OaAEP1-Mediated Enzymatic Synthesis and Immobilization of Polymerized Protein for Single-Molecule Force Spectroscopy
Summary
Here, we present a protocol to conjugate protein monomer by enzymes forming protein polymer with a controlled sequence and immobilize it on the surface for single-molecule force spectroscopy studies.
Abstract
Chemical and bio-conjugation techniques have been developed rapidly in recent years and allow the building of protein polymers. However, a controlled protein polymerization process is always a challenge. Here, we have developed an enzymatic methodology for constructing polymerized protein step by step in a rationally-controlled sequence. In this method, the C-terminus of a protein monomer is NGL for protein conjugation using OaAEP1 (Oldenlandia affinis asparaginyl endopeptidases) 1) while the N-terminus was a cleavable TEV (tobacco etch virus) cleavage site plus an L (ENLYFQ/GL) for temporary N-terminal protecting. Consequently, OaAEP1 was able to add only one protein monomer at a time, and then the TEV protease cleaved the N-terminus between Q and G to expose the NH2-Gly-Leu. Then the unit is ready for next OaAEP1 ligation. The engineered polyprotein is examined by unfolding individual protein domain using atomic force microscopy-based single-molecule force spectroscopy (AFM-SMFS). Therefore, this study provides a useful strategy for polyprotein engineering and immobilization.
Introduction
Compared with synthetic polymers, natural multi-domain proteins have a uniform structure with a well-controlled number and type of subdomains1. This feature usually leads to improved biological function and stability2,3. Many approaches, such as cysteine-based disulfide bond coupling and recombinant DNA technology, have been developed for building such a polymerized protein with multiple domains4,5,6,7. However, the former method always results in a random and uncontrolled sequence, and the latter one leads to other problems, including the difficulty for the overexpression of toxic and large-size proteins and the purification of complex protein with cofactor and other delicate enzymes.
To meet this challenge, we develop an enzymatic method that conjugates protein monomer together for polymer/polyprotein in a stepwise fashion using a protein ligase OaAEP1 combined with a protease TEV8,9. OaAEP1 is a strict and efficient endopeptidase. Two proteins can be linked covalently as Asn-Gly-Leu sequence (NGL) through two termini by OaAEP1 in less than 30 min if the N-terminus is Gly-Leu residues(GL) and the other of which the C-terminus is NGL residues10. However, the use of OaAEP1 only to link protein monomer leads to a protein polymer with an uncontrolled sequence like the cysteine-based coupling method. Therefore, we design the N-terminus of the protein unit with a removable TEV protease site plus a leucine residue as ENLYFQ/G-L-POI. Before the TEV cleavage, the N-terminal would not participate in OaAEP1 ligation. And then the GL residues at N-terminus, which are compatible with further OaAEP1 ligation, is exposed after the TEV cleavage. Thus, we have achieved a sequential enzymatic biosynthesis method of polyprotein with a relatively well-controlled sequence.
Here, our stepwise enzymatic synthesis method can be used in polyprotein sample preparation, including sequence-controlled and uncontrolled, and protein immobilization for single-molecule studies as well, especially for the complex system such as metalloprotein.
Moreover, AFM-based SMFS experiments allow us to confirm the protein polymer construction and stability at the single-molecule level. Single-molecule force spectroscopy, including AFM, optical tweezer and magnetic tweezer, is a general tool in nanotechnology to manipulate biomolecule mechanically and measure their stability11,12,13,14,15,16,17,18,19,20. Single-molecule AFM has been widely used in the study of protein (un)folding21,22,23,24,25, the strength measurement of receptor-ligand interaction26,27,28,29,30,31,32,33,34,35, inorganic chemical bond20,36,37,38,39,40,41,42,43 and metal-ligand bond in metalloprotein44,45,46,47,48,49,50. Here, single-molecule AFM is used to verify the synthesized polyprotein sequence based on the corresponding protein unfolding signal.
Protocol
1. Protein production
- Gene clone
- Purchase genes coding for the protein of interest (POI): Ubiquitin, Rubredoxin (RD)51, the cellulose-binding module (CBM), dockerin-X domain (XDoc) and cohesion from Ruminococcus flavefacience, tobacco etch virus (TEV) protease, elastin-like polypeptides (ELPs).
- Perform polymerase chain reaction and use three-restriction digestion enzyme system BamHI-BglII-KpnI for recombining the gene from different protein fragments.
- Confirm all genes by direct DNA sequencing.
- Proteins expression and production
- Transform E. coli BL21(DE3) with the pQE80L-POI or pET28a-POI plasmid for expression.
- Pick one single colony into 15 mL of LB medium with respective antibiotics (e.g., 100 μg/mL ampicillin sodium salt or 50 μg/mL, kanamycin). Keep shaking the cultures at 200 rpm at 37 °C for 16-20 h.
- Dilute the overnight cultures into 800 mL of LB medium (1:50 dilution). For rubredoxin, centrifuge the culture at 1,800 x g, then resuspend in 15 mL of M9 medium (supplemented with 0.4% glucose, 0.1 mM CaCl2, 2 mM MgSO4), and then dilute it into 800 mL of M9 medium.
- Incubate the culture at 37 °C while shaking at 200 rpm, until the culture reaches an optical density at 600 nm (OD600) of 0.6. Save 100 μL sample of the culture as the pre-induction control for testing protein expression.
- Induce protein expression by adding IPTG to a final concentration of 1 mM and shake the culture at 37 °C for 4 h at 200 rpm. Reserve a 100 μL sample of the culture as the post-induction control for testing protein expression.
- Centrifuge the culture at 13,000 x g for 25 min at 4 °C and store at -80 °C before purification.
NOTE: The protocol can be paused here.
- Purification of protein of interest
- Resuspend the cells in 25 mL of lysis buffer (50 mM Tris, 150 mM NaCl, pH 7.4 containing DNase, RNase, PMSF) and use a sonicator (15% amplitude) to lyse it for 30 min on ice.
- Clarify the cell lysate at 19,000 x g for 40 min at 4 °C.
- Pack 1 mL (bed volume) of Co-NTA or Ni-NTA affinity column and wash the column with 10 column volumes (CV) of ultrapure water and then 10 CVs of wash buffer (50 mM Tris, 150 mM NaCl, 2 mM imidazole, pH 7.4) by gravity flow.
- Pass the protein supernatant through the column by gravity flow for three times.
- Pour wash buffer on the column with 50 CVs to move away contaminant proteins.
- Elute the bound protein with 3 CVs of ice-cold elution buffer (20 mM Tris, 400 mM NaCl, 250 mM imidazole, pH 7.4). When it comes to rubredoxin proteins, further anion exchange purification using anion exchange column at pH 8.5 at 4 °C is necessary.
- Analyze the sample by SDS-PAGE.
2. Functionalization of coverslip and cantilever surface
- Functionalized coverslip surface preparation
- Dissolve 20 g of potassium chromate in 40 mL of ultrapure water. Slowly add 360 mL of concentrated sulfuric acid to the potassium chromate solution with glass rod stir gently and to prepare the chromic acid.
CAUTION: The chemical used here and the final chromic acid is strongly corrosive and acidic. Work with proper protective equipment. The solution releases heat when add concentrated sulfuric acid, which means slow adding and proper pause for cooling down. - Clean and activate a glass coverslip at 80 °C for 30 min by chromic acid treatment. Completely immerse the coverslips in 1% (v/v) APTES toluene solution for 1 h at room temperature while protecting them from light.
- Wash the coverslip with toluene and absolute ethyl alcohol and dry the coverslip with a stream of nitrogen.
- Incubate the coverslip at 80 °C for 15 min and then cool down to room temperature.
- Add 200 μL of sulfo-SMCC (1 mg/mL) in dimethyl sulfoxide (DMSO) solution between two immobilized coverslips and incubate for 1 h protected from light.
- Wash the coverslip with DMSO first and then with absolute ethyl alcohol to remove residual sulfo-SMCC.
- Dry the coverslip under a stream of nitrogen.
- Pipet 60 μL of 200 μM GL-ELP50nm-C protein solution onto a functionalized coverslip and incubate for about 3 h.
- Wash the coverslip with ultrapure water to remove the unreacted GL-ELP50nm-C.
NOTE: Functionalized coverslips are capable for about two weeks under storage at 4 °C.
- Dissolve 20 g of potassium chromate in 40 mL of ultrapure water. Slowly add 360 mL of concentrated sulfuric acid to the potassium chromate solution with glass rod stir gently and to prepare the chromic acid.
- Functionalized cantilever surface preparation
- Clean the cantilevers at 80 °C for 10 min by chromic acid treatment.
- Functionalize the cantilever by amino-silanization with 1% (v/v) APTES toluene solution and then bake the cantilever at 80 °C for 15 min before conjugating to sulfo-SMCC.
- Link the C-ELP50nm-NGL to the surface with the maleimide group of sulfo-SMCC for 1.5 h.
- Wash away the unreacted C-ELP50nm-NGL on the coverslip by ultrapure water.
- Immerse a functionalized cantilever in 200 μL of 50 μM GL-CBM-XDoc protein solution containing 200 nM OaAEP1 at 25 °C for 20-30 min. Then use AFM buffer (100 mM Tris, 100 mM NaCl, pH 7.4) to wash away unreacted protein.
NOTE: The surface chemistry of the cantilevers and the coverslip are similar.
3. Stepwise polyprotein preparation with controlled sequences
- Link the ligation unit Coh-tev-L-POI-NGL to the GL-ELP50nm immobilized on the coverslip surface by OaAEP1 for 30 min.
- Use 15-20 mL of AFM buffer (100 mM Tris, 100 mM NaCl, pH 7.4) to wash away any unreacted proteins.
- Add 100 μL of TEV protease (0.5 mM EDTA, 75 mM NaCl, 25 mM Tris-HCl 10% [v/v] glycerol, pH 8.0) to cleave the protein unit at the TEV recognize site for 1 h at 25 °C.
- Use 15-20 mL of AFM buffer to wash away residual proteins.
- Link the ligation unit Coh-tev-L-POI-NGL to the GL-Ub-NGL-Glass by OaAEP1 for 30 min.
- Repeat steps 3.3 to 3.5 N-1 times to build protein construct GL-(Ub)n-NGL on the glass surface. Omit the last TEV cleavage reaction to reserve cohesin on the protein-polymer as Coh-tev-L-(Ub)n-NGL-Glass.
4. AFM Experiment measurement and data analysis
- AFM measurements
- Add 1 mL of AFM buffer to the chamber with 10 mM CaCl2 and 5 mM Ascorbic Acid.
- Choose the D tip of the functionalized AFM probe for the experiment. Use the equipartition theorem to calibrate the cantilever in AFM buffer with an accurate spring constant (k) value before each experiment.
- Attach the cantilever tip to the sample surface to form the Cohesin/Dockerin pair.
- Retract the cantilever at a constant velocity of 400 nm·s−1 from the surface. In the meantime, record the force-extension curve at a sample rate of 4000 Hz.
- Data analysis
- Use JPK data processing select force-extension traces.
- Use software to analyze the traces. Fit the curves with the worm-like-chain (WLC) model of polymer elasticity and obtain unfolding force and contour length increment for individual protein unfolding peak.
- Fit the histograms of unfolding forces with the Gaussian model to obtain the most probable values of unfolding force (<Fu>) and contour length increment (<ΔLc>).
Representative Results
The NGL residues introduced between adjacent proteins by OaAEP1 ligation will not affect protein monomer stability in the polymer as the unfolding force (<Fu>), and contour length increment (<ΔLc>) is comparable with the previous study (Figure 1). The purification result of the rubredoxin protein is shown in Figure 2. To prove the protein after TEV cleavage is compatible with the following OaAEP1 ligation to construct protein polymer with a control sequence and the construction is high-efficiency, Figure 3 provides an SDS-PAGE image as a reference. The steps of the functionalized cantilever and coverslip preparation are described in Figure 4. The stepwise enzymatic biosynthesis and immobilization of polyprotein on the coverslip are shown in Figure 5. Use this protocol, a protein polymer with the controlled sequence can be built and suitable for AFM-based SMFS experiments.
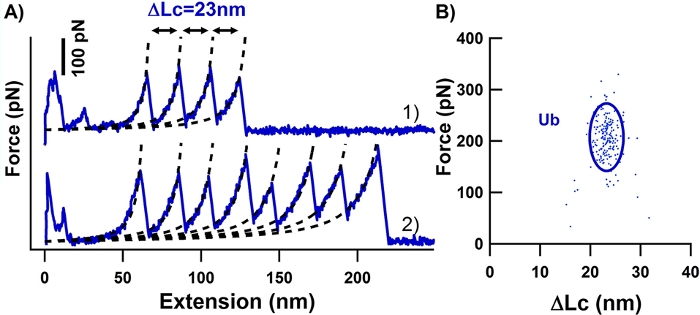
Figure 1: AFM-based SMFS measurements of polyprotein built by OaAEP1. (A) Typical sawtooth-like force-extension curves of Ub (curve 1 in blue) were shown with expected ΔLc of ~23 nm. (B) The scatter plot presents the relationship between Ub unfolding force (202 ± 44 pN, average ± s.d., n = 198) and ΔLc (23 ± 2 nm, average ± s.d.). This figure has been modified from Ref.8. Please click here to view a larger version of this figure.
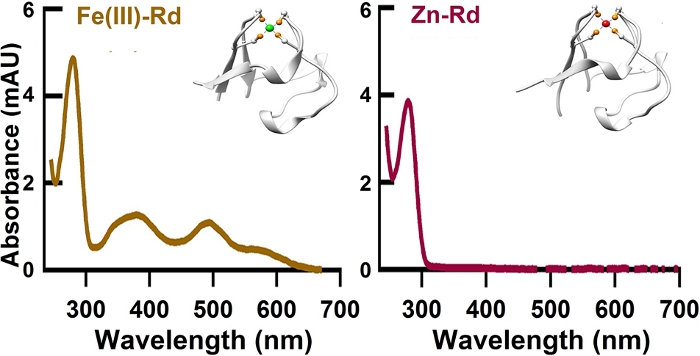
Figure 2: The UV-Vis absorbance spectra of GL-GB1-Fe(III)-Rd-NGL and GL-GB1-(Zn)-Rd-NGL. The Fe(III)-form Rd (Left spectrum, colored in brown, PDB code:1BRF) presented typical UV-Vis absorption peaks at 495 nm and 579 nm while the Zn-form did not (Right spectrum, colored in wine, PDB code: 1IRN). This figure has been modified from Ref.8. Please click here to view a larger version of this figure.
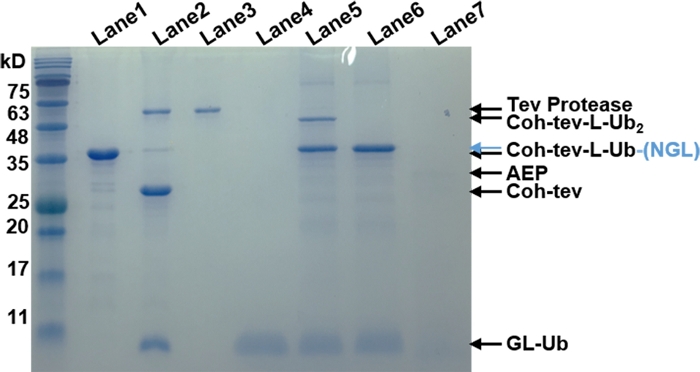
Figure 3: SDS-PAGE gel results of stepwise digestion and ligation using TEV protease and OaAEP1 to build the Ub dimer. Lanes 1–4 showed Coh-tev-L-Ub, the result protein mixture of TEV cleavage, pure sfGFP-TEV protease and purified product (GL-Ub). Lanes 5–7 showed the cleaved GL-Ub and Coh-tev-L-Ub-NGL ligation mixture with (Lane 5) or without (Lane 6) OaAEP1 and pure OaAEP1. This figure has been modified from Ref.8. Please click here to view a larger version of this figure.
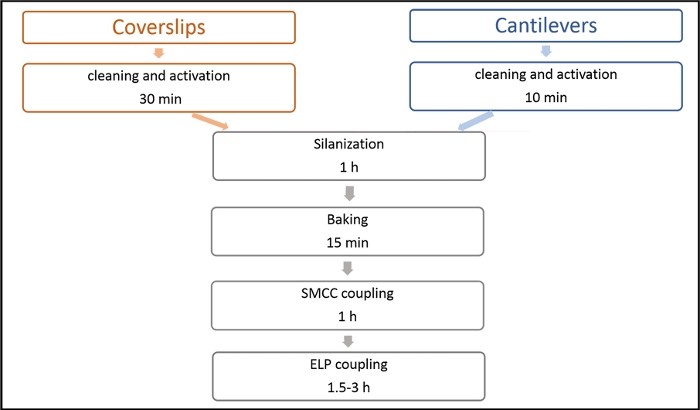
Figure 4: Process chart describing each step for functionalizing glass coverslips and cantilever. After cleaning and activation by chromic acid, coverslip and cantilever share similar functionalization process, except the last step in which GL-ELP50nm-C couples with coverslip while C-ELP50nm-NGL couples with cantilever. Please click here to view a larger version of this figure.
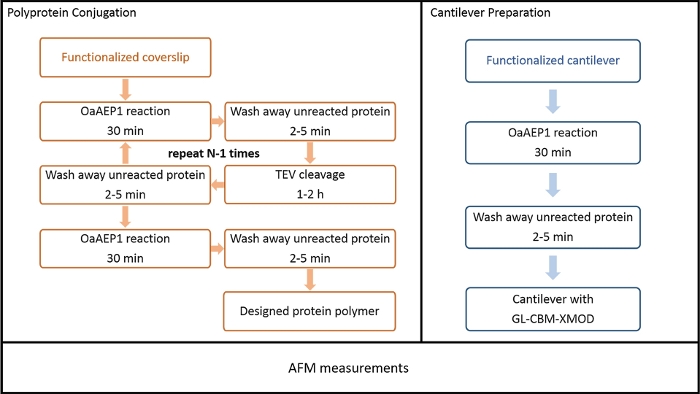
Figure 5: Process chart describing each step for polyprotein immobilization on the surface. Top left process flow diagram shows the stepwise building of polyprotein with controlled sequences on the coverslip. Top right diagram shows the preparation of the functionalized cantilever used in the AFM measurements. Please click here to view a larger version of this figure.
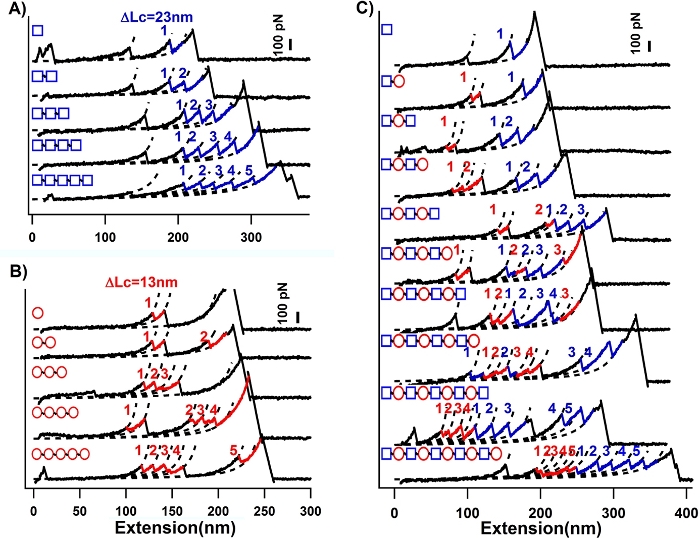
Figure 6: Typical unfolding traces of the protein polymers with a rationally controlled sequence by AFM-based SMFS. (A) Typical sawtooth-like force-extension curves of Ub presented ΔLc of ~23 nm as expected. (B) Typical force-extension curves of Rd presented ΔLc of ~13 nm as expected. (C) Typical force-extension curves of (Ub-Rd)n protein mixture in which the blue peak means the unfolding events of Ub while the red means Rd. This figure has been modified from Ref.8. Please click here to view a larger version of this figure.
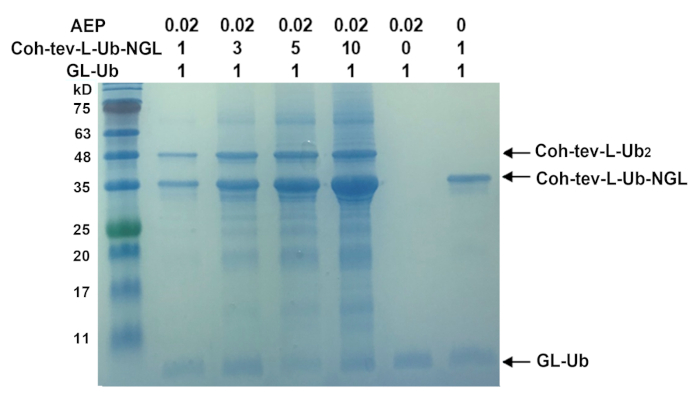
Supplementary Figure 1: SDS-PAGE gel results of the protein ligation efficiency under different ratio between two reactants. The ligation efficiency was 20% when the ratio is 1 to 1 and reached 75% at the ratio of 10 to 1. This figure has been modified from Ref.8. Please click here to view a larger version of this figure.
Discussion
We have described a protocol for enzymatic biosynthesis and immobilization of polyprotein and verified the polyprotein design by AFM-based SMFS. This methodology provides a novel approach to building the protein-polymer in a designed sequence, which complements previous methods for polyprotein engineering and immobilization4,6,52,53,54,55,56,57,58,59,60,61.
Compared with the classic recombinant DNA methodology for polyprotein construction7,62, our method bases on the ligation between small protein monomers. Thus, it allows the expression of large-sized or toxic protein molecules for polyprotein construction. Additionally, it allows the purification of the protein monomer before conjugation.
Compared with the widely-used bi-cysteine method forming intermolecular disulfide bond for protein polymerization4, our enzymatic method using both OaAEP1 and TEV protease results in a polyprotein with a relatively controlled sequence and defined connection geometry. And it does not use cysteine, which is an essential functional residue for many proteins.
Our method is mostly similar to sortase-based protein conjugation59. The unique feature of our method is that the OaAEP1-based protein ligation is much more efficient, thus allows the construction of protein pentamer with a reasonable yield10,53. It also needs fewer residues for ligation and results in a shorter three-residues NGL linker. As a result, it shows no "linker effect" as the newly formed NGL linker does not affect the stability of individual protein monomer or induce any unnatural protein-protein interaction. Nevertheless, we believe that all methods have their own advantages and disadvantages. For example, the classic recombinant DNA method does not add any residue between protein monomer and not require the use of any enzyme for ligation. And the bi-cysteine method is simple and easy for protein polymerization. Thus, they can all be useful under different experimental requirements.
For our stepwise construction of polyprotein, it is crucial to remove the unreacted protein completely. Take enough volume of AFM buffer and enough time to clean the reacted surface carefully. Otherwise, the residual protein or protease will affect further synthesis reactions.
The efficiency of OaAEP1 ligation is a critical limit to our method as the TEV cleavage efficiency is almost complete (96%). It is critical to raise the ratio between the two reactants, GL-protein, and protein-NGL, to improve the ligation efficiency. Our study shows that when protein-NGL is tenfold to GL-protein, the efficiency increases from 20% (the ratio is 1 to 1) to 75% (Supplementary Figure 1). It is critical to consume the reactant, which was immobilized onto the surface as the free reactant can be moved away by washing with buffer. Additionally, whether the N- or C- terminus is exposed to the solution is also a crucial factor to ligation. It is an optional approach to expose the terminal by adding a linker containing the recognized site to the respective terminal.
In the end, our protocol is an enzymatic way to conjugate proteins in a designed sequence. It also provides an alternative approach to couple and immobilize protein samples in single-molecule studies.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21771103, 21977047), Natural Science Foundation of Jiangsu Province (Grant No. BK20160639) and Shuangchuang Program of Jiangsu Province.
Materials
| iron (III) chloride hexahydrate | Energy chemical | 99% | |
| Zinc chloride | Alfa Aesar | 100.00% | |
| calcium chloride hydrate | Alfa Aesar | 99.9965% crystalline aggregate | |
| L-Ascorbic Acid | Sigma Life Science | Bio Xtra, ≥99.0%, crystalline | |
| (3-Aminopropyl) triethoxysilane | Sigma-Aldrich | ≥99% | |
| sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate | Thermo Scientific | 90% | |
| Glycerol | Macklin | 99% | |
| 5,5'-dithiobis(2-nitrobenzoic acid) | Alfa Aesar | ||
| Genes | Genscript | ||
| Equipment | |||
| Nanowizard 4 AFM | JPK Germany | ||
| MLCT cantilever | Bruker Corp | ||
| Mono Q 5/50 GL | GE Healthcare | ||
| AKTA FPLC system | GE Healthcare | ||
| Glass coverslip | Sail Brand | ||
| Nanodrop 2000 | Thermo Scientific | ||
| Avanti JXN-30 Centrifuge | Beckman Coulter | ||
| Gel Image System | Tanon |
References
- Rief, M., Gautel, M., Oesterhelt, F., Fernandez, J. M., Gaub, H. E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276 (5315), 1109-1112 (1997).
- Yang, Y. J., Holmberg, A. L., Olsen, B. D. Artificially Engineered Protein Polymers. Annual Review of Chemical and Biomolecular Engineering. 8 (1), 549-575 (2017).
- Yang, J., et al. Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology. Proceedings of the National Academy of Sciences of the United States of America. 115 (36), 8509-8517 (2018).
- Dietz, H., et al. Cysteine engineering of polyproteins for single-molecule force spectroscopy. Nature Protocols. 1 (1), 80-84 (2006).
- Carrion-Vazquez, M., et al. Mechanical and chemical unfolding of a single protein: A comparison. Proceedings of the National Academy of Sciences of the United States of America. 96 (7), 3694-3699 (1999).
- Hoffmann, T., et al. Rapid and Robust Polyprotein Production Facilitates Single-Molecule Mechanical Characterization of beta-Barrel Assembly Machinery Polypeptide Transport Associated Domains. ACS Nano. 9 (9), 8811-8821 (2015).
- Hoffmann, T., Dougan, L. Single molecule force spectroscopy using polyproteins. Chemical Society Reviews. 41 (14), 4781-4796 (2012).
- Deng, Y., et al. Enzymatic biosynthesis and immobilization of polyprotein verified at the single-molecule level. Nature Communications. 10 (1), 2775 (2019).
- Yuan, G., et al. Single-Molecule Force Spectroscopy Reveals that Iron-Ligand Bonds Modulate Proteins in Different Modes. The Journal of Physical Chemistry Letters. 10 (18), 5428-5433 (2019).
- Yang, R., et al. Engineering a Catalytically Efficient Recombinant Protein Ligase. Journal of the American Chemical Society. 139 (15), 5351-5358 (2017).
- Woodside, M. T., Block, S. M. Reconstructing Folding Energy Landscapes by Single-Molecule Force Spectroscopy. Annual Review of Biophysics. 43, 19-39 (2014).
- Sen Mojumdar, S., et al. Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nature Communications. 8 (1), 1881 (2017).
- Singh, D., Ha, T. Understanding the Molecular Mechanisms of the CRISPR Toolbox Using Single Molecule Approaches. ACS Chemical Biology. 13 (3), 516-526 (2018).
- You, H., Le, S., Chen, H., Qin, L., Yan, J. Single-molecule Manipulation of G-quadruplexes by Magnetic Tweezers. Journal of Visualized Experiments. (127), e56328 (2017).
- Suren, T., et al. Single-molecule force spectroscopy reveals folding steps associated with hormone binding and activation of the glucocorticoid receptor. Proceedings of the National Academy of Sciences of the United States of America. 115 (46), 11688-11693 (2018).
- Tapia-Rojo, R., Eckels, E. C., Fernández, J. M. Ephemeral states in protein folding under force captured with a magnetic tweezers design. Proceedings of the National Academy of Sciences of the United States of America. 116 (16), 7873-7878 (2019).
- Chen, H., et al. Dynamics of Equilibrium Folding and Unfolding Transitions of Titin Immunoglobulin Domain under Constant Forces. Journal of the American Chemical Society. 137 (10), 3540-3546 (2015).
- Fu, L., Wang, H., Li, H. Harvesting Mechanical Work From Folding-Based Protein Engines: From Single-Molecule Mechanochemical Cycles to Macroscopic Devices. Chinese Chemical Society. 1 (1), 138-147 (2019).
- Scholl, Z. N., Li, Q., Josephs, E., Apostolidou, D., Marszalek, P. E. Force Spectroscopy of Single Protein Molecules Using an Atomic Force Microscope. Journal of Visualized Experiments. (144), e55989 (2019).
- Zhang, S., et al. Towards Unveiling the Exact Molecular Structure of Amorphous Red Phosphorus by Single-Molecule Studies. Angewandte Chemie International Edition. 58 (6), 1659-1663 (2019).
- Yu, H., Siewny, M. G., Edwards, D. T., Sanders, A. W., Perkins, T. T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science. 355 (6328), 945-950 (2017).
- Thoma, J., Sapra, K. T., Müller, D. J. Single-Molecule Force Spectroscopy of Transmembrane β-Barrel Proteins. Annual Review of Analytical Chemistry. 11 (1), 375-395 (2018).
- Chen, Y., Radford, S. E., Brockwell, D. J. Force-induced remodelling of proteins and their complexes. Current Opinion in Structural Biology. 30, 89-99 (2015).
- Takahashi, H., Rico, F., Chipot, C., Scheuring, S. alpha-Helix Unwinding as Force Buffer in Spectrins. ACS Nano. 12 (3), 2719-2727 (2018).
- Borgia, A., Williams, P. M., Clarke, J. Single-molecule studies of protein folding. Annu. Rev. Biochem. 77, 101-125 (2008).
- Florin, E., Moy, V., Gaub, H. Adhesion forces between individual ligand-receptor pairs. Science. 264 (5157), 415-417 (1994).
- Zakeri, B., et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proceedings of the National Academy of Sciences of the United States of America. 109 (12), 690-697 (2012).
- Ott, W., Jobst, M. A., Schoeler, C., Gaub, H. E., Nash, M. A. Single-molecule force spectroscopy on polyproteins and receptor-ligand complexes: The current toolbox. Journal of Structural Biology. 197 (1), 3-12 (2017).
- Stahl, S. W., et al. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proceedings of the National Academy of Sciences of the United States of America. 109 (50), 20431-20436 (2012).
- Oh, Y. J., et al. Ultra-Sensitive and Label-Free Probing of Binding Affinity Using Recognition Imaging. Nano Letters. 19 (1), 612-617 (2019).
- Vera Andrés, M., Carrion-Vazquez, M. Direct Identification of Protein-Protein Interactions by Single-Molecule Force Spectroscopy. Angewandte Chemie International Edition. 55 (45), 13970-13973 (2016).
- Yu, H., Heenan, P. R., Edwards, D. T., Uyetake, L., Perkins, T. T. Quantifying the Initial Unfolding of Bacteriorhodopsin Reveals Retinal Stabilization. Angewandte Chemie International Edition. 58 (6), 1710-1713 (2019).
- Jobst, M. A., Schoeler, C., Malinowska, K., Nash, M. A. Investigating Receptor-ligand Systems of the Cellulosome with AFM-based Single-molecule Force Spectroscopy. Journal of Visualized Experiments. (82), e50950 (2013).
- Stetter, F. W. S., Kienle, S., Krysiak, S., Hugel, T. Investigating Single Molecule Adhesion by Atomic Force Spectroscopy. Journal of Visualized Experiments. (96), e52456 (2015).
- Nadler, H., et al. Deciphering the Mechanical Properties of Type III Secretion System EspA Protein by Single Molecule Force Spectroscopy. Langmuir. , (2018).
- Giganti, D., Yan, K., Badilla, C. L., Fernandez, J. M., Alegre-Cebollada, J. Disulfide isomerization reactions in titin immunoglobulin domains enable a mode of protein elasticity. Nature Communications. 9 (1), 185 (2018).
- Huang, W., et al. Maleimide-thiol adducts stabilized through stretching. Nature Chemistry. 11 (4), 310-319 (2019).
- Li, Y. R., et al. Single-Molecule Mechanics of Catechol-Iron Coordination Bonds. ACS Biomaterials Science, Engineering. 3 (6), 979-989 (2017).
- Popa, I., et al. Nanomechanics of HaloTag Tethers. Journal of the American Chemical Society. 135 (34), 12762-12771 (2013).
- Xue, Y., Li, X., Li, H., Zhang, W. Quantifying thiol-gold interactions towards the efficient strength control. Nature Communications. 5, 4348 (2014).
- Wiita, A. P., Ainavarapu, S. R. K., Huang, H. H., Fernandez, J. M. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proceedings of the National Academy of Sciences of the United States of America. 103 (19), 7222-7227 (2006).
- Pill, M. F., East, A. L. L., Marx, D., Beyer, M. K., Clausen-Schaumann, H. Mechanical Activation Drastically Accelerates Amide Bond Hydrolysis, Matching Enzyme Activity. Angewandte Chemie International Edition. 58 (29), 9787-9790 (2019).
- Conti, M., Falini, G., Samori, B. How strong is the coordination bond between a histidine tag and Ni-nitrilotriacetate? An experiment of mechanochemistry on single molecules. Angew. Chem. Int. Ed. 39 (1), 215-218 (2000).
- Beedle, A. E. M., Lezamiz, A., Stirnemann, G., Garcia-Manyes, S. The mechanochemistry of copper reports on the directionality of unfolding in model cupredoxin proteins. Nature Communications. 6, 7894 (2015).
- Li, H., Zheng, P. Single molecule force spectroscopy: a new tool for bioinorganic chemistry. Current Opinion in Chemical Biology. 43, 58-67 (2018).
- Zheng, P., Takayama, S. i. J., Mauk, A. G., Li, H. Hydrogen bond strength modulates the mechanical strength of ferric-thiolate bonds in rubredoxin. Journal of the American Chemical Society. 134 (9), 4124-4131 (2012).
- Lei, H., et al. Reversible Unfolding and Folding of the Metalloprotein Ferredoxin Revealed by Single-Molecule Atomic Force Microscopy. Journal of the American Chemical Society. 139 (4), 1538-1544 (2017).
- Yuan, G., et al. Multistep Protein Unfolding Scenarios from the Rupture of a Complex Metal Cluster Cd3S9. Scientific Reports. 9 (1), 10518 (2019).
- Zheng, P., Arantes, G. M., Field, M. J., Li, H. Force-induced chemical reactions on the metal centre in a single metalloprotein molecule. Nature Communications. 6, 7569 (2015).
- Arantes, G. M., Bhattacharjee, A., Field, M. J. Homolytic cleavage of Fe-S bonds in rubredoxin under mechanical stress. Angewandte Chemie International Edition. 52 (31), 8144-8146 (2013).
- Blake, P. R., et al. Determinants of protein hyperthermostability: purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochimie. 30 (45), 10885-10895 (1991).
- Ott, W., Durner, E., Mediated Gaub, H. E. Enzyme-Mediated, Site-Specific Protein Coupling Strategies for Surface-Based Binding Assays. Angewandte Chemie International Edition. 57 (39), 12666-12669 (2018).
- Garg, S., Singaraju, G. S., Yengkhom, S., Rakshit, S. Tailored Polyproteins Using Sequential Staple and Cut. Bioconjugate Chemistry. 29 (5), 1714-1719 (2018).
- Veggiani, G., et al. Programmable Polyproteams Built Using Twin Peptide Superglues. Proceedings of the National Academy of Sciences of the United States of America. 113 (5), 1202-1207 (2016).
- Pelegri-O’Day, E. M., Maynard, H. D. Controlled Radical Polymerization as an Enabling Approach for the Next Generation of Protein-Polymer Conjugates. Accounts of Chemical Research. 49 (9), 1777-1785 (2016).
- Zheng, P., Cao, Y., Li, H. Facile method of constructing polyproteins for single-molecule force spectroscopy studies. Langmuir. 27 (10), 5713-5718 (2011).
- Zimmermann, J. L., Nicolaus, T., Neuert, G., Blank, K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nature Protocols. 5 (6), 975-985 (2010).
- Becke, T. D., et al. Covalent Immobilization of Proteins for the Single Molecule Force Spectroscopy. Journal of Visualized Experiments. (138), e58167 (2018).
- Liu, H. P., Ta, D. T., Nash, M. A. Mechanical polyprotein assembly using sfp and sortase-mediated domain oligomerization for single-molecule studies. Small Methods. 2 (6), (2018).
- Zhang, Y., Park, K. Y., Suazo, K. F., Distefano, M. D. Recent progress in enzymatic protein labelling techniques and their applications. Chemical Society Reviews. 47 (24), 9106-9136 (2018).
- Luo, Q., Hou, C., Bai, Y., Wang, R., Liu, J. Protein Assembly: Versatile Approaches to Construct Highly Ordered Nanostructures. Chemical Reviews. 116 (22), 13571-13632 (2016).
- Valle-Orero, J., Rivas-Pardo, J. A., Popa, I. Multidomain proteins under force. Nanotechnology. 28 (17), 174003 (2017).

