Humanized NOG Mice for Intravaginal HIV Exposure and Treatment of HIV Infection
Summary
We have developed a protocol for the generation and evaluation of a humanized and human immunodeficiency virus-infected NOG mouse model based on stem cell transplant, intravaginal human immunodeficiency virus exposure, and droplet digital PCR RNA quantification.
Abstract
Humanized mice provide a sophisticated platform to study human immunodeficiency virus (HIV) virology and to test antiviral drugs. This protocol describes the establishment of a human immune system in adult NOG mice. Here, we explain all the practical steps from isolation of umbilical cord blood derived human CD34+ cells and their subsequent intravenous transplantation into the mice, to the manipulation of the model through HIV infection, combination antiretroviral therapy (cART), and blood sampling. Approximately 75,000 hCD34+ cells are injected intravenously into the mice and the level of human chimerism, also known as humanization, in the peripheral blood is estimated longitudinally for months by flow cytometry. A total of 75,000 hCD34+ cells yields 20%–50% human CD45+ cells in the peripheral blood. The mice are susceptible to intravaginal infection with HIV and blood can be sampled once weekly for analysis, and twice monthly for extended periods. This protocol describes an assay for quantification of plasma viral load using droplet digital PCR (ddPCR). We show how the mice can be effectively treated with a standard-of-care cART regimen in the diet. The delivery of cART in the form of regular mouse chow is a significant refinement of the experimental model. This model can be used for preclinical analysis of both systemic and topical pre-exposure prophylaxis compounds as well as for testing of novel treatments and HIV cure strategies.
Introduction
Human immunodeficiency virus (HIV) is a chronic infection with more than 37 million infected individuals worldwide1. Combination antiviral therapy (cART) is a life-saving therapy, but a cure is still warranted. Thus, there is a need for animal models that mirror the human immune system and its responses in order to facilitate continued research in HIV. Multiple types of humanized mice that are capable of supporting cell and tissue engraftment have been developed by transplanting human cells into severely immunodeficient mice2. Such humanized mice are susceptible to HIV infection and provide an important alternative to nonhuman primate simian immunodeficiency virus models, as they are cheaper and simpler to use than nonhuman primates. Humanized mice have facilitated research in HIV viral transmission, pathogenesis, prevention, and treatment3,4,5,6,7,8,9,10,11.
We present a flexible humanized model system for HIV research developed by transplanting cord blood derived human stem cells into mice of the NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) background. Besides being of non-fetal origin, the practical bioengineering of these mice is less technically demanding compared to the microsurgical procedures involved in the transplant of the blood-liver-thymus (BLT) construct.
We show how to establish HIV infection through intravaginal transmission and how to monitor the plasma viral load with a sensitive droplet digital PCR (ddPCR)-based setup. Subsequently, we describe the establishment of standard cART given as part of the daily mouse diet. The aim of these combined methods is to reduce stress to the animals and facilitate large-scale experiments where time spent handling each animal is limited12.
In humans, a CCR5Δ32/wt or CCR5Δ32/ Δ32 genotype causes reduced susceptibility to HIV infection with transmitter/founder viruses13, and some precautions must be taken when bioengineering humanized mice with stem cells for the purpose of HIV studies. This is especially true in our region because naturally occurring variants in the CCR5 gene, particularly Δ32 deletions, are more prevalent in Scandinavian and Baltic native populations compared to rest of the world14,15. Thus, our protocol includes an easy, high-throughput assay for screening donor hematopoietic stem cells for CCR5 variants prior to transplantation.
For the intravaginal exposure we chose the transmitter/founder R5 virus RHPA4259, isolated from a woman in an early stage of infection who was infected intravaginally16. We exposed the mice to a viral dose that was sufficient to yield successful transmission in the majority of mice, but below a 100% transmission rate. Choosing such a dose enables a sufficient dynamic range in transmission rate such that antiviral effects of a drug candidate can result in protected animals in HIV prevention experiments and decreased viral load for treatment studies.
Protocol
All cord blood samples were obtained in strict accordance with locally approved protocols, including informed consent of anonymous donation by the parents. All animal experiments were approved and performed in strict accordance with Danish national regulations under the license 2017-15-0201-01312.
CAUTION: Handle HIV exposed mice and blood with extreme caution. Decontaminate all surfaces and liquids that have been in contact with HIV with a confirmed HIV disinfectant (Table of Materials).
1. Isolation of human CD34+ stem cells
- Collect cord blood samples in EDTA-coated blood collection tubes after planned Caesarean sections or vaginal births and according to local ethical approvals.
- Isolate PMBCs from cord blood by density-gradient separation according to the manufacturer's protocol.
- Isolate CD34+ cells from the PBMC population by first pre-enriching with antibodies against common markers for mature cells, which induces crosslinking of cells of undesired lineages with red blood cells. This is followed by CD34+ cell enrichment using magnetic beads, according to the manufacturer's protocol.
- Determine live cell count by standard trypan blue exclusion by resuspending 10 µL of cell suspension in 90 µL of trypan blue. Add 10 µL of this solution to a hemacytometer and count non-blue cells, according to the manufacturer's protocol.
- Viably cryopreserve CD34+ cells in 1 mL of 10% DMSO in fetal bovine serum (FBS) until the day of mouse transplantation.
- Viably cryopreserve a small fraction of both isolated (CD34+) and flow-through cells (CD34-) separately for assessing CD34+ stem cell purity (~30,000 cells of each sample). Alternatively, test the purity on freshly enriched cells (see step 2 below).
- Freeze a fraction of non-pelleted flow-through (CD34-) for determination of CCR5Δ32 status. Cells can be frozen directly without conditioned freezing solution, but the presence of red blood cells in the pellet can inhibit the subsequent PCR if the flow-through is pelleted.
2. Assessing CD34+ stem cell purity via flow cytometry
- Thaw the isolated cells (CD34+) and flow-through cells (CD34-). Wash the cells by resuspending the cells from each vial in 9 mL of room temperature (RT) FACS buffer, consisting of 2% fetal bovine serum (FBS) in phosphate-buffered saline (PBS).
- Centrifuge for 5 min at 300 x g at RT to pellet cells.
- Pour off the supernatant, resuspend the cells in the remaining liquid and transfer to FACS tubes. Repeat the washing step with 3 mL of FACS buffer. After the second centrifugation, pour off the supernatant and resuspend the cells in the remaining liquid.
- Add 5 µL of Fc Receptor blocking solution (Table of Materials) and leave for 10 min at RT. Do not wash off the Fc Receptor blocking solution.
- Add the mix containing predetermined volumes of antibodies against human CD3 (clone SK7) BUV395, CD34 (clone AC136), FITC, and CD45 (clone 2D1) APC (Table 1). Leave the cells for 30 min at RT in the dark. The fluorophores must be chosen based on parameters that can be assessed with available flow cytometers without requiring a compensation matrix.
- Wash the cells with 3 mL of FACS buffer.
- Centrifuge for 5 min at 300 x g at RT to pellet the cells.
- Pour off the supernatant and resuspend the cells in the remaining liquid.
- Repeat this washing step 2x to ensure all unbound antibodies have been removed.
- Record the samples on the flow cytometer (Table of Materials) and perform data analysis with appropriate software. The gating strategy is presented in Figure 1A–F.
3. Genetic screening for CCR5Δ32 variants in cord blood
- Incubate 1.25 µL of non-pelleted flow-through with 11.25 µL of PCR mix containing 200 µM of dNTP mix, 0.01 U/µL high fidelity DNA polymerase, and the forward and reverse primers detailed in Table 2.
- Adjust the volume with nuclease-free water to approximately 12.5 µL for each PCR reaction.
- Amplify the genomic fragments with the PCR cycling program detailed in Table 3.
- Separate the PCR products on a 2% agarose gel13.
- PCR products from the wild type alleles and the Δ32 alleles yield PCR fragments of 196 base pairs and 164 base pairs bands respectively, making them easily distinguishable by gel electrophoresis13 (Figure 1G).
4. Intravenous stem cell transplant
NOTE: Having one person prepare the cells in the laboratory and another person prepare the mice and workspace for transplants is an efficient approach.
- In an animal facility, 4–6 h before the planned transplantation of the stem cells, irradiate 6–7 week-old female NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) mice (Table of Materials) with 0.75 Gy with a Cs137 source. The best preconditioning dose may vary based on mouse age, source of radiation, and other factors. This process conditions the animals for successful engraftment with human stem cells.
- In an animal facility, prepare the flow bench workspace and all reagents before bringing the mice or cells into the workspace.
- Place a sterile blue pad to cover the working surface of the flow bench. Prepare sterile gauze and a sharps container.
- Place a heating lamp disinfected with 70% ethanol in the flow bench with an empty sterile mouse cage underneath the heat.
- In the laboratory, thaw isolated CD34+ cells and dilute them in 9 mL of 37 °C plain RPMI.
- Centrifuge the cells at 350 x g for 5 min at RT, discard the supernatant by aspiration, and resuspend the pellet in 1 mL of plain RPMI at 37 °C.
- Determine the cell count by trypan blue exclusion, and adjust the volume to 200 µL per mouse. Make extra to take into account possible loss due to the subsequent handling steps.
- Prepare to transplant 75,000 CD34+ cells per 200 µL into each mouse.
- The cells can be kept at 4 °C during transport to the animal facility before the transplant. Avoid keeping the cells on ice to reduce aggregation or clumping.
- In the animal facility, bring the cage with the mice into the flow bench and transfer the mice to the cage under the heating lamp to dilate the blood vessels. Leave one end of the cage away from the heat source so that the mice can move away from the heat upon becoming warm. Mice that have moved to the end of the cage, away from the heat source, are sufficiently warm for a successful tail vein injection.
- Load a 1 mL lubricated syringe to above the 800 µL mark with suspended CD34+ cells. Using a lubricated 1 mL syringe will dramatically ease the intravenous injection and increase the precision of this technique.
- Attach a 30 G 13 mm needle and prepare the needle and syringe for injection. This order of operation allows for the syringe to be loaded quicker while protecting the integrity of the cells to be transplanted given the possible damage that can occur during the rapid aspiration of cells through such a small gauge needle. Fill the needle hub with liquid by pressing the plunger and remove liquid down to the 200 µL mark of the needle.
- Place a heated mouse (step 4.6) in a restrainer used for giving IV injections. Carefully inject 200 µL of the cell suspension into the tail vein of the mouse. Spend 2 s performing the plunge and keep the needle inserted for approximately 2 s after completion of the injection. This ensures that the cells have migrated adequately far from the injection site prior to removal of the needle.
- As necessary, wipe the mouse tail with sterile gauze to remove any visible blood. Put the mouse back into its non-heated home cage. Repeat the injection procedure with the remaining mice.
5. Blood collection and processing for analysis
NOTE: Human cell engraftment in the peripheral blood can be evaluated via flow cytometry 3–5 months after human stem cell transplantation.
- Draw blood samples from the mice using local IACUC-approved techniques.
- Collect a maximum of 70–100 µL of total blood into sterile PCR microcentrifuge tubes containing 10 µL of 0.5 M EDTA (pH = 8.0) to avoid coagulation of blood.
6. Evaluation of human engraftment via flow cytometry
- Transfer 40–50 µL of blood to FACS tubes.
- Add 5 µL of Fc Receptor blocking solution to prevent nonspecific binding of antibodies and leave for 10 min at RT.
- Add the mouse anti-human antibody mix containing CD4 (clone SK3) BUV 496, CD8 (clone RPA-T8) BV421, CD3 (clone OKT3) FITC, CD19 (clone sj25c1) PE-Cy7, CD45 (clone 2D1) APC (Table 4) and leave to stain in the dark at RT for 30 min. Fluorophores must be chosen based on parameters that can be assessed with the available flow cytometers without requiring a compensation matrix.
- Add 2 mL of an appropriate red blood cell lysing buffer to each tube to lyse the red blood cells. Use a lysis buffer specifically formulated for antibody staining prior to red blood cell lysis (a suitable example is given in the Table of Materials). Vortex briefly to ensure equal distribution of the cells in the lysing solution and leave for 10 min at RT. Equal distribution of the cells is important.
- Add 2 mL of FACS buffer to stop the lysis reaction.
- Centrifuge for 5 min at 300 x g at RT to pellet the cells.
- Pour off the supernatant and vortex gently until the cells are resuspended.
- Add 3 mL of FACS buffer and centrifuge for 5 min at 300 x g at RT.
- Pour off the supernatant and resuspend the cells.
- Record the samples on an appropriate flow cytometer and analyze using appropriate software (Table of Materials). Representative analysis and results are depicted in Figure 2 and Figure 3.
7. Intravaginal HIV exposure
NOTE: The virus used for intravaginal exposure of the mice can be produced using previously published protocols17. The virus is kept at -80 °C and transported between locations while stored on dry ice following locally approved protocols. The virus is stored on dry ice until immediately before the exposure of the mice. The virus can be diluted into plain RPMI (avoid RPMI that has antibiotics or serum additives) to achieve the appropriate concentration immediately prior to exposure (21,400 IUs were used for this IVAG exposure). Once generated, keep the diluted stock on wet ice throughout the procedure to avoiding freeze-thaw cycles that would occur if the diluted virus was placed back on dry ice once thawed.
- Prepare all the equipment and flow bench workspace as presented in Figure 4 before bringing the mice or virus into the flow bench (similar to step 4.2).
- Place the heating lamp focus in the center of the workspace where the mouse will be located during the HIV exposure procedure. The heating lamp will ensure no decrease in body temperature of the mice. Other equipment that controls temperature can also be used, (e.g. a heated gel pad or a circulating warm water blanket, according to local IACUC regulations18).
- Bring sterile 20 µL pipette tips and an appropriate pipette into the bench. Place a container with liquid disinfectant (Table of Materials) in the bench for immediate inactivation of materials and liquids that have been in contact with the virus.
- Place a mouse into a chamber with 3% isoflurane gas and paper towels. This percentage of gas will anesthetize the animals within 2–4 min. As with all other materials used with immunodeficient mice, the anesthesia apparatus must be properly disinfected prior to use.
- Once anesthetized, transfer the mouse to a sterile blue pad under the heating lamp. Insert the mouse snout into a mask supplying continuous 3% isoflurane gas to maintain anesthesia. Hold the mouse at the base of the tail, stomach facing up, with your hand supporting the mouse back as depicted in Figure 4.
- With a sterile pipette tip, stimulate the genital area by gently stroking upwards towards the anus to induce emptying of the rectum, relieving pressure on the vagina.
- Carefully bare the vaginal opening by wrapping the mouse tail across your fingers such that the vulva naturally opens, perhaps with slight nudging using a sterile pipette tip.
- Change the pipette tip and pipette 20 µL of virus atraumatically into the mouse vagina without creating bubbles. Do not insert the tip deep into the vagina. Rather, with the vulva opened, place the pipette tip at the level of the vaginal opening, avoid going deeper to eliminate the potential for abrasions during the inoculation process, release the virus, and allow gravity to pull the virus into the vagina. Alternatively, use a 22 G 1.25 mm blunt-end, straight needle, as described in Veselinovic et al.6.
- Retain the mouse in this position with the vagina facing up for 5 min after exposure to avoid gravity-induced leakage of the virus suspension.
- Carefully place the mouse into the home cage, taking care to place the mouse on its back.
8. Processing of blood samples prior to viral load analysis
- Collect blood as described in section 5 above.
- Centrifuge the blood samples for 5 min at 500 x g at RT to separate the plasma and cells.
- Collect 40 µL of plasma for viral load measurement into a new sterile PCR-approved microcentrifuge tube and store at -80 °C for at least 1 h until further processing. It is important to freeze all samples before RNA extraction to avoid the risk of bias from comparing RNA levels in samples that have not been frozen prior to RNA isolation to samples frozen prior to RNA isolation.
- Adjust the volume of blood back to the original volume by adding 40 µL of suspension media (PBS with 2.5% bovine serum albumin, 50 U/mL penicillin G and streptomycin, and 10 U/mL DNase, sterile-filtered at 0.22 µm).
- Transfer 15 µL of the adjusted blood volume to a new PCR-approved microcentrifuge tube.
- Add 1 mL of 1x RBC lysis solution (Table of Materials), vortex, and incubate for 10 min at RT.
- Centrifuge for 1 min at 9,600 x g at RT to pellet cells.
- Aspirate supernatant and leave only the tiny white cell pellet, because red blood cell contamination can inhibit PCR.
- Store pellet at -80 °C for at least 1 h until further processing.
NOTE: Optionally, any remaining blood from step 8.4 can be used for flow cytometry analysis, as described above in step 6.
9. DNA extraction using a proteinase K extraction method
- Extract DNA from peripheral cell pellets (generated in step 8.8) using a proteinase K extraction method as described below. This method has been demonstrated to extract the highest DNA yield from a small volume of blood such as that required for the serial blood collections utilized in this study19.
- Add 25 μL of proteinase K (20 μg/mL) to 1 mL of 0.1M Tris buffer.
- Vortex the proteinase K solution briefly.
- Add 50 μL of the proteinase K solution to each cell pellet to be digested.
- Mix by pipetting up and down and confirm the resuspension of the cell pellet.
- Shake on a thermoshaker (Table of Materials) at 400 rpm (depending on the instrument) at 56 °C for 1 h. Tape tubes down to hold them in place, if necessary.
- Immediately and in the same thermoshaker, inactivate proteinase K with a temperature shift to 95 °C while the shaking continues for an additional 20 min.
- Vortex each sample.
- Place each sample at -80 °C for a minimum of 30 min.
- Thaw, then centrifuge the samples at 17,000 x g for 1 min at RT to pellet unwanted cellular fragments.
- Place the DNA-containing supernatant into a new microcentrifuge tube.
- The DNA template is ready for PCR. The DNA templates can be stored at -80 °C.
10. RNA extraction, cDNA synthesis, and ddPCR quantification of viral RNA
- Isolate RNA from thawed mouse plasma with a virus RNA isolation kit following the manufacturer's protocol (Table of Materials).
- After addition of the sample to the column, add an on-column DNase treatment step to ensure removal of all DNA in the plasma sample.
- For each sample, add 95 µL of RNase-free DNase solution (mix 2 µL RNAse-free DNase and 98 µL reaction buffer) to the column and incubate for 15 min at RT.
- Store the RNA samples at -80 °C for at least 1 h before further processing. It is important to freeze all samples after RNA extraction do avoid the risk of bias when comparing samples that have not been frozen to samples that were frozen.
- Synthesize cDNA using a reverse transcriptase step using reagents as described previously20. Make sure to add 0.5 µL of an RNase inhibitor to the cDNA reaction to avoid degradation of the RNA.
- Perform cDNA synthesis with the program detailed in Table 5.
- Store the cDNA samples at -80 °C for at least 1 h. It is important to freeze all samples after cDNA synthesis do avoid the risk of bias when comparing samples that have not been frozen to samples that were frozen.
- Prepare samples for ddPCR as follows20:
- Mix 3 µL of the cDNA sample with 11 µL of the ddPCR probe mixture (no dUTP)20, 250 nM minor groove-binding probe, and 900 nM of each of the forward and reverse primers as detailed in Table 6.
- Adjust the total PCR volume to 22 µL with nuclease-free water.
- Emulsify the PCR mixes with Droplet Generation Oil for Probes on a droplet generator according to the manufacturer's protocol and previous descriptions20.
- Run the PCR program as detailed in Table 7.
NOTE: The primer/probe sequences and PCR programs displayed here have been specifically designed and optimized for sensitive detection of the HIV strain RHPA4259. Primer and probe sequences can easily be adjusted to detect any other HIV strain of choice. - Detect droplet fluorescence from the samples on a droplet reader, and analyze the results with appropriate software according to the manufacturer's protocol.
11. Treatment with cART-containing chow
- Feed mice with pellets containing a standard cART regimen containing 4,800 mg/kg raltegravir (RAL), 720 mg/kg tenofovir disoproxil fumarate (TDF), and 520 mg/kg emtricitabine (FTC)21 (Table 8). These doses were determined assuming that a mouse weighs 25 g and eats 4 g of chow per day. This corresponds to a daily dose of 768 mg/kg RAL, 2.88 mg/kg TDF, and 83 mg/kg FTC21.
- Use a cART diet that has been prepared by an external vendor (See Table of Materials) of prescription drugs. Other companies could potentially also produce this regimen. Use a cART diet colored red to easily distinguish it from ordinary mouse chow.
- Produce a control chow diet without cART in a standard brown color for easy distinction.
- For initiation of cART, prepare sterile mouse cages with the addition of cART-containing chow diet, and then transfer the mice from the old cage to the new cage.
- Monitor the weights of the mice and consumption of cART-containing chow by visual inspection to ensure that the mice are adjusting to the change.
Representative Results
The gating strategy for the analysis of stem cell purity is depicted in Figure 1. Figure 1A–C shows the purified CD34+ population and Figure 1D–F the CD34- flow-through used to illustrate that the minimal amount of the CD34+ population is lost in the isolation process. The purity of isolated CD34+ stem cells was between 85%–95% with less than 1% T-cell contamination. Figure 1G depicts CCR5 bands from one adult human control donor with the CCR5Δ32/wt genotype, followed by bands from two CCR5wt/wt and one CCR5Δ32/wt stem cell donors. The frequency of the genotype CCR5Δ32/wt in a group of 19 donors was 15.8% (Figure 1H). This is in agreement with larger epidemiological studies14,15 reporting the genotype in up to 23.6% of investigated persons in Denmark.
Human CD45+ levels in mice peripheral blood was assessed via flow cytometry 3–5 months after transplantation of human CD34+ stem cells. The gating strategy is presented in Figure 2A–E. Figure 3A and Figure 3B illustrate the variability between 10 and 16 individual mice receiving stem cells from two different donors. Transplantation of 75,000 hCD34+ cells yielded 20%–50% human CD45+ in the peripheral blood. All mice developed human B and T cells, including both CD4- and CD8+ T cells.
For atraumatic intravaginal exposures, the setup depicted in Figure 4 was used. Mice were anaesthetized in a closed chamber and kept under anesthesia during the exposure. Mice were held with the vagina facing up for 5 min after exposure to ensure virus solution engagement with mucosal surfaces.
Figure 5A shows the 64% HIV transmission success rate observed using this model. Mice were challenged with 21,400 infectious units (IU) of RHPA4259 intravaginally. This dose resulted in 64% of mice becoming HIV infected following vaginal exposure. For comparison, data from two different cohorts of mice exposed through an intravenous route are included. As expected, 100% of the mice became HIV+ with similar doses of RHPA and an additional strain (YU2) using this route.
Figure 5B depicts representative results from three mice that were infected with HIV and switched to a diet containing standard cART. Mice were switched back to regular mouse chow after 40 days of cART. In this assay setup, the limit for viral load detection was 725 copies/mL. Viral loads were all below the detection limit after 4 weeks of cART. After cessation of cART, the virus rebounded, mirroring clinical data22. Mice on cART tolerated the change in diet well as indicated in Figure 5C.
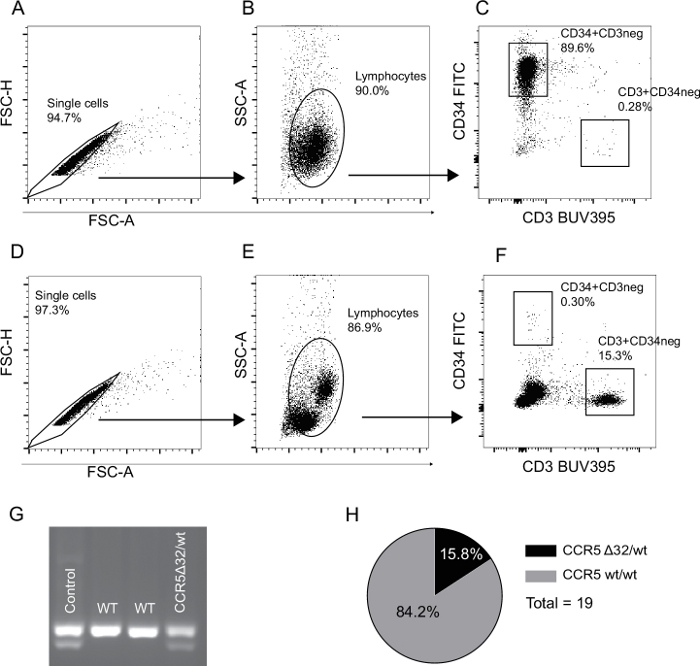
Figure 1: Representative flow cytometry gating strategy for validation of stem cell purity and CCR5 donor variant status. (A–C) The gating strategy used for the isolated CD34+ cell population. Doublets and debris are excluded in panel A and B respectively (FSC-A vs. FSC-H and FSC-A vs. SSC-A). (C) The frequency of CD34+ stem cells and CD3+ T cell contamination. (D–F) The CD34- flow-through gating strategy. Percentages in gates are calculated as a fraction of the parent population. (G) The results of a CCR5Δ32/wt PCR analysis. Lane 1: DNA from a human CCR5Δ32/wt donor, lanes 2 and 3: two CCR5wt/wt human stem cell donors, lane 4: A CCR5Δ32/wt human stem cell donor. (H) Frequency of the genotype CCR5Δ32/wt in our group of 19 stem cell samples is 15.8%. Please click here to view a larger version of this figure.
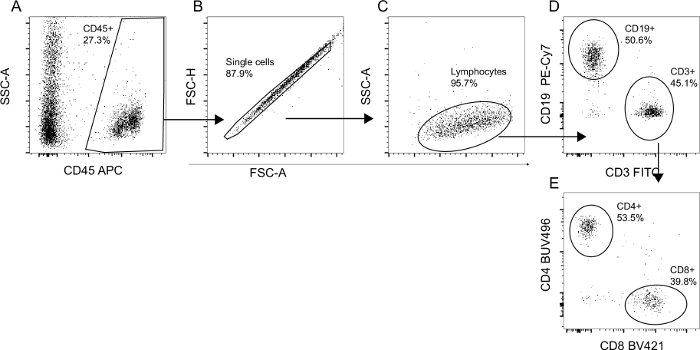
Figure 2: Flow cytometry gating strategy for validation of human cell engraftment and differentiation. The total mononuclear cell population from humanized mice was analyzed via flow cytometry. (A) The percentage of human CD45+ cells was determined as a fraction of the total recorded events. (B) Doublets were subsequently excluded based on FSC-A/FSC-H gating. (C) The true lymphocyte population was defined based on size and granularity. (D) Lymphocytes were then characterized as either CD3+ (T cells) or CD19+ (B cells). (E) CD3+ T cells were either CD4+ T cells or CD8+ T cells. Percentages in gates were calculated as a fraction of the parent population. Please click here to view a larger version of this figure.
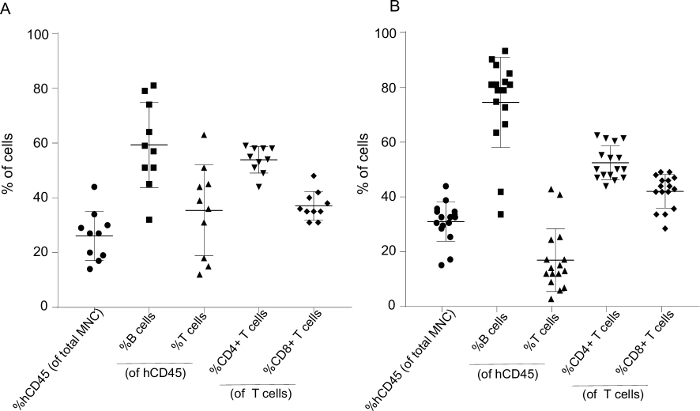
Figure 3: Representative humanization levels 4–5 months after stem cell transplantation with cell subtype fractions for 10 and 16 mice generated from two different human donors. (A) The mononuclear cell population (MNC) from 10 and (B) 16 humanized mice were analyzed via flow cytometry and gated as presented in Figure 2. The fraction of human CD45+ cells is presented as %hCD45 (of total MNC), and %B and %T cells as a fraction of hCD45. T cells were subsequently divided into %CD4 and %CD8. Each data point represents one mouse. Data is presented as mean ± SD. Please click here to view a larger version of this figure.
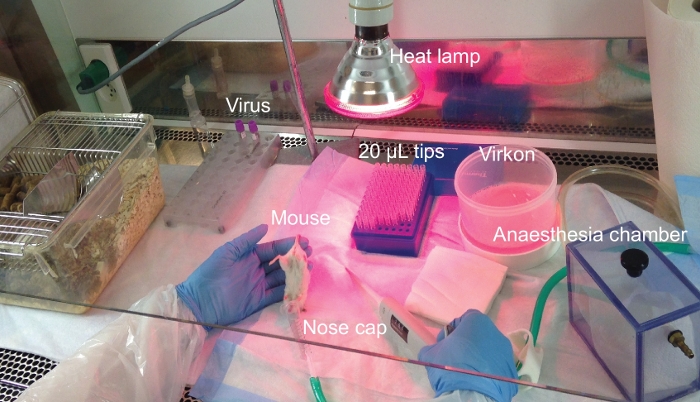
Figure 4: Experimental lab bench setup for intravaginal exposure of mice. Experimental setup for HIV exposure of humanized mice through the intravaginal route. The procedure is performed in a flow bench where all reagents and surfaces have been sterilized prior to use. Please click here to view a larger version of this figure.
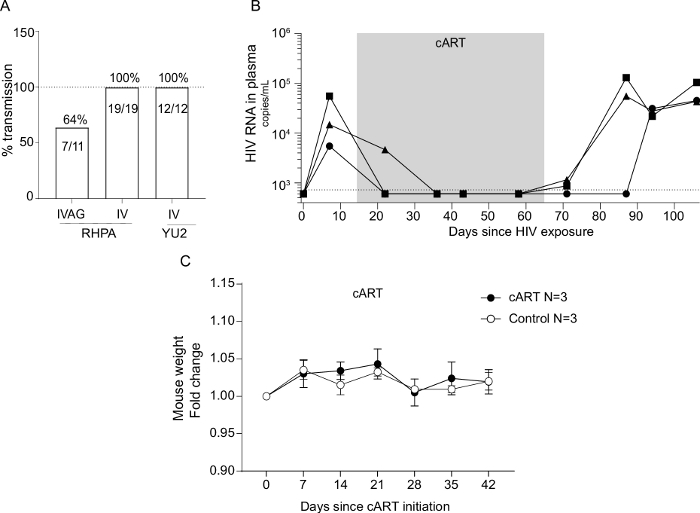
Figure 5: Rate of HIV strain transmission through different exposure routes and efficacy and safety of cART-containing chow in viral suppression. (A) Humanized NOG mice were successfully infected with two different strains of HIV through either the intravaginal or the intravenous route. Mice were exposed with 21,400 IUs of RHPA4259 intravaginally, 5,157 IUs IV with RHPA4259, or 3,000 IUs IV with YU2. Details regarding IV exposure of humanized mice are not included in this protocol. HIV infections were successfully treated with a cART regimen delivered through mouse chow. (B) The viral load decreased to below detection for all three mice on cART and rebound emerged after the cessation of cART. The dotted line indicates limit of quantification at 725 copies/mL. Mice fed with cART chow had similar weight development as mice housed on non-cART chow during the same time period, indicating no taste-preference or side effects of the cART diet. (C) Weights are presented as fold change compared to the start of cART. Each data point represents the mean of three animals ± SD. Please click here to view a larger version of this figure.
| Antibody target | Clone | Fluorophore |
| CD3 | clone SK7 | BUV395 |
| CD34 | clone AC136 | FITC |
| CD45 | clone 2D1 | APC |
Table 1: Antibodies used for determination of stem cell purity. Suggested multicolor flow cytometry panel for evaluation of stem cell purity. Listed are the antibody target, the clone, and the fluorophore.
| CCR5Δ32 detection | Primers |
| Forward primer | 5'CTTCATTACACCTGCAGCT'3 |
| Reverse primer | 5'TGAAGATAAGCCTCACAGCC'3 |
Table 2: CCR5Δ32 variant detection PCR primers. Forward and reverse primers used for detection of the 32 bp deletion in the CCR5 gene.
| No. of Cycles | 1 | 45 | 1 | ∞ |
| Temperature (°C) | 98 | 98/63/72 | 72 | 10 |
| Time | 30 s | 10 s/30 s/15 s | 5 min | ∞ |
Table 3: CCR5Δ32 variant detection PCR program. PCR cycling program used for amplification of the CCR5 gene.
| Antibody target | Clone | Fluorophore |
| CD4 | SK3 | BUV 496 |
| CD8 | RPA-T8 | BV421 |
| CD3 | OKT3 | FITC |
| CD19 | sj25c1 | PE-Cy7 |
| CD45 | 2D1 | APC |
Table 4: Antibodies used for determination of mouse humanization. Suggested multicolor flow cytometry panel for humanization. Listed are the antibody target, the clone, and the fluorophore.
| No. of Cycles | 1 | 1 | ∞ |
| Temperature (°C) | 51 | 80 | 4 |
| Time | 45 min | 15 min | ∞ |
Table 5: cDNA amplification program. Program used for amplification of complementary strand DNA to the viral RNA.
| HIV quantification | Primers |
| Forward primer | 5'AGGGCAGCATAGAGCAAAAA'3 |
| Reverse primer | 5'CAAAGGAATGGGGGTTCTTT'3 |
| FAM probe | 5'ATCCCCACTTCAACAGATGC'3 |
Table 6: HIV ddPCR primers. Primers and probes used for ddPCR amplification of viral cDNA.
| No. of Cycles | 1 | 39 | 1 | ∞ |
| Temperature (°C) | 95 | 95/54.5 | 98 | 4 |
| Time | 10 min | 30 s/1 min | 10 min | ∞ |
Table 7: HIV ddPCR program. PCR cycling program used for amplification of viral RNA.
| Raltegravir (RAL) | 4800 mg/kg |
| Tenofovir disoproxil fumarate (TDF) | 720 mg/kg |
| Emtricitabine (FTC) | 520 mg/kg |
Table 8: Mouse cART chow diet. Mouse chow diet was formulated as previously published21. The chow diet was made on a base of standard mouse chow, and after production, the food was γ-irradiated with 25 kGy and double-bagged. The chow was stored at -20 °C until use.
Discussion
The severely immunocompromised mouse strain NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) is extremely well suited for transplantation of human cells and tissues. Both innate and adaptive immune pathways in these mice are compromised. NOG and NSG mice harbor a Prkdcscid mutation that results in defective T and B cell function. Furthermore, these mice lack a functional interleukin-2 receptor γ-chain (common gamma chain, IL2rg) which is indispensable in the binding complexes of many key cytokines such as IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21. Immunodeficient mice, such as the NOG, transplanted with a human immune system are a powerful tool for the study of HIV transmission and immunology. Contributions in these fields made using humanized mice have been extensively reviewed2,23,24,25,26. The use of these mice to study human innate immune responses are also gaining increased attention27,28.
The aim of this manuscript is to supply a comprehensive protocol of mouse and ddPCR procedures to go from a naive mouse to HIV transmission and treatment data. Our system utilized ddPCR for quantification of viral RNA and DNA. In a ddPCR reaction, the reactants are partitioned into up to 20,000 droplets, each containing a single, separate micro PCR reaction. The amplification of a target inside a droplet leads to a positive fluorescent signal for that droplet. Thus, the readout is binary and by applying Poisson statistical analyses, the number of positive reactions can be directly translated to a number of template copies in the original sample. The benefit of ddPCR lies in its ability to directly quantify a target independent of a standard curve. This is particularly attractive when analyzing RNA samples that are challenging to utilize as PCR standard curves due to their labile nature29. Moreover, by analyzing multiple replicas of the same sample and merging the individual data points for the final sample quantification, the binary nature of ddPCR makes it possible to lower the detection limit of template copies per mL of sample29. This is especially important in a humanized mouse setting, where only limited sample material is available and high sensitivity is required.
Administration of cART to humanized mice can be done either by oral gavage or intraperitoneal injections with solutions of cART30,31,32, and as shown recently, by formulation into the diet21. One of our major aims was the implementation of a cART regimen in the mouse diet to reduce potential stress on the animals due to the extra handling steps inherent in other drug delivery methods. The dose of medicine that a mouse will eat can be accurately estimated based on the average daily food intake of the mice33. Oral delivery through the diet serves as the easiest delivery route with both minimal stress for the animal and minimal workload for the handler. We based our combination of antiviral drugs on previous published studies in humanized mice21,30. Furthermore, our cART strategy is clinically relevant given that the drug combination utilized clinically is orally administered by patients around the globe.
Certain limitations are noted regarding the use of NOG mice. Importantly, human T cells in these mice are cultivated in a mouse thymic environment, as opposed to a human environment. The recent focus is on generating xenorecipient strains that have a favorable environment for the development of robust human immune responses. These new strains include immunodeficient mice that are transgenic for human MHC molecules, such as A2. These models enable HLA-restricted antigen T-cell responses that result in better maturation and effector functions of the adaptive immune system34. Another approach is to replace mouse genes with key human cytokines for IL-3/GM-CSF35, IL-636, IL-1537, TPO, M-CSF38, and IL-7/TSLP31. Such models have gained increased attention for their ability to generate better differentiation of innate cell types. Our protocol is easily adaptable for the humanization and HIV infection of mice using any such enhanced genetic background immunodeficient strain.
In summary, the ease and utility of the described approach facilitates research in HIV-related fields in vivo. Humanized mice can be a very powerful tool in guiding research towards generating better research hypotheses. Along with the generation of more "human" humanized mice with human transgenes, we believe our standardized protocol will contribute to the streamlining of experimental procedures across different research environments.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank the Biomedicine Animal Facility staff at Aarhus University, particularly Ms. Jani Kær for colony maintenance efforts and for tracking mouse weights. The authors would like to thank Professor Florian Klein for developing standard-of-care cART and for guidance. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pRHPA.c/2635 (cat# 11744) from Dr. John Kappes and Dr. Christina Ochsenbauer.
Materials
| Blue pad | VWR | 56616-031 | Should be sterilized prior to use |
| Bovine serum albumin (BSA) | Sigma | A8022 | |
| CD19 (clone sj25c1) PE-Cy7 | BD Bioscience | 557835 | |
| CD3 (clone OKT3) FITC | Biolegend | 317306 | |
| CD3 (clone SK7) BUV395 | BD Bioscience | 564001 | |
| CD34 (clone AC136) FITC | Miltenyi | 130-113-740 | |
| CD4 (clone SK3) BUV 496 | BD Bioscience | 564652/51 | |
| CD45 (clone 2D1) APC | Biolegend | 368511/12 | |
| CD8 (clone RPA-T8) BV421 | BD Bioscience | 562428 | |
| ddPCR Supermix for probes (no dUTP) | Bio-Rad | 1863025 | |
| DMSO | Merck | 10,02,95,21,000 | |
| DNAse | Sigma | D4263 | For suspension buffer |
| dNTP mix | Life Technologies | R0192 | |
| Dulbeccos phosphate-buffered saline (PBS) | Biowest | L0615-500 | |
| EasySep Human Cord Blood CD34 Positive Selection Kit II | Stemcell | 17896 | |
| EDTA | Invitrogen | 15575-038 | |
| FACS Lysing solution 10X | BD | 349202 | Dilute 1:10 in dH20 immediately before use |
| FACS tubes (Falcon 5 mL round-botton) | Falcon | 352052 | |
| Fc Receptor blocking solution (Human Trustain FcX) | Biolegend | 422302 | |
| Fetal bovine serum | Sigma | F8192-500 | |
| Ficoll-Paque PLUS | GE Healthcare | 17144002 | |
| Flowjo v.10 | |||
| Gauze | Mesoft | 157300 | Should be sterilized prior to use |
| Heating lamp | Custom made | ||
| Hemacytometer (Bürker-Türk) | VWR | DOWC1597418 | |
| Isoflurane gas | Orion Pharma | 9658 | |
| LSR Fortessa X20 flow cytometer | BD | ||
| Microcentrifuge tubes, PCR-PT approved | Sarstedt | 72692405 | |
| Mouse cART food | ssniff Spezialdiäten GmbH | Custom made product | |
| Mouse restrainer | Custom made product | ||
| Needle, Microlance 3, 30G ½" | BD | 304000 | |
| NOG mice NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac | Taconic | NOG-F | |
| Nuclease-free water | VWR chemicals | 436912C | |
| Nucleospin 96 Virus DNA and RNA isolation kit | Macherey-Nagel | 740691 | |
| PCR-approved microcentrifuge tubes | Sarstedt | 72.692.405 | |
| Penicillin-Streptomycin solution 100X | Biowest | L0022-100 | |
| Phusion Hot Start II DNA polymerase | Life Technologies | F549S | |
| Pipette tips, sterile, ART 20P Barrier | ThermoScientific | 2149P | |
| Proteinase K | NEB | 100005398 | |
| QuantaSoft software | Bio-Rad | ||
| QX100 Droplet Generator | Bio-Rad | 1886-3008 | |
| QX100 Droplet Reader | Bio-Rad | 186-3003 | |
| RBC lysis solution | Biolegend | 420301 | |
| RNase-free DNAse size F + reaction buffer | Macherey-Nagel | 740963 | |
| RNAseOUT Recombinant Ribonuclease inhibitor | ThermoScientific | 10777-019 | |
| RPMI | Biowest | L0501-500 | Dissolve in H20 |
| Softject 1 mL syringe | Henke Sass Wolf | 5010-200V0 | |
| Superscript III Reverse Transcriptase | ThermoFisher Scientific | 18080044 | |
| Thermoshaker | VWR | 89370-910 | |
| Trypane blue | Sigma | T8154 | |
| Ultrapure 0.5 EDTA, pH 8.0 | ThermoFisher Scientific | 15575-020 | |
| Virkon S (virus disinfectant) | Dupont | 7511 |
References
- Skelton, J. K., Ortega-Prieto, A. M., Dorner, M. A Hitchhiker’s guide to humanized mice: new pathways to studying viral infections. Immunology. 154 (1), 50-61 (2018).
- Denton, P. W., Krisko, J. F., Powell, D. A., Mathias, M., Kwak, Y. T. Systemic Administration of Antiretrovirals Prior to Exposure Prevents Rectal and Intravenous HIV-1 Transmission in Humanized BLT Mice. PLoS ONE. 5 (1), 8829 (2010).
- Zou, W., et al. Nef functions in BLT mice to enhance HIV-1 replication and deplete CD4 + CD8 + thymocytes. Retrovirology. 9 (1), 44 (2012).
- Berges, B. K., Akkina, S. R., Folkvord, J. M., Connick, E., Akkina, R. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2 -/- γc -/- (RAG-hu) mice. Virology. 373 (2), 342-351 (2008).
- Veselinovic, M., Charlins, P., Akkina, R. Modeling HIV-1 Mucosal Transmission and Prevention in Humanized Mice. Methods Mol Biol. , 203-220 (2016).
- Neff, C. P., Kurisu, T., Ndolo, T., Fox, K., Akkina, R. A topical microbicide gel formulation of CCR5 antagonist maraviroc prevents HIV-1 vaginal transmission in humanized RAG-hu mice. PLoS ONE. 6 (6), 20209 (2011).
- Neff, P. C., Ndolo, T., Tandon, A., Habu, Y., Akkina, R. Oral pre-exposure prophylaxis by anti-retrovirals raltegravir and maraviroc protects against HIV-1 vaginal transmission in a humanized mouse model. PLoS ONE. 5 (12), 15257 (2010).
- Veselinovic, M., et al. HIV pre-exposure prophylaxis: Mucosal tissue drug distribution of RT inhibitor Tenofovir and entry inhibitor Maraviroc in a humanized mouse model. Virology. 464-465, 253-263 (2014).
- Akkina, R., et al. Humanized Rag1-/-γc-/- mice support multilineage hematopoiesis and are susceptible to HIV-1 infection via systemic and vaginal routes. PLoS ONE. 6 (6), 20169 (2011).
- Zhou, J., et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Molecular Therapy. 19 (12), 2228-2238 (2011).
- Balcombe, J. P., Barnard, N. D., Sandusky, C. Laboratory routines cause animal stress. Contemporary Topics in Laboratory Animal Science. 43 (6), 42-51 (2004).
- Trecarichi, E. M., et al. Partial protective effect of CCR5-Delta 32 heterozygosity in a cohort of heterosexual Italian HIV-1 exposed uninfected individuals. AIDS Research and Therapy. 3 (1), (2006).
- Novembre, J., Galvani, A. P., Slatkin, M. The geographic spread of the CCR5 Δ32 HIV-resistance allele. PLoS Biology. 3 (11), 1954-1962 (2005).
- Solloch, U. V., et al. Frequencies of gene variant CCR5-Δ32 in 87 countries based on next-generation sequencing of 1.3 million individuals sampled from 3 national DKMS donor centers. Human Immunology. 78 (11-12), 710-717 (2017).
- Ochsenbauer, C., et al. Generation of Transmitted/Founder HIV-1 Infectious Molecular Clones and Characterization of Their Replication Capacity in CD4 T Lymphocytes and Monocyte-Derived Macrophages. Journal of Virology. 86 (5), 2715-2728 (2012).
- Andersen, A. H. F., et al. Long-Acting, Potent Delivery of Combination Antiretroviral Therapy. ACS Macro Letters. 7 (5), 587-591 (2018).
- Caro, A. C., Hankenson, F. C., Marx, J. O. Comparison of thermoregulatory devices used during anesthesia of C57BL/6 mice and correlations between body temperature and physiologic parameters. Journal of the American Association for Laboratory Animal Science JAALAS. 52 (5), 577-583 (2013).
- Gatlin, J., Padgett, A., Melkus, M. W., Kelly, P. F., Garcia, J. V. Long-term engraftment of nonobese diabetic/severe combined immunodeficient mice with human CD34+ cells transduced by a self-inactivating human immunodeficiency virus type 1 vector. Human Gene Therapy. 12 (9), 1079-1089 (2001).
- Leth, S., et al. HIV-1 transcriptional activity during frequent longitudinal sampling in aviremic patients on antiretroviral therapy. AIDS. 30 (5), 713-721 (2016).
- Halper-Stromberg, A., et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 158 (5), 989-999 (2014).
- Rothenberger, M. K., et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proceedings of the National Academy of Sciences. 112 (10), 1126-1134 (2015).
- Rongvaux, A., et al. Human Hemato-Lymphoid System Mice: Current Use and Future Potential for Medicine. Annual Review of Immunology. 31 (1), 635-674 (2013).
- Walsh, N. C., et al. Humanized Mouse Models of Clinical Disease. Annual Review of Pathology: Mechanisms of Disease. 12 (1), 187-215 (2017).
- Denton, P. W., García, J. V. Humanized mouse models of HIV infection. AIDS Reviews. 13 (3), 135-148 (2011).
- Denton, P. W., Søgaard, O. S., Tolstrup, M. Using animal models to overcome temporal, spatial and combinatorial challenges in HIV persistence research. Journal of Translational Medicine. 14 (1), (2016).
- Andersen, A. H. F., et al. cAIMP administration in humanized mice induces a chimerization-level-dependent STING response. Immunology. 157 (2), 163-172 (2019).
- Tanaka, S., et al. Development of Mature and Functional Human Myeloid Subsets in Hematopoietic Stem Cell-Engrafted NOD/SCID/IL2r KO Mice. The Journal of Immunology. 188 (12), 6145-6155 (2012).
- Quan, P. L., Sauzade, M., Brouzes, E. DPCR: A technology review. Sensors (Switzerland). 18 (4), (2018).
- Denton, P. W., et al. Generation of HIV Latency in Humanized BLT Mice. Journal of Virology. 86 (1), 630-634 (2012).
- Li, Y., et al. A human immune system mouse model with robust lymph node development. Nature Methods. 15 (8), 623-630 (2018).
- Satheesan, S., et al. HIV Replication and Latency in a Humanized NSG Mouse Model during Suppressive Oral Combinational Antiretroviral Therapy. Journal of Virology. 92 (7), 02118 (2018).
- Bachmanov, A. A., Reed, D. R., Beauchamp, G. K., Tordoff, M. G. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behavior Genetics. 32 (6), 435-443 (2002).
- Shultz, L. D., et al. Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r null humanized mice. Proceedings of the National Academy of Sciences. 107 (29), 13022-13027 (2010).
- Willinger, T., et al. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proceedings of the National Academy of Sciences. 108 (6), 2390-2395 (2011).
- Hanazawa, A., et al. Generation of human immunosuppressive myeloid cell populations in human interleukin-6 transgenic NOG mice. Frontiers in Immunology. 9, (2018).
- Huntington, N. D., et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. The Journal of Experimental Medicine. 206 (1), 25-34 (2009).
- Rongvaux, A., et al. Development and function of human innate immune cells in a humanized mouse model. Nature Biotechnology. 32 (4), 364-372 (2014).

