them on the transmission electron microscope.
- Image samples at 80 kV on a TEM.
8. Determination of endotoxin levels in the samples using a kinetic limulus amoebocyte lysate (LAL) assay
- Remove the kit and samples from the refrigerator and allow to equilibrate to RT.
- To perform this assay, a plate reader with a heat block and the ability to read the samples for 40 reads at a wavelength of 405 nm at 37 °C is required. Write a program template so that wells are read every 150 s to identify the onset time point (the OD increased by 0.2 in comparison with the first read).
- Dilute the sample to the immunization dose concentration in PBS.
NOTE: The pH of the refolding buffer is outside the range of the LAL assay. The dilution of the sample in PBS will set the pH to the acceptable range. - Resuspend the control endotoxin in the appropriate volume of endotoxin-free LAL water as determined by the certificate of analysis to generate 50 EU/mL. Vigorously vortex the vial for 15 min to ensure complete resuspension of the endotoxin.
- Generate the endotoxin standard curve by preforming a 10-fold serial dilution in glass vials in the range of 50 EU to 0.005 EU/mL. For each dilution, add 0.1 mL of the previous dilution to 0.9 mL of LAL water. Vortex vigorously after combination for 1 min.
- Add standard curve dilutions and SHB-SAPN samples in duplicate to a 96-well plate. Use LAL water as a negative control in duplicate as well. Preincubate the plate at 37 °C for 15 min.
- Towards the end of the incubation, resuspend the assay reagent vial with 2.6 mL of LAL water. Gently mix content with a serological pipette.
- Add 100 µL of the assay reagent to each well of the 96-well plate. Quickly move the plate to the plate reader and run the program template written in step 8.2.
- Once the program is completed, generate a standard curve using the log value of the controls versus the log value of the onset time. Use the formula generated from this curve to calculate the endotoxin concentration in the samples.
Representative Results
The fully assembled SHB-SAPN shown here is built upon protein sequences (Figure 1A) that are predicted to fold into a particle that contains 60 copies of the monomer (Figure 1B). Figure 2 provides an outline of the method for the production, purification, and identification of the SHB-SAPN core. E. coli from a glycerol stock that contained a pPep-T expression vector with gene sequence of the SHB-SAPN core were induced in BL21 (DE3) E. coli. Bacterial cells were successfully grown and lysed under denaturing and reducing conditions.
Total cell lysate was used to purify SHB-SAPN monomers by FPLC using a Ni2+ column (Figure 3A). The FPLC chromatograph demonstrates that protein eluted both at 150 mM and 500 mM imidazole (Figure 3A). The chromatogram also shows two other peaks at 185 mL and 210 mL total volume corresponding to the isopropanol wash and the imidazole-free wash, respectively. The fractions and the purity of the recombinant protein were identified by gradient SDS-PAGE gels (Figure 3B). The protein of interest was primarily located in fractions 68‒79 (278‒300 mL total volume). These fractions were combined for further analyses. Western blot with anti-His antibody (N-terminal) and 167-D-IV antibody (C-terminus) indicated that the pooled fractions were indeed the protein of interest (Figure 4A,B). These blots also demonstrated the presence of the SHB-SAPN multimers. Earlier washes and elution fractions tended to contain a higher concentration of multimerized protein and were therefore excluded.
The samples that contained the protein monomers of interest were folded into the fully assembled SHB-SAPN by dialysis. Particle size distribution was determined by DLS and nanoparticle tracking analysis (Figure 5A,B). The DLS identified particles with a Z-average hydrodynamic diameter of 67 nm while the NTA system measured a mean size of 81 nm. The slight size differences were due to the particle sizing techniques, however the size from both analyses were in the expected range of 20‒100 nm8,24,25. SHB-SAPNs were visualized by TEM and the images showed well-formed individual particles with the size distribution obtained from the two particle sizing techniques (Figure 5C).
During the purification of the protein, the column was washed with isopropanol to decrease the LPS contamination in the final SHB-SAPN product. To verify if the endotoxin level was acceptable for immunization, the concentration of LPS in SHB-SAPN samples purified with or without the isopropanol wash step was determined by a kinetic LAL assay. The results indicated that the isopropanol wash decreased the endotoxin levels from >0.25 EU/µg to 0.010 EU/µg of SHB-SAPN protein (Table 1).
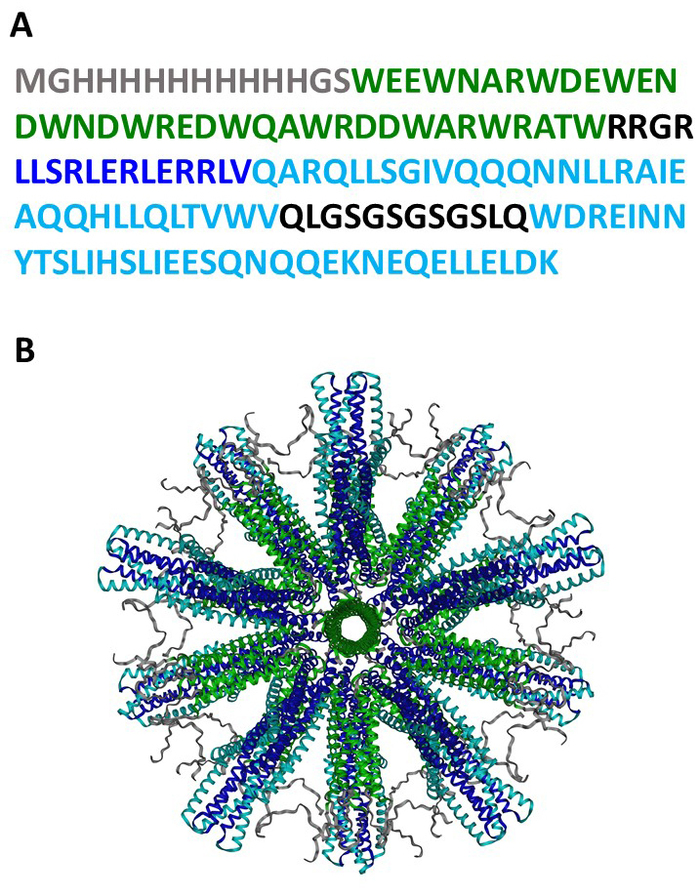
Figure 1: SHB-SAPN protein sequence and structure. (A) The amino acid sequence of the SHB-SAPN monomer. (B) Computer model of the structure of the fully assembled SHB-SAPN core consisting of 60 protein monomers. Color scheme for amino acid sequences: Grey = HisTag; Green = pentamer; Dark blue = de novo designed trimer; Light blue = six-helix bundle. Please click here to view a larger version of this figure.
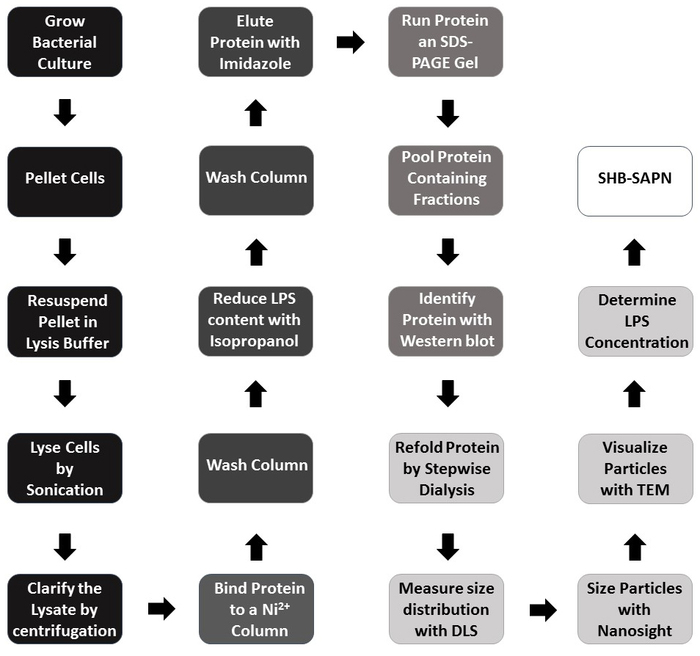
Figure 2: Flowchart of the protocol for SHB-SAPN production. Color scheme: Black = expression of the protein monomer in E. coli; Dark grey = monomer purification; Medium grey = monomer identification; Light grey = refolding and characterization; White = fully assembled SHB-SAPN product. In steps labeled with dark and medium grey, the protein is under denaturing and reducing conditions. Please click here to view a larger version of this figure.
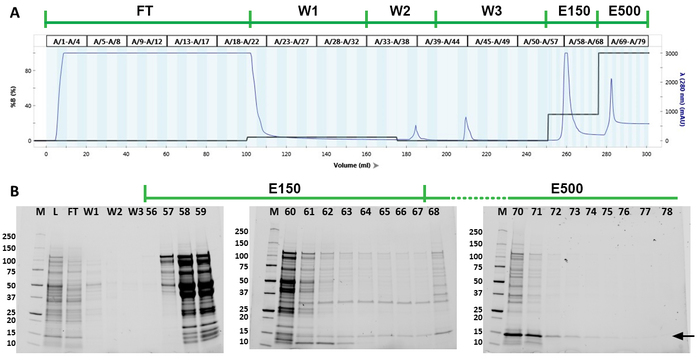
Figure 3: Protein purification of the SHB-SAPN monomers. (A) Chromatograph from the FPLC purification. Green line above the chromatogram indicates the purification steps. Blue line in the chromatogram represents the optical density of the fractions at 280 nm wavelength. The black line shows what percent of buffer B (8 M urea, 20 mM Tris, 50 mM sodium phosphate monobasic, 5 mM TCEP, 500 mM Imidazole, pH 8.5) that was used in each stage of the purification. (B) SDS-PAGE gels of the pooled fractions from the lysate (L), flow through (FT), first wash (W1), isopropanol wash (W2), third wash (W3), and individual fractions from the 150 mM imidazole (E150) and 500 mM imidazole (E500) elution steps of the purification. Molecular markers in the first lane (M) identify bands between 10 and 250 kDa. Target protein is indicated by a black arrow. Please click here to view a larger version of this figure.
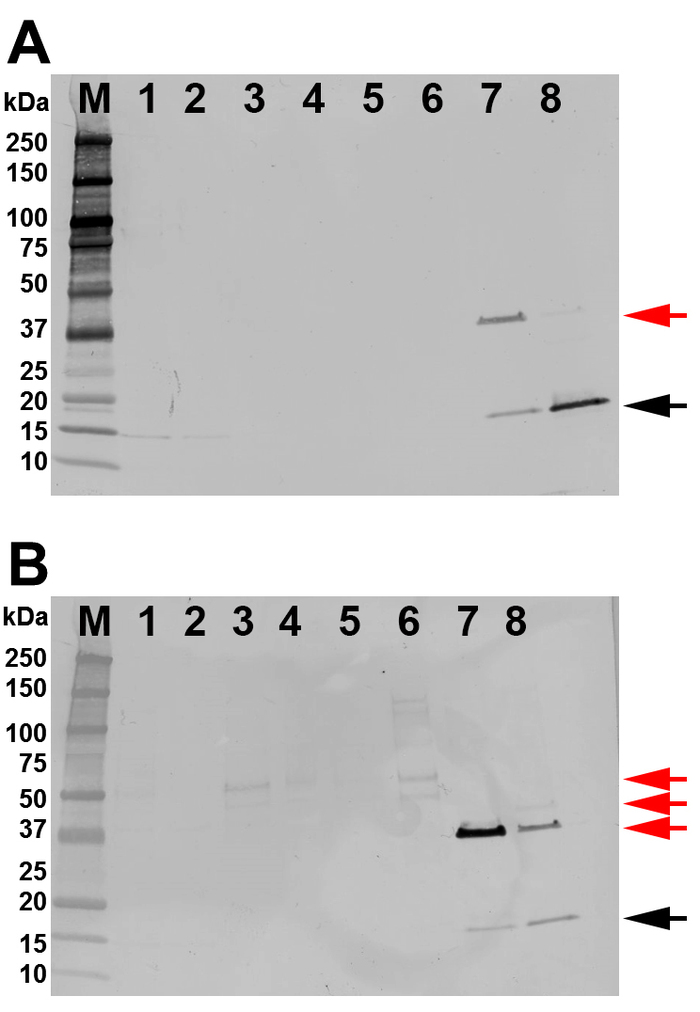
Figure 4: Identification of the protein by western blot. All lanes are loaded with 100 ng of protein. (A) Results of a western blot with anti-6x HisTag. (B) Results of a western blot with a 167-D-IV HIV-1 monoclonal antibody. Lanes are labeled as: M = molecular weight marker; 1 = lysate; 2 = Flow through; 3 = first wash; 4 = isopropanol wash (second wash); 5 = third wash; 6 = pooled volume fractions 56‒61 (first elution peak), 7 = pooled volume factions 62‒67 (between the two peaks), 8 = pooled volume fractions 68‒78 (second elution peak). Target protein with the expected band size of 18.07 kDa as the monomeric SHB-SAPN band is indicated by a black arrow. Extra banding in lanes 7 and 8 are dimers, trimers, and multimers of the SHB-SAPN (red arrow). Please click here to view a larger version of this figure.
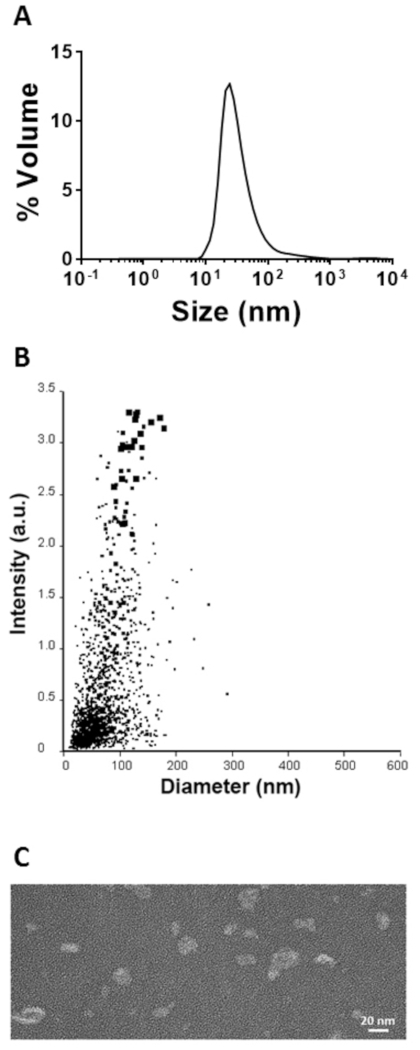
Figure 5: Characterization of the refolded SHB-SAPN. (A). Particle size distribution as determined by DLS. (B) Particle size as determined by nanoparticle tracking (system). (C) Visualization of the SHB-SAPN particles by TEM. Please click here to view a larger version of this figure.
| Sample | Endotoxin (EU/mL) | Endotoxin (EU/µg of Protein) |
| SAPN with Isopropanol Wash | 2.02 | 0.01 |
| SAPN without Isopropanol Wash | >50 | >0.25 |
| Negative Control | Below detection level | N/A |
Table 1: Endotoxin levels of refolded SHB-SAPNs. Endotoxin levels in SHB-SAPN samples purified with or without an isopropanol wash presented both as endotoxin units/mL and endotoxin units/µg of SHB-SAPN protein.
Discussion
Nanotechnology provides many advantages and solutions for subunit vaccine development. Nanovaccines can repeatedly present antigens as particulates to the host immune system increasing immunogenicity26. While there are many different types of nanovaccines, we believe that ones composed of de novo designed protein seem to be the strongest approach for vaccine development1. They can be engineered without any sequence homology to the host proteins and present the antigen of interest in close to native-like conformation while providing low production cost and high product yields. A prime example of this approach is the SAPN technology, which we have applied to vaccines against multiple infectious diseases7. Addressing the difficulties in HIV-1 vaccine development, we have engineered a unique SHB-SAPN core to effectively present the V1V2 antigen in a native-like trimeric conformation9. Many vaccine targets, particularly for viral diseases, are present as trimers27. This phenomenon indicates that our SAPN design has wide implications for the development of subunit vaccines.
In this method, we demonstrate how to produce SHB-SAPNs in an E. coli expression system. We expressed high yields of protein (about 6 mg/100 mL of culture). The protein contained 10 histidines and was easily purified using an immobilized metal affinity chromatography with Ni2+ column. This length of the His-Tag was found to be the optimal for the highest protein yield. The purified protein contained the full-length of the designed protein as indicated by the presence of both the N-terminal HisTag and the C terminal heptad repeat. We utilized widely accepted techniques and optimized them for the expression, production, and characterization of the SHB-SAPN core. Lack of the production of the full-length protein during the development of a SAPN containing a new protein epitope could indicate an expression problem of the gene in the host cell. If it happens, the gene and the expression system must be redesigned and adapted to the described protocol. Modification of sonication time or intensity may also increase the concentration of the predicted full-length protein.
Refolded particles were in the expected size range (20 to 100 nm)8,24,25 as determined by DLS and nanoparticle tracking analysis. These results were further confirmed by using TEM. If there are problems in this step, it is normally due to a problem with the pH or ionic strength of the refolding buffer. When large size particles are detected on the particle sizing techniques, it indicates aggregation, which can be avoided by increasing the pH of the refolding buffer. If the particles are not detected by DLS, verify the concentration of the protein and check the pH of the buffer. The final protein concentration for DLS should be at least 100 µg/mL. If the concentration is not the problem, it indicates the abundance of small, incompletely formed particles, whose concentration can be reduced by decreasing the pH. Alternatively, the sodium chloride concentration can be adjusted to the optimum range to minimize the presence of particles with unwanted size.
Finally, by using an isopropanol wash step during purification we were able to reduce contaminating LPS from the host E. coli to 0.01 EU/µg of SAPN which is below the Food and Drug Administration (FDA) limit of 5 EU/kg of body weight for injectable products28. This level can be further reduced by using an anion exchange column also known as Q column. If high levels of endotoxin are still present, check all materials that were used for buffer preparation. Remember to use only depyrogenated glassware and endotoxin free plasticware in this method.
These results indicate that we have successfully developed a method to produce the SHB-SAPN core that can be used for pre-clinical immunization studies. This method with only slight modifications, if any, can be applied to the purification of SHB-SAPNs when an antigen of interest is added. Using this method as a starting point one of the major changes is in the elution step. Different proteins elute at different imidazole concentrations that must be determined experimentally. The other major difference might be the composition of the refolding buffer. Optimization would require testing different pH conditions as well as ionic strengths.
In consideration of future work, only two slight modifications are needed to allow human application of the SHB-SAPN. The first is that the expression vector needs to be changed to a kanamycin resistance selectable marker due to the ampicillin allergy in humans29. The other major requirement of the protein manufacturing for human use is to produce the SHB-SAPN in animal product-free media. A small-scale study already indicated a reasonable yield of protein in a plant-based media. The work presented here is easily scalable for ultimate GMP production as demonstrated with a malaria vaccine candidate, FMP01416. This large scale FMP014-SAPN production included both the anion exchange and the cation exchange steps to further reduce LPS and Ni2+ content from the final product. This bacterial-expressed SAPN has been already scaled up for an upcoming Phase 1/2a clinical trial.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by a cooperative agreement (W81XWH-11-2-0174) between the Henry M Jackson Foundation for the Advancement of Military Medicine, Inc., and the US Department of Defense. The anti-HIV-1 gp41 mAb 167-D IV antibody was received from Dr. Susan Zolla-Pazner through the NIH AIDS Reagent Program.
Materials
| 10x Tris/Glycine/SDS | BioRad | 1610732 | 1 L |
| 2-Mercaptoethanol | BioRad | 1610710 | 25 mL |
| 2-propanol | Fisher | BP26181 | 4 L |
| 2x Laemmli Sample Buffer | BioRad | 1610737 | 30 mL |
| 40ul Cuvette Pack of 100 with Stoppers | Malvern Panalytical | ZEN0040 | 100 pack |
| 4–20% Mini-PROTEAN TGX Precast Protein Gels, 10-well, 30 µl | BioRad | 4561093 | 10 pack |
| Ampicillin | Fisher | BP1760-25 | 25 g |
| Anti-6X His tag antibody [HIS.H8] | AbCam | ab18184 | 100 mg |
| Anti-HIV-1 gp41 Monoclonal (167-D IV) | AIDS Reagent Repository | 11681 | 100 mg |
| BCIP/NBT Substrate, Solution | Southern Biotech | 0302-01 | 100 mL |
| Corning Disposable Vacuum Filter/Storage Systems | Fisher | 09-761-108 | A variety of sizes |
| Formvar/Carbon 400 mesh, Copper approx. grid hole size: 42µm | Ted Pella, Inc | 01754-F | 25 pack |
| GE Healthcare 5 mL HisTrap HP Prepacked Columns | GE HealthCare | 45-000-325 | 5 pack |
| Glycerol | Fisher | BP229-4 | 4 L |
| Goat Anti-Mouse IgG H&L (Alkaline Phosphatase) | ABCam | ab97020 | 1 mg |
| Imidazole | Fisher | O3196-500 | 500 g |
| Instant NonFat Dry Milk | Quality Biological | A614-1003 | 10 pack |
| Kinetic-QCL Kinetic Chormogenic LAL Assay | Lonza Walkersville | 50650U | 192 Test Kit |
| LAL Reagent Grade Multi-well Plates | Lonza Walkersville | 25-340 | 1 plate |
| Magic Media E. coli Expression Medium | ThermoFisher | K6803 | 1 L |
| MilliporeSigma Millex Sterile Syringe 0.22 mm Filters | Millipore | SLGV033RB | 250 pack |
| Mouse Anti-Human IgG Fc-AP | Southern Biotech | 9040-04 | 1.0 mL |
| One Shot BL21 Star (DE3) Chemically Competent E. coli | ThermoFisher | C601003 | 20 vials |
| Precision Plus Protein Unstained Protein Standards, Strep-tagged recombinant, | BioRad | 1610363 | 1 mL |
| Slide-A-Lyzer Dialysis Cassettes, 10K MWCO, 12 mL | ThermoFisher | 66810 | 8 pack |
| Sodium Chloride | Fisher | BP358-212 | 2.5 kg |
| Sodium Phosphate Monobasic | Fisher | BP329-500 | 500 g |
| Tris Base | Fisher | BP152-1 | 1 kg |
| Tris-(2-carboxyethyl)phosphine hydrochloride | Biosynth International | C-1818 | 100 g |
| Uranyl Acetate, Reagent, A.C.S | Electron Micoscopy Services | 541-09-3 | 25 g |
| Urea | Fisher | BP169-500 | 2.5 kg |
| Whatman qualitative filter paper | Sigma Aldrich | WHA10010155 | pack of 500 |
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| ChromLab Software ver 4 | BioRad | 12009390 | Software |
| Epoch 2 Microplate Spectrophotometer | BioTek | EPOCH2 | Plate Reader |
| Fiberlite F14-14 x 50cy Fixed-Angle Rotor | ThermoFisher | 096-145075 | Rotor |
| Gel Doc EZ Gel Documentation System | BioRad | 1708270 | Gel Imager for Stain free Gels |
| JEOL TEM | JEOL | 1400 | Transmission Electron Microscope |
| Mini-PROTEAN Tetra Vertical Electrophoresis Cell for Mini Precast Gels | BioRad | 1658004 | To run gels |
| NanoDrop One Microvolume UV-Vis Spectrophotometer | ThermoFisher | ND-ONE-W | For Protein Concentration |
| NanoSight NS300 | Malvern Panalytical | Particle Sizing | |
| NanoSight NTA software NTA | Malvern Panalytical | Particle Sizing | |
| New Brunswick Innova 44/44R | Eppendorf | M1282-0000 | Incubator/Shaker |
| NGC Quest 10 Chromatography System | BioRad | 7880001 | FPLC to aid in protein purification |
| PELCO easiGlow Glow Discharge Cleaning System | Ted Pella, INC | 91000S | To clean grids |
| PowerPac Universal Power Supply | BioRad | 1645070 | To run gels |
| Rocker Shaker | Daigger | EF5536A | For Western |
| Sonifer 450 | Branson | also known as 096-145075 | Sonicator |
| Thermo Scientific Sorvall LYNX 4000 Superspeed Centrifuge | ThermoFisher | 75-006-580 | Centrifuge |
| Trans-Blot Turbo Mini Nitrocellulose Transfer Packs | BioRad | 1704158 | For Western |
| Trans-Blot Turbo Transfer System | BioRad | 1704150 | For Western |
| Vortex-Genie 2 | Daigger | EF3030A | Vortex |
| Zetasizer Nano ZS | Malvern Panalytical | Particle Sizing | |
| Zetasizer Software | Malvern Panalytical | Particle Sizing |
References
- Karch, C. P., Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochemical Pharmacology. 120, 1-14 (2016).
- Snapper, C. M. Distinct Immunologic Properties of Soluble Versus Particulate Antigens. Frontiers in Immunology. 9, 598 (2018).
- Kelly, H. G., Kent, S. J., Wheatley, A. K. Immunological basis for enhanced immunity of nanoparticle vaccines. Expert Review of Vaccines. , 1-12 (2019).
- Yeates, T. O. Geometric Principles for Designing Highly Symmetric Self-Assembling Protein Nanomaterials. Annual Review of Biophysics. 46, 23-42 (2017).
- Marcandalli, J., et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell. 176 (6), 1420-1431 (2019).
- Ross, J. F., et al. Decorating Self-Assembled Peptide Cages with Proteins. ACS Nano. 11 (8), 7901-7914 (2017).
- Karch, C. P., Matyas, G. R., Burkhard, P., Beck, Z. Self-Assembling Protein Nanoparticles: implications for HIV-1 vaccine development. Nanomedicine (Lond). 13 (17), 2121-2125 (2018).
- Raman, S., Machaidze, G., Lustig, A., Aebi, U., Burkhard, P. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles). Nanomedicine. 2 (2), 95-102 (2006).
- Karch, C. P., et al. Design and characterization of a self-assembling protein nanoparticle displaying HIV-1 Env V1V2 loop in a native-like trimeric conformation as vaccine antigen. Nanomedicine. , (2018).
- Karch, C. P., et al. The use of a P. falciparum specific coiled-coil domain to construct a self-assembling protein nanoparticle vaccine to prevent malaria. Journal of Nanobiotechnology. 15 (1), 62 (2017).
- Li, J., et al. A self-adjuvanted nanoparticle based vaccine against infectious bronchitis virus. PLoS One. 13 (9), e0203771 (2018).
- Wahome, N., et al. Conformation-specific display of 4E10 and 2F5 epitopes on self-assembling protein nanoparticles as a potential HIV vaccine. Chemical Biology & Drug Design. 80 (3), 349-357 (2012).
- El Bissati, K., et al. Effectiveness of a novel immunogenic nanoparticle platform for Toxoplasma peptide vaccine in HLA transgenic mice. Vaccine. 32 (26), 3243-3248 (2014).
- Kaba, S. A., et al. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. Journal of Immunology. 183 (11), 7268-7277 (2009).
- Kaba, S. A., et al. Protective antibody and CD8+ T-cell responses to the Plasmodium falciparum circumsporozoite protein induced by a nanoparticle vaccine. PLoS One. 7 (10), e48304 (2012).
- Seth, L., et al. Development of a self-assembling protein nanoparticle vaccine targeting Plasmodium falciparum Circumsporozoite Protein delivered in three Army Liposome Formulation adjuvants. Vaccine. 35 (41), 5448-5454 (2017).
- Kaba, S. A., et al. Self-assembling protein nanoparticles with built-in flagellin domains increases protective efficacy of a Plasmodium falciparum based vaccine. Vaccine. 36 (6), 906-914 (2018).
- El Bissati, K., et al. Protein nanovaccine confers robust immunity against Toxoplasma. NPJ Vaccines. 2, 24 (2017).
- Karch, C. P., et al. Vaccination with self-adjuvanted protein nanoparticles provides protection against lethal influenza challenge. Nanomedicine. 13 (1), 241-251 (2017).
- Haynes, B. F., et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. New England Journal of Medicine. 366 (14), 1275-1286 (2012).
- Rerks-Ngarm, S., et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. New England Journal of Medicine. 361 (23), 2209-2220 (2009).
- O' Connell, R. J., Kim, J. H., Excler, J. L. The HIV-1 gp120 V1V2 loop: structure, function and importance for vaccine development. Expert Review of Vaccines. 13 (12), 1489-1500 (2014).
- Babapoor, S., et al. A Novel Vaccine Using Nanoparticle Platform to Present Immunogenic M2e against Avian Influenza Infection. Influenza Research and Treatment. 2011, 126794 (2011).
- Indelicato, G., Burkhard, P., Twarock, R. Classification of self-assembling protein nanoparticle architectures for applications in vaccine design. Royal Society Open Science. 4 (4), 161092 (2017).
- Indelicato, G., et al. Principles Governing the Self-Assembly of Coiled-Coil Protein Nanoparticles. Biophysical Journal. 110 (3), 646-660 (2016).
- Doll, T. A., Raman, S., Dey, R., Burkhard, P. Nanoscale assemblies and their biomedical applications. Journal of the Royal Society Interface. 10 (80), 20120740 (2013).
- Rey, F. A., Lok, S. M. Common Features of Enveloped Viruses and Implications for Immunogen Design for Next-Generation Vaccines. Cell. 172 (6), 1319-1334 (2018).
- . Bacterial Endotoxins. United States Pharmacopeia (USP). , (2011).
- . Points to Consider (PTC) in the Characterization of Cell Lines Used to Produce Biologicals Available from: https://www.fda.gov/media/76255/download (1993)

