Isolation of Region-specific Microglia from One Adult Mouse Brain Hemisphere for Deep Single-cell RNA Sequencing
Summary
We provide a protocol for isolation of microglia from different dissected regions of an adult mouse brain hemisphere, followed by semi-automated library preparation for deep single-cell RNA sequencing of full-length transcriptomes. This method will help to elucidate functional heterogeneity of microglia in health and disease.
Abstract
As resident macrophages in the central nervous system, microglia actively control brain development and homeostasis, and their dysfunctions may drive human diseases. Considerable advances have been made to uncover the molecular signatures of homeostatic microglia as well as alterations of their gene expression in response to environmental stimuli. With the advent and maturation of single-cell genomic methodologies, it is increasingly recognized that heterogenous microglia may underlie the diverse roles they play in different developmental and pathological conditions. Further dissection of such heterogeneity can be achieved through efficient isolation of microglia from a given region of interest, followed by sensitive profiling of individual cells. Here, we provide a detailed protocol for the rapid isolation of microglia from different brain regions in a single adult mouse brain hemisphere. We also demonstrate how to use these sorted microglia for plate-based deep single-cell RNA sequencing. We discuss the adaptability of this method to other scenarios and provide guidelines for improving the system to accommodate large-scale studies.
Introduction
Microglia, representing 5%−10% of all neural cells, are resident macrophages scattered throughout the central nervous system (CNS)1. Protected behind blood-brain barrier, typical microglia in a healthy adult brain contain many fine processes that rapidly extend and retract to interact with neurons and other glial cells in the parenchyma. Microglia can also adopt the amoeboid morphology associated with increased phagocytic function during specific developmental stages or upon immune challenges in injury and disease1,2,3,4. Recent exciting discoveries have clearly demonstrated that microglia are by no means passive bystanders to brain-derived or pathological signals, but play pivotal roles in controlling brain development and homeostasis, for instance, by supporting neuronal survival, pruning immature synapses, promoting oligodendrocyte lineage cells differentiation as well as angiogenesis1. As more functions of microglia are elucidated, the excitement is further fueled by human genetics studies, which showed that many neurodegenerative disease risk genes, such as TREM2, are predominantly or exclusively expressed by microglia5,6,7. Given their significance in development and plausible disease-driving roles, tremendous effort has recently been put towards our understanding of microglial gene regulation and function in hope of finding new therapeutic targets for neurodegenerative diseases1,8.
RNA sequencing (RNA-seq) allows unbiased characterization of cell type-specific gene expression, which in turn guides scientists to investigate gene functions in dense cellular networks7. RNA-seq had been mostly done on bulk samples, leading to the discovery of a homeostatic microglial gene signature that distinguishes them from other neural and immune cells9. However, such an approach could overlook molecular and functional differences among microglia, especially those transiently present in development, or associated with aging and disease. Indeed, single-cell RNA-seq (scRNA-seq) offers the sensitivity and resolution that have revolutionized the field by revealing previously underappreciated heterogeneity of microglia in a variety of contexts2,3,10. In addition, due to the presence of other similar immune cells at the CNS-circulation interface, scRNA-seq provides information aiding the design of new tools to separate and functionally dissect these related cells with little prior knowledge2,11.
A diverse array of scRNA-seq platforms have been invented, each suitable for certain applications12. In general, droplet-based methods, such as 10x Genomics, are higher in throughput with (tens of) thousands of cells sequenced in each run, and they are less selective for the input which may contain mixed cell populations requiring broad categorization. Plate-based methods provide higher sensitivity and read depth13,14, usually targeting specific populations from cell sorting to reveal subtle differences or rare transcripts. Given the small percentage of microglial cells, particularly those development- or disease-associated subpopulations, among all CNS cell types, it is often desirable to isolate microglia from a specific region of interest and obtain deep and full-length transcriptomic information in order to understand their heterogeneity.
Here, we provide details on how to isolate microglia from different mouse brain regions dissected from a single hemisphere, which are used for single-cell (or bulk) RNA-seq following a semi-automated plate-based library preparation procedure. The other hemisphere can then be used for histological validation. Streamlined from a previously published method9, this isolation protocol aims to maximize the yield from small amount of starting materials, and meanwhile maintain endogenous microglial gene expression profiles. We use fluorescence-activated cell sorting (FACS) to enrich microglia (or other related immune cells of interest) into 96-well plates and miniaturize the volumes of reagents for library preparation in order to increase throughput. We highlight this sensitive scRNA-seq platform, although other plate-based strategies may be applied. This method can be easily adapted to isolate microglia from other dissected tissues, such as injury or disease foci, and the age of the mouse can vary across almost any postnatal stages. Efficient isolation of regional microglia for single-cell transcriptomics studies will facilitate better understanding of their functions in health and disease.
Protocol
All procedures involving rodents conformed to Stanford University guidelines, which comply with national and state laws and policies. All animal procedures were approved by Stanford University’s Administrative Panel on Laboratory Animal Care.
NOTE: All solution and buffer compositions are provided in Table of Materials.
1. Preparation on the Day of Cell Isolation
- Prepare the following reagents and chill them on ice: medium A (50 mL), magnetic-activated cell sorting (MCS) buffer (30 mL), FACS buffer (25 mL), phosphate-buffered saline (PBS; 30 mL), DNase (320 µL), and RNase inhibitor (30 µL).
NOTE: The volumes provided here are sufficient to isolate microglia from 4 brain regions (e.g., cortex, cerebellum, hippocampus and striatum) of a single mouse brain hemisphere. Scale up if more tissues are used. - Place four clean 2 mL Dounce homogenizers on ice and add 2 mL of medium A with 80 µL of DNase (12,500 units/mL) and 5 µL of RNase inhibitor into each Dounce homogenizer. Chill the pistons in 15 mL tubes.
- Label four 6 cm Petri dishes with brain regions for tissue collection and add 200 µL of cold medium A into each dish on ice. Add about 5 mL of medium A into a 6 cm Petri dish for dissection.
NOTE: Prior to use, make sure all reagents are sterile and suitable for RNA applications. Bench and dissection tools need to be clean and sprayed with RNase decontamination solution (Table of Materials).
2. Brain Region Dissection
NOTE: This step should take ~30 min.
- Inject 400−500 µL of ketamine/xylazine (24 mg/mL ketamine and 2.4 mg/mL xylazine) into the peritoneum of a juvenile to adult stage mouse (>1 month old).
NOTE: A male mouse is used here for illustration, but either gender is suitable. - Wait for 5 min and pinch a hind paw to ensure lack of retraction.
- Immediately use a 26 G x 3/8 needle to perform transcardial perfusion with 20−30 mL of ice-cold PBS until buffer runs out with no visible blood.
- Decapitate the mouse with a pair of surgical scissors. Use small scissors to cut open the skin to expose the underneath skull, and then cut through the sagittal suture, lambdoidal suture and coronal suture. Use forceps to pull off both sides of parietal bone and interparietal bone without damaging the tissue, and carefully move the brain into the dissection Petri dish with medium A.
- Use a pre-chilled blade to cut the brain through midline into two hemispheres.
NOTE: The four brain regions described here from a single hemisphere yield sufficient numbers of microglia for single-cell or bulk RNA-seq applications. The other hemisphere can be used for immunohistochemistry or RNA in situ validation. - Separate the cerebellum from the cortical lobe and the brain stem with #55 forceps and transfer the tissue into a collection Petri dish.
- Use #55 forceps to carefully dissect out the hippocampus and the striatum from the cortex and transfer each tissue into a collection Petri dish.
3. Mechanical Tissue Dissociation
NOTE: Keep cells and reagents cool all the time except during staining steps. This step should take ~30 min.
- Chop each brain tissue with a razor blade into <1 mm3 fine pieces.
- Use 1 mL pipette (with tips cut off) to transfer tissue pieces into the pre-chilled Dounce homogenizers, each containing 2 mL of medium A with DNase and RNase inhibitor.
- Homogenize the tissue by slowly twisting the piston in and out of the Dounce homogenizer for 6−10 full strokes, until no visible chunks are present.
NOTE: Douncing kills neurons and most other glial cells but leave microglial cells rather intact. Both insufficient and over-douncing can lead to low yield of cells. - Transfer dissociated tissues into 50 mL tubes through 70 µm strainers.
- Rinse each Dounce homogenizer and piston with a total of 6 mL of cold medium A and filter the rinsing solution through the same strainer into the corresponding tube.
- Transfer the single cell suspensions (about 8 mL each) into 15 mL conical tubes and centrifuge at 400 x g for 5 min at 4 °C with brake = 5.
4. Myelin Removal
NOTE: This step should take ~60 min.
- Prepare 1 large depletion (LD) column (for cortex) and 3 large selection (LS) columns (for the other 3 regions) in a magnetic separator (Table of Materials). Rinse each column with 3 mL of MCS buffer.
- Once centrifugation is finished, pipette out and discard the supernatant without disturbing the pellet. For the cortex and cerebellum tissues, resuspend cells in 850 µL of MCS buffer with 1.8 µL of RNase inhibitor. For hippocampus and striatum tissues, resuspend cells in 400 µL of MCS buffer with 0.9 µL of RNase inhibitor.
NOTE: The columns (LD vs. LS) and the volume used for resuspension are optimized based on the amount of myelin present in the tissue. If other brain regions are assayed, these conditions may need to be adjusted depending on the size of the tissue and how much myelin may exist. Effectiveness of myelin removal can be estimated in step 4.9. - Add 100 µL of myelin removal beads each into cells from cortex and cerebellum and add 50 µL of myelin removal beads each into cells from hippocampus and striatum.
- Gently mix the cells with beads and incubate the tubes on ice for 10 min.
- Bring the volume of the tube with cortical cells to 2 mL with MCS buffer, and all others to 1 mL (i.e., add 1 mL for cortical cells; 500 µL for hippocampal and striatal cells; no need to add buffer to cerebellar cells).
- Once columns are empty of rinsing buffer, place a 15 mL tube below each column. Load cortical cells (2 mL) onto the LD column, and all others (1 mL each) onto the LS columns. Immediately use 1 mL of MCS buffer each to wash the original tubes and load the solution onto the corresponding columns.
- Wash the LD column once with 1 mL of MCS buffer and wash each LS column twice with 1 mL of MCS buffer each wash. Continue to collect the flow-through solution during washing.
- Filter cells into round bottom FACS tubes through 35 µm strainer caps.
NOTE: Each tube should collect about 4 mL of single cell suspension depleted of myelin. - Optionally, take 10 µL of the cell suspension and mix it with 10 µL of 0.4% trypan blue solution. Examine the cells under a 10x bright field microscope to estimate the yield, survival rate and the level of residual myelin.
NOTE: A successful preparation should generate over 90% live cells (round in shape and excluding the blue dye) with little to no myelin debris. - Pellet cells in the FACS tubes at 400 x g for 5 min at 4 °C, with brake = 5. Slowly pour out supernatant and dab the edge of the tube on tissue paper. Resuspend cells in each tube with 300 µL of FACS buffer.
NOTE: If MCS sorting is preferred, CD11b beads can be used to select microglia and other myeloid cells following myelin removal. These cells can then be used for non-plate-based scRNA-seq as well as bulk RNA-seq. The caveat of this alternative approach is that CD11b beads do not separate microglia from other related immune cells which are also positive for this antigen.
5. Staining for Fluorescence-activated Cell Sorting
NOTE: This step should take ~40 min.
- Add 5 µL of mouse Fc receptors block reagent (Table of Materials) into each tube. Incubate for 5 min on ice.
- Add 1 µL of CD45-PE-Cy7 and 1 µL of CD11b-BV421 into each tube.
NOTE: Antibodies with other conjugated fluorophores may be used. Antibodies against TMEM119, a specific marker for homeostatic microglia9, can also be included, but it is recommended to be used together with CD45 and CD11b, because certain microglia subpopulations may lose TMEM119 surface expression. - Incubate the tubes on a shaker for 10 min at room temperature (RT).
- Add 2 mL of FACS buffer to wash.
- Pellet cells at 4 °C, 400 x g for 5 min. Slowly pour out supernatant and dab the edge of the tube on tissue paper. Resuspend cells in each tube using 400 µL of FACS buffer with 1 µL of RNase inhibitor and 0.5 µL of propidium iodide (PI, 1:1,000 dilution).
6. Index FACS Sorting
NOTE: This step should take ~1 h.
- Following standard FACS procedure, draw gates based on scatter (excluding debris), single cells, live cells (PI negative), microglia and myeloid cells (CD45+, CD11b+).
NOTE: Typical microglial cells express CD45 at lower levels compared with other border-associated macrophages, such as perivascular and meningeal macrophages, and therefore gating on CD45 low CD11b+ should be sufficient if the focus of the research is gene expression profiles in these classical microglia1,2. However, certain subsets of microglia may have higher CD45 expression and this is particularly true during development or in disease conditions. To ensure unbiased analysis of microglial gene expression, CD45 high and CD45 low immunophenotypes can be recorded through index sorting setting and both populations collected for sequencing and downstream analysis. In addition, TMEM119 surface expression may be used to target the homeostatic microglial population (see step 5.2) and analyzed together with CD45 levels and the sequencing results. - Sort single microglia (with a 100 µm nozzle) into 96-well polymerase chain reaction (PCR) plates, containing 4 µL of lysis buffer each well (see the published protocol14 for details).
NOTE: The External RNA Control Consortium (ERCC) RNA spike-in mix (Table of Materials) can be added at 1:2.4 x 107 in the lysis buffer for quality control and normalization purposes2. - Briefly vortex the plates and spin down using a bench-top centrifuge.
- Immediately freeze the samples on dry ice. Store the plates at -80 °C until library preparation.
NOTE: Alternatively, more cells can be collected into 1.5 mL tubes with RNA extraction buffer for bulk RNA-seq. According to authors’ experience, 3,000 cells are sufficient to generate good quality libraries following the published protocol2,14.
7. Single-cell RNA-sequencing Library Preparation
NOTE: Here, the published protocol14 is followed to generate scRNA-seq libraries with the aid of liquid handling robotics and a few modifications. In this article, the procedure is only briefly described, and the differences are highlighted. Processing 4 plates simultaneously takes about 2.5 days.
- Thaw plates on ice and perform reverse transcription with Oligo-dT30VN primer (in the lysis buffer) to generate cDNA with thermal cyclers (Table 1): 42 °C 90 min; 70 °C 5 min; 4 °C hold. Then amplify cDNA with an additional exonuclease digestion step at the beginning (Table 2) using the PCR master mix and in situ PCR (ISPCR) primers (Table of Materials) and the following PCR condition: 37 °C 30 min; 95 °C 3 min; 23 cycles of 98 °C 20 s, 67 °C 15 s, 72 °C 4 min; 72 °C 5 min.
- Purify cDNA using 18 µL of magnetic beads per well (0.7:1 ratio). Incubate the plate on a magnetic stand for 5 min and wash samples twice with freshly made 80% ethanol, 80 µL per well each time. After drying the plate for 15-20 min, elute cDNA with 20 µL of elution buffer per well.
NOTE: For quality control, use a fragment analyzer (high sensitivity next-generation sequencing [NGS] fragment analysis, 1−6,000 bp) to check size distributions and concentrations of cDNA, and only further process samples with a smear between 500−5,000 bp, and higher than 0.05 ng/µL. - To generate libraries, mix 0.4 µL of each cDNA sample with 1.2 µL of Tn5 tagmentation reagents from the library preparation kit (Table of Materials) in a 384-well plate with the aid of a nanoliter pipetting machine, at 55 °C 10 min.
NOTE: Here, 1/12.5 of the suggested reaction volume (5 µL sample and 15 µL of tagmentation reagents) is added in order to reduce cost and meanwhile increase throughput. A nanoliter pipetting machine (Table of Materials) is used to transfer reagents (this and following steps) from and to 384-well plates. - Add 0.4 µL of neutralization buffer to stop the tagmentation reaction at RT for 5 min.
- Add 384 indexes (Table of Materials, 0.4 µL forward and 0.4 µL reverse), 1.2 µL of PCR mix to samples (2 µL each), and amplify the libraries using the following condition: 72 °C 3 min; 95 °C 30 s; 10 cycles of 95 °C 10 s, 55 °C 30 s, 72 °C 1 min; 72 °C 5 min.
- Pool all individual libraries (take 1 µL each) from the same 384-well plate together, and use magnetic beads (Table of Materials, 0.7:1 ratio) to purify the final pooled libraries.
- Use a fluorometer (Table of Materials) to measure the concentrations and a bioanalyzer (Table of Materials) to examine the size distributions of pooled libraries before sequencing. Target the sequencing depth at 1 million raw reads per cell.
- Follow standard bioinformatic procedures to filter and trim sequencing reads and perform alignment2. Use the count table and meta data as inputs to do clustering analysis with the Seurat package15.
Representative Results
This protocol describes a method to isolate and sort microglia from different brain regions in one adult perfused brain hemisphere, followed by scRNA-seq. We use douncing to create single cell suspension and also as a first step to enrich microglia. Insufficient or over-douncing reduces the yield. In addition, adult mouse brains contain high levels of myelin, which can also reduce sorting efficiency and yield if not removed properly. Therefore, we examine the cell suspension under microscope by using trypan blue and a hemocytometer to estimate the yield, cell viability and efficacy of myelin removal (step 4.9) before performing antibody staining (Figure 1). Total cell counts at this point should be over 30,000 for cortex, and over 5,000 for other tissues. Over 90% of cells should be viable with little myelin debris.
We use a FACS machine to sort microglia (or myeloid cells), which are typically CD45 low and CD11b positive. At least for the cortical tissue, successful isolation should generate over 80% microglia out of all live single cells (Figure 2). The dying/dead population represents only a small fraction of the preparation (around 10%).
Once individual microglia are captured into the lysis buffer, RNA is released and subsequently reverse transcribed to cDNA, which is then amplified for 23 cycles. It is important to check the quality of these cDNA samples—at least a portion of them—before making libraries. As a capillary electrophoresis platform, the fragment analyzer and high-sensitive NGS fragment kits (1−6,000 bp) provide quick and accurate information about size distribution as well as quantity of cDNA molecules present in each well of a 96-well plate (Figure 3A). Samples showing a smear (500−5,000 bp) and above certain concentration threshold (e.g., 0.05 ng/µL) can be used to make libraries. Similarly, the pooled libraries should be tested on a bioanalyzer before sequencing (Figure 3B).
We sequence the samples to a depth of over 1 million raw reads per cell, which saturates the detection power of this scRNA-seq methodology16. With about 60% mapping rate, over 2,000 genes per microglial cell can be detected. We obtained published data that were generated using this isolation method17, and demonstrate its reproducibility from independent experiments and sensitivity for detecting microglia-specific genes across the sequenced population (Figure 4).
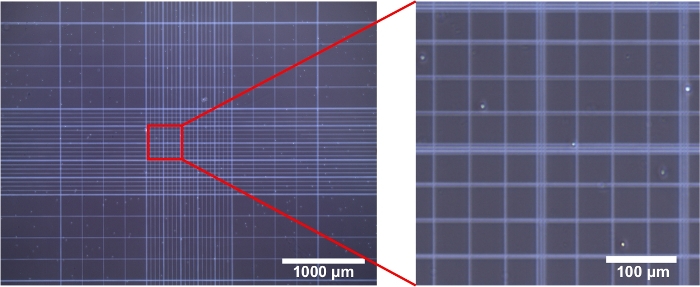
Figure 1: Estimation of microglia isolation yield, cell viability and efficiency of myelin removal. The left panel showed the bright field image (4x magnification) of trypan blue stained cells (from cortex) after passing through myelin removal columns. Results for other regions would look similar but with fewer cells. The vast majority (if not all) of cells appeared bright (non-stained) and round due to loss of processes. The right image was the zoom-in (20x) of the boxed area and little myelin debris was present. Please click here to view a larger version of this figure.
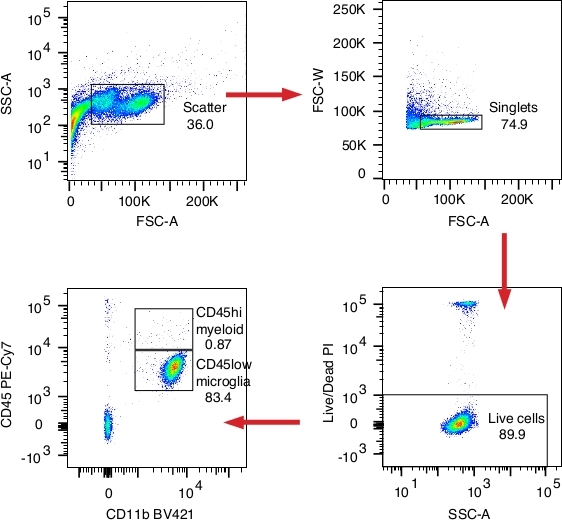
Figure 2: Gates used for sorting microglia. Data showed the gating strategy for sorting microglia from the cortex, and the same strategy was used for other regions. Cell debris was first excluded from the scatter plot, and single cells were gated by forward scatter-area (FSC-A)/forward scatter-width (FSC-W). Live cells were gated by PI negative staining, which were from about 90% of all single cells. Microglia, representing roughly 80% of live cells, were sorted from the CD45 low CD11b+ gate. A total of ~30,000 microglia could be isolated and sorted from the cortex tissue (from one hemisphere of a 3-month old mouse). Index sorting was performed on a FACS machine. Please click here to view a larger version of this figure.
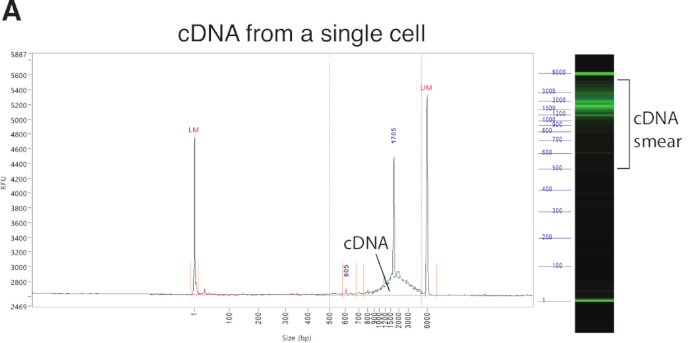
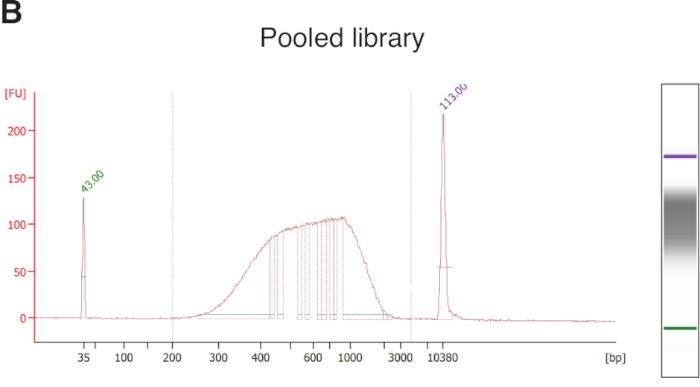
Figure 3: Representative quality control results for amplified cDNA from a single microglial cell and an eventual pooled library. (A) Using a fragment analyzer, all fragments are plotted with their sizes on the X axis, and relative fluorescence intensity on the Y axis, signifying the abundance of a given sized cDNA. LM (lower marker, 1 bp) and UM (upper marker, 6,000 bp) are loading markers with known concentrations used for measuring cDNA quantity. Successfully amplified cDNA from a representative microglial cell forms a curve (green dotted line) on the size distribution graph or a smear (labeled bracket) on the gel graph between 500 bp and 5,000 bp. Other labeled peaks were amplified from ERCC spike-in molecules or ribosomal RNA. The concentration of cDNA between 500 bp and 5,000 bp can be quantified, and those samples with concentrations higher than 0.05 ng/μL and showing such typical curves are retained for library preparation. RFU, relative fluorescence unit. (B) Representative quality control result on a bioanalyzer showing size distribution of a final pooled library (with about 380 cells). Typically, it ranges from 200 bp to 2,000 bp with an average size of 400−600 bp. The two sharp peaks were loading markers. FU, fluorescence unit. Please click here to view a larger version of this figure.
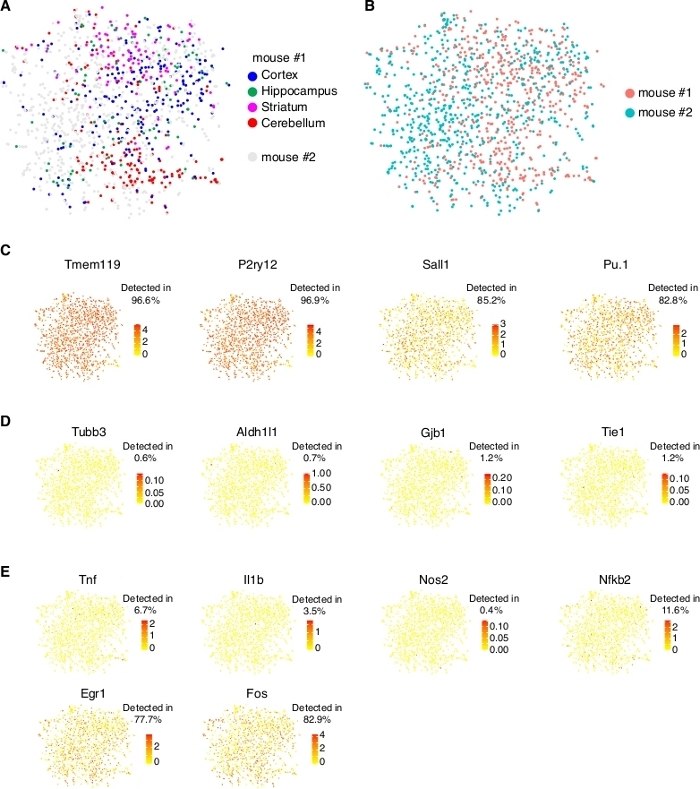
Figure 4: scRNA-seq analysis of microglia isolated from 4 brain regions of two male mice. (A) tSNE plot showing intermingled pattern of regional microglia (only cells from mouse #1 were highlighted). They do not form distinct clusters according to region origins beyond subtle shift into concentrated areas on the tSNE plot (possibly due to batch effects). This observation is consistent with limited regional heterogeneity of microglia based on global gene expression in the adult mice (see a recent publication2 for further discussion on this topic). (B) tSNE plot showing overlapping pattern of microglia from two individual mice that were processed independently. Although small batch effects may exist (cells concentrating in certain areas of the plot), these cells do not form distinct clusters according to animal origins. This result suggests the reproducibility of the protocol for comparing data between experiments. (C) Expression of microglia signature genes detected by scRNA-seq showing over 95% detection rate for specific markers, such as Tmem119 and P2ry12, and over 80% detection rate for known transcription factors such as Sall1 and Pu.1. The data were re-analyzed from published literature17. (D) Vast majority of isolated microglia lack expression of genes specific to other cell types, such as Tubb3 (neurons), Aldh1l1 (astrocytes), Gjb1 (oligodendrocytes), and Tie1 (endothelial cells). (E) Expression of genes related microglial activation or stress. Classical markers, Tnf, Il1b, Nos2, and Nfkb2, which are lowly expressed, are shown on the top. Early response genes, Egr1 and Fos, are shown at the bottom (see discussion). Please click here to view a larger version of this figure.
| Reagent | Volume (µL) |
| Cell lysis | 4 |
| Reverse transcriptase (100 U/µL) | 0.95 |
| Rnase inhibitor | 0.25 |
| 5x First strand buffer | 2 |
| Dithiothreitol (DTT; 100 mM) | 0.5 |
| Betaine (5 M) | 2 |
| MgCl2 (1 M) | 0.06 |
| Template switch oligo (TSO; 100 µM) | 0.1 |
| H2O | 0.14 |
| Total | 10 |
Table 1: Reverse transcription condition. Reagent volumes for one reverse transcription reaction are provided.
| Reagent | Volume (µL) |
| Reverse transcription product | 10 |
| 2x PCR master mix | 12.5 |
| ISPCR primer (10 µM) | 0.25 |
| Lambda exonuclease | 0.1125 |
| H2O | 2.1375 |
| Total | 25 |
Table 2: PCR amplification condition. Reagent volumes for one PCR amplification of cDNA from a single cell are provided.
| Plate-based (this protocol) | Droplet-based (10x Genomics) | |
| Sensitivity | More genes detected | Fewer genes detected |
| Full length | Yes | No (5' or 3' end) |
| Flexibility for cell numbers/populations | Suitable for characterization of small or rare subpopulations | Suitable for broad categorization of large cell populations |
| Throughput | Up to several thousands of cells | Hundreds to tens of thousands of cells |
| Unique molecular identifier | No | Yes |
| Cost per cell | $1−$5 | less than $1 |
| Experimental difficulty | More steps and usually require liquid handling robotics | Simple to perform with commercialized machines |
| Cell populations | Targeted by FACS sorting | Unbiased |
Table 3: Comparison between plate-based and droplet-based scRNA-seq methods. In this article, the plate-based full-length scRNA-seq procedure is provided, which has the advantages of higher sensitivity, full-length sequencing and flexibility for small numbers of cells as inputs. Droplet-based methods offer the advantages of higher throughput, lower cost, easy to perform and data in unique molecular identifiers. They are complementary depending on the purpose of the experiments.
Discussion
Microglia actively interact with other cell types in the CNS, and they are very sensitive to environmental stimuli. In order to minimize inflammatory responses and aberrant changes in their gene expression during the isolation process, this protocol has been streamlined from a previously published method9, and it is now suitable to isolate microglia from multiple regions of a single mouse brain hemisphere in parallel. The tissues and reagents are kept at cold temperature and experiments are performed in a timely manner (about 3.5 h from dissection to sorting) with fewer washes and less reagents based on limited tissue sizes. We choose douncing over other homogenization methods, such as enzymatic digestion, because mechanical dissociation can kill and thus remove neurons and other glial cells while causing little damage or activation to microglia9,18. Over-douncing, however, may reduce the yield of microglia and should be avoided. It has been previously shown that mechanical dissociation and beads-based myelin removal followed by FACS sorting introduce less activation to microglia, based on minimal expression of classical inflammatory genes, e.g., Tnf, Il1b, Nos2, and Nfkb29 (Figure 4E). Nonetheless, microglia could still respond to the ex vivo conditions during dissociation and sorting by upregulating early response genes such as Fos and Egr1 (Figure 4E), which are not normally expressed by microglia in vivo2,19, and such changes from transcriptomic studies should always be validated on histology sections.
Here we provide procedures for isolating microglia from cortex, cerebellum, hippocampus and striatum from an adult mouse brain hemisphere, and this protocol can be easily adapted to other regions or stages. This is useful for situations, for example, when disease-affected microglia are limited to certain areas of the brain which can be dissected out, or in aging studies, where pooling samples is inappropriate due to significant individual variations. With the availability of antibodies against microglial (or pan innate immune) epitopes, this protocol could also be adapted for isolating microglia in rat or human tissues9. When adjusting for larger or older tissues, it is important to consider and test the volume of myelin-removal beads and the type of columns used. An alternative approach for myelin removal is by density gradient centrifugation in commercial low-viscosity media20. Overall these two methods are comparable in their efficiency, although the density gradient centrifugation method may provide slightly higher yields, and on the other hand, the magnetic beads method is less likely to introduce endotoxins, which may activate microglia21,22.
Due to the small percentage of microglia among total brain cells, it is often essential to enrich or purify microglia before performing single-cell transcriptomic studies. We use FACS as a sensitive approach to capture lowly represented cell types, and select microglia based on CD11b and CD45 surface expression. From one hemisphere, we routinely obtain ~30,000 microglia for cortex, and ~5,000 microglia for cerebellum, hippocampus or striatum. These yields are sufficient for most single-cell applications as well as bulk RNA-seq (3,000 or more cells can be used). It is worth mentioning that microglia may upregulate CD45 immunoreactivity during development or disease2, in which case cells with both low and high levels of CD45 should be recorded and index sorted for analysis. Another option for enriching microglia is to use CD11b beads-mediated magnetic-activated cell sorting following myelin removal, but this would limit scRNA-seq to droplet methods.
To study microglial gene expression at single-cell resolution, we usually sort cells into at least 2−3 96-well plates per sample and perform library preparation with liquid handling machines. It has been shown that this plate-based full-length library preparation approach is one of the most sensitive scRNA-seq methods in detection limit, allowing accurate quantification of rare transcripts, such as transcription factors13,14,16. Because of full-length sequencing, this approach is not subject to 5′ or 3′ bias and can be used to analyze splicing variants (Table 3). In addition, unlike droplet-based methods, which usually require hundreds or thousands of cells in a reaction, this plate-based protocol has the flexibility to include a small number of cells in each experiment, and gradually expand the population size by adding more plates into the design later on. This is advantageous when studying small subsets of microglia in certain conditions such as embryonic development.
While this protocol reproducibly generates high-quality scRNA-seq libraries for brain regional microglia, it is often only suitable for small-scale studies due to the relatively high cost (about $5/cell for library preparation, still cheaper than $23/cell if using the off the shelf SmartSeq v4 kit). Several strategies can help reduce the cost and increase the throughput. First, cells can be sorted into 384-well plates with as little as 0.5 µL of lysis buffer, and this only requires 1/8 volumes of reagents for the initial reverse transcription and PCR amplification steps. Second, it is possible to produce in-house Tn5 transposase that has similar quality as the one in a commercially available kit23. These two modifications could reduce the library preparation cost down to $1/cell. Third, using customized indexes, thousands of cells can be pooled together for sequencing on a system without sacrificing the sequencing depth (>1 million reads/cell)17. These improvements along with the incorporation of automated liquid handling robots, will enable high-throughput deep scRNA-seq of microglia isolated from almost any defined regions.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Mariko L. Bennett, Liana Nicole Bonanno, and Spyros Darmanis for their help during the development of this protocol. We also thank the Stanford Shared FACS Facility, particularly Meredith Weglarz and Lisa Nichols; Yen Tran, Michael Eckart from Stanford Protein and Nucleic Acid Facility (PAN) for their great support for the filming. This work is funded by the JPB Foundation and Vincent J. Coates Foundation.
Materials
| 5 M Betaine | Sigma-Aldrich | Cat# B0300-5VL | |
| 10 mM dNTP mix | Thermo Fisher Scientific | Cat# R0192 | |
| 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat# 15575020 | |
| 10X Hanks’ Balanced Salt Solution | Thermo Fisher Scientific | Cat# 14185-052 | |
| 1 M HEPES | Thermo Fisher Scientific | Cat# 15630080 | |
| 1X KAPA HIFI Hotstart Master Mix | Kapa Biosciences | Cat# KK2602 | |
| 5 mL Round Bottom Polystyrene Tube, with Cell Strainer Cap | Corning | Cat# 352235 | |
| AATI, High Sensitivity NGS Fragment Analysis Kit (1 bp – 6,000 bp) | Advanced Analytical | Cat# DNF-474-1000 | |
| Bovine Serum Albumin | Sigma Aldrich | Cat# A8806 | |
| DNase I | Worthington | Cat# LS002007 | Working solution: 12500 units/ml |
| DTT, Molecular Grade | Promega | Cat# P1171 | |
| ERCC RNA Spike-In Mix | Thermo Fisher Scientific | Cat# 4456740 | |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10437-028 | |
| Illumina XT Index Kit v2 Set A (96 indexes) | Illumina | Cat# FC-131-2001 | |
| Illumina XT Index Kit v2 Set B (96 indexes) | Illumina | Cat# FC-131-2002 | |
| Illumina XT Index Kit v2 Set C (96 indexes) | Illumina | Cat# FC-131-2003 | |
| Illumina XT Index Kit v2 Set D (96 indexes) | Illumina | Cat# FC-131-2004 | |
| Lambda Exonuclease (5 U/μl) | New England BioLabs | Cat# M0262S | |
| Mouse Fc block | BD Pharmingen | Cat# 553142 | |
| Myelin removal beads | Miltenyl Biotec | Cat# 130-096-433 | |
| Nextera XT DNA Sample Prep Kit | Illumina | Cat# FC-131-1096 | |
| NextSeq 500/550 High Output Kit v2.5 (150 Cycles) | Illumina | Cat# 20024907 | |
| PBS (10X), pH 7.4 | Thermo Fisher Scientific | Cat# 70011044 | |
| PCRClean DX beads | Aline Biosciences | Cat# C-1003-50 | |
| Propidium Iodide | Thermo Fisher Scientific | Cat# P3566 | Staining: 1:1000 |
| Qubit dsDNA HS Assay Kit | Thermal Fisher Scientific | Cat# Q32851 | |
| Rat monoclonal anti mouse/human CD11b, Brilliant Violet 421 (clone M1/70) | BioLegend | Cat# 101236; RRID: AB_11203704 | Staining: 1:300 |
| Rat monoclonal anti mouse CD45, PE/Cy7 (clone 30-F11) | Thermo Fisher Scientific | Cat# 25-0451-82; RRID: AB_469625 | Staining: 1:300 |
| Recombinant RNase Inhibitor | Takara Bio | Cat# 2313B | |
| SMARTScribe Reverse Transcriptase (100 U/μl) | Clontech | Cat# 639538 | Containing 5x First strand buffer |
| Oligonucleotides | |||
| 0.1 μM ISPCR Oligo: 5' – AAGCAGTGGTATCAA CGCAGAGT-3' |
(Picelli et al., 2014) | ||
| Oligo-dT30VN primer: 5' – AAGCAGTGGTATCAACGCA GAGTACT 30 VN-3' |
(Picelli et al., 2014) | ||
| TSO 5' – AAGCAGTGGTATCAACGCAGA GTACATrGrG+G-3' ("r" is forribobases and "+" is for an LNA base) |
(Picelli et al., 2014) | ||
| Solutions | |||
| FACS buffer | Recipe: sterile-filtered 1% FBS, 2 mM EDTA, 25 mM HEPES in 1X PBS | ||
| MCS buffer | Recipe: sterile-filtered 0.5% BSA, 2 mM EDTA in 1X PBS | ||
| Medium A | Recipe: 15 mM HEPES, 0.5% glucose in 1X HBSS without phenol red | ||
| Plates | |||
| 384-well Rigi-Plate PCR Microplates, Axygen Scientific | VWR | 89005-556 | |
| Hard-shell 96-well PCR plates | Bio-Rad | HSP9631 | |
| Others | |||
| Dumont #55 forceps | Fine Science Tools | 11295-51 | |
| Dounce homogenizer, 2 ml | Wheaton | 357422 | |
| Large depletion column | Miltenyi Biotec | 130-042-901 | |
| Large selection column | Miltenyi Biotec | 130-042-401 | |
| MACS MultiStand | Miltenyi Biotec | 130-042-303 | |
| QuadroMACS Separator | Miltenyi Biotec | 130-090-976 | |
| RNAzap | Thermo Fisher Scientific | AM9780 | |
| Strainer (70 μm) | Falcon | 352350 | |
| Equipment | |||
| BD FACSAria II | BD Biosciences | http://www.bdbiosciences.com/ | |
| Bioanalyzer | Agilent | 2100 | |
| Fragment Analyzer | Agilent | 5300 | |
| Mosquito HTS nanoliter pipetting robot | TTP Labtech | https://www.ttplabtech.com/ | |
| Qubit 4 Fluorometer | Thermo Fisher Scientific | Q33226 | |
References
- Li, Q., Barres, B. A. Microglia and macrophages in brain homeostasis and disease. Nature Reviews Immunology. 18 (4), 225-242 (2018).
- Li, Q., et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron. 101 (2), 207-223 (2019).
- Keren-Shaul, H., et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169 (7), 1276-1290 (2017).
- Bohlen, C. J., et al. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron. 94 (4), 759-773 (2017).
- Guerreiro, R., et al. TREM2 variants in Alzheimer’s disease. New England Journal of Medicine. 368 (2), 117-127 (2013).
- Jonsson, T., et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. New England Journal of Medicine. 368 (2), 107-116 (2013).
- Zhang, Y., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience. 34 (36), 11929-11947 (2014).
- Colonna, M., Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annual Review of Immunology. 35, 441-468 (2017).
- Bennett, M. L., et al. New tools for studying microglia in the mouse and human CNS. Proceedings of the National Academy of Sciences of the United States of America. 113 (12), 1738-1746 (2016).
- Hammond, T. R., et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 50 (1), 253-271 (2019).
- Van Hove, H., et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nature Neuroscience. 22 (6), 1021-1035 (2019).
- Chen, G., Ning, B., Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Frontiers in Genetics. 10, 317 (2019).
- Svensson, V., et al. Power analysis of single-cell RNA-sequencing experiments. Nature Methods. 14 (4), 381-387 (2017).
- Picelli, S., et al. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols. 9 (1), 171-181 (2014).
- Macosko, E. Z., et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 161 (5), 1202-1214 (2015).
- Ziegenhain, C., et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular Cell. 65 (4), 631-643 (2017).
- Tabula Muris, C., et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 562 (7727), 367-372 (2018).
- Bohlen, C. J., Bennett, F. C., Bennett, M. L. Isolation and Culture of Microglia. Current Protocols in Immunology. 125 (1), 70 (2018).
- Haimon, Z., et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nature Immunology. 19 (6), 636-644 (2018).
- Collins, H. Y., Bohlen, C. J. Isolation and Culture of Rodent Microglia to Promote a Dynamic Ramified Morphology in Serum-free Medium. Journal of Visualized Experiments. (133), e57122 (2018).
- Nikodemova, M., Watters, J. J. Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. Journal of Neuroinflammation. 9, 147 (2012).
- Swartzlander, D. B., et al. Concurrent cell type-specific isolation and profiling of mouse brains in inflammation and Alzheimer’s disease. Journal of Clinical Investigation Insight. 3 (13), 121109 (2018).
- Hennig, B. P., et al. Large-Scale Low-Cost NGS Library Preparation Using a Robust Tn5 Purification and Tagmentation Protocol. G3. 8 (1), 79-89 (2018).

