Synthesis of Aptamer-PEI-g-PEG Modified Gold Nanoparticles Loaded with Doxorubicin for Targeted Drug Delivery
Summary
In this protocol, doxorubicin-loaded AS1411-g-PEI-g-PEG modified gold nanoparticles are synthesized via three-step amide reactions. Then, doxorubicin is loaded and delivered to target cancer cells for cancer therapy.
Abstract
Due to drug resistance and toxicity in healthy cells, use of doxorubicin (DOX) has been limited in clinical cancer therapy. This protocol describes the designing of poly(ethylenimine) grafted with polyethylene glycol (PEI-g-PEG) copolymer functionalized gold nanoparticles (AuNPs) with loaded aptamer (AS1411) and DOX through amide reactions. AS1411 is specifically bonded with targeted nucleolin receptors on cancer cells so that DOX targets cancer cells instead of healthy cells. First, PEG is carboxylated, then grafted to branched PEI to obtain a PEI-g-PEG copolymer, which is confirmed by 1H NMR analysis. Next, PEI-g-PEG copolymer coated gold nanoparticles (PEI-g-PEG@AuNPs) are synthesized, and DOX and AS1411 are covalently bonded to AuNPs gradually via amide reactions. The diameter of the prepared AS1411-g-DOX-g-PEI-g-PEG@AuNPs is ~39.9 nm, with a zeta potential of -29.3 mV, indicating that the nanoparticles are stable in water and cell medium. Cell cytotoxicity assays show that the newly designed DOX loaded AuNPs are able to kill cancer cells (A549). This synthesis demonstrates the delicate arrangement of PEI-g-PEG copolymers, aptamers, and DOX on AuNPs that are achieved by sequential amide reactions. Such aptamer-PEI-g-PEG functionalized AuNPs provide a promising platform for targeted drug delivery in cancer therapy.
Introduction
Being the major public health problem worldwide, cancer is widely characterized as having a low cure rate, high recurrence rate, and high mortality rate1,2. Current conventional anti-cancer methods include surgery, chemotherapy, and radiotherapy3, among which chemotherapy is the primary treatment for cancer patients in the clinic4. Clinical used anticancer drugs mainly include paclitaxel (PTX)5 and doxorubicin (DOX)6,7. DOX, an antineoplastic drug, has been broadly applied in clinical chemotherapy, due to the advantages of cancer cytotoxicity and inhibition of cancer cell proliferation8,9. However, DOX causes cardiotoxicity10,11, and the short half-life of DOX restricts its application in the clinic12. Therefore, degradable drug carriers are needed to load DOX and subequently release in a controlled fashion to a targeted area.
Nanoparticles have been widely used in targeted drug delivery systems and have several advantages in cancer treatment (i.e., sizeable surface-to-volume ratio, small size, ability to encapsulate various drugs, and tunable surface chemistry, etc.)13,14,15. In particular, gold nanoparticles (AuNPs) have been widely used in biological and biomedical applications, such as photothermal cancer therapy16,17. The unique properties of AuNPs, such as facile synthesis and general surface functionalization, have excellent prospects in the clinical field of cancer therapy18. Also, AuNPs have been used to identify drug delivery strategies, diagnose tumors, and overcome resistance in many studies19,20.
Notwithstanding, AuNPs need to be further tailored to overcome drug resistance via high local release at tumor lesions through enhanced permeation and retention (EPR), such as the targeting and accessibility properties. Polymer functionalized AuNPs have exhibited unique advantages, such as improved water solubility of hydrophobic anticancer drugs and prolonged circulation time21,22. Various biocompatible polymers have been used for AuNP coatings, such as polyethylene glycol (PEG), polyethyleneimine (PEI), hyaluronic acid, heparin, and xanthan gum. Then the stability, as well as the payload, of AuNPs is improved well23. Specifically, PEI is a highly branched polymer that is composed of many repeating units of primary, secondary, and tertiary amines24. PEI has excellent solubility, low viscosity, and a high degree of functionality, which is suitable for coating on AuNPs.
On the other hand, anti-cancer drugs need to be delivered to cancer cells directly with improved loading efficiency, and with lower toxicity for treating primary and advanced metastatic tumors25. Targeted ligands have great potential for anti-cancer drug targeted delivery systems26. Its selectivity for target molecule binding confers anti-cancer drug targeting specificity and increases drug enrichment in diseased tissues27. More ligands include antibodies, polypeptides, and small molecules. Compared to other ligands, nucleic acid aptamers can be synthesized in vitro and are easy to modify. AS1411 is an unmodified 26 bp phosphodiester oligonucleotide that forms a stable dimeric G-tetramer structure to specifically bind to an overexpressed target nuclear protein receptor on cancer cells28,29,30. AS1411 inhibits the proliferation of many cancer cells but does not affect the growth of healthy cells31,32. As a result, AS1411 has been used to fabricate an ideal targeted drug delivery system.
In this study, a PEI-g-PEG copolymer is synthesized via an amide reaction, then PEI-g-PEG copolymer coated gold nanoparticles (PEI-g-PEG@AuNPs) are fabricated. Additionally, DOX and AS1411 are sequentially linked to the prepared PEI-g-PEG@AuNPs, as shown in Figure 1. This detailed protocol is intended to help researchers avoid many of the common pitfalls associated with the fabrication of new PEI-g-PEG@AuNPs loaded with DOX and AS1411.
Protocol
CAUTION: Make sure to consult all relevant material safety data sheets (MSDS) before using all chemicals. Several of the chemicals used for preparing copolymer and nanoparticles are acutely toxic. Nanoparticles also have potential hazards. Make sure to use all appropriate safety practices and personal protective equipment, including gloves, lab coat, hoods, full-length pants, and close-toed shoes.
1. Synthesis of double-carboxyl polyethylene glycol (CT-PEG)33
- Add 1.46 g (14.6 mmol) of succinic anhydride (SA) and 209 mg (1.71 mmol) of 4-dimethylaminopyridine (DMAP) to a 100 mL round bottom flask.
- Add 15 mL of anhydrous tetrahydrofuran (THF) to the flask used in step 1.1 and fit a glass stopper. Keep the flask at 0 °C for 30 min.
- Add 4.28 g (4.28 mmol) of polyethylene glycol (PEG) and 1.8 mL (12.8 mmol) of triethylamine (TEA) to a new flask.
- Add 15 mL of anhydrous THF to the flask used in step 1.3 and fit a glass stopper. Transfer the solution slowly to the flask used in step 1.2, using a syringe under nitrogen atmosphere.
- Stir the solution at 0 °C for 2 h, then continue the reaction at room temperature (RT) overnight.
- Using a rotary evaporator (40 °C, 0.1 MPa), concentrate the reaction solution and remove the THF solvent.
- At RT, dissolve the reaction solution from step 1.6 in 15 mL of 1.325 g/mL dichloromethane (DCM), then add 15 mL of cold diethyl ether (Et2O) to obtain the precipitation product (polyethylene glycol diacid). Remove the solvent via filter paper.
NOTE: The precipitation step can be repeated 3x. - Dry the precipitates under vacuum at RT for 48 h.
2. Synthesis of PEI-g-PEG copolymer
- Add 305.47 mg of CT-PEG from step 1.8 and 5 mL of dimethyl sulfoxide (DMSO) to a flask and stir at RT to ensure that CT-PEG is fully dissolved in DMSO.
- Dissolve 49.46 mg of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) in 5 mL of DMSO, then add the solution to the flask used in step 2.1 and stir for 30 min at RT.
- Dissolve 29.69 mg of N-hydroxysuccinimide (NHS) in 5 mL of DMSO and add the solution to the flask used in step 2.1. Continue to stir at RT for 3 h.
- Dissolve 28.6 μL of polyethyleneimine (PEI) in 10 mL of DMSO and add the solution dropwise to the flask used in step 2.1. Stir for 3 days, at least.
- Transfer the reacted solution from step 2.4 to a dialysis bag (1,000 molecular weight cut-off [MWCO]). Place the dialysis bag into a 1 L beaker with 500 mL of ultrapure water as the dialysate. Change the ultrapure water every 12 h for 3 days.
- Transfer the solution in step 2.5 to another dialysis bag (10,000 MWCO). Place the dialysis bag into a 1 L beaker with 500 mL of ultrapure water as the dialysate. Change the ultrapure water every 12 h for 3 days.
- Concentrate the solution from step 2.6 using a rotary evaporator (40 °C, 0.1 MPa) and freeze-dry the sample to obtain the PEI-g-PEG powder.
3. Synthesis of PEI-g-PEG@AuNPs
- Dissolve 5 mg of prepared PEI-g-PEG (step 2.7) in 5 mL of ultrapure water in a new flask and fit with a glass stopper.
- Add 100 mL of 0.3 mM HAuCl4 solution to the flask and stir the solution for 3 h at RT.
NOTE: The color of the solution should change immediately from yellow to orange. - Add 1 mL of 1 mg/mL NaBH4 solution to the flask and stir the solution for 3 h at RT.
NOTE: The reaction solution should instantly turn burgundy. - Dialyze the reaction product using a dialysis bag (1,000 MWCO) for 3 days as described in step 2.5 to obtain the PEI-g-PEG@AuNPs solution.
4. Synthesis of DOX-g-PEI-g-PEG@AuNPs
- Add 1 mL of 2.2 mg/mL DOX solution and 20 mL of PEI-g-PEG@AuNPs solution to a new flask and fit with a glass stopper.
- Dissolve 0.727 mg of EDC in 1 mL of ultrapure water and add the EDC solution to the flask used in step 4.1.
- Dissolve 0.437 mg of NHS in 1 mL of ultrapure water. Add the NHS solution to the flask and stir at RT for 1 h.
- Dialyze the reaction product by using a dialysis bag (1,000 MWCO) for 3 days as described in step 2.5 to obtain the DOX-g-PEI-g-PEG@AuNPs solution.
5. Synthesis of AS1411-g-DOX-g-PEI-g-PEG@AuNPs
- Add 20 mL of DOX-g-PEI-g-PEG@AuNPs solution and 4OD of AS1411 (OD = optical density; 1OD ≈ 33 μg) to a new flask.
- Dissolve 28.76 mg of EDC in 1 mL of ultrapure water and add the EDC solution to the flask used in step 5.1.
- Dissolve 17.27 mg of NHS in 1 mL of ultrapure water. Add the NHS solution to the flask used in step 5.1 and stir the reaction for 1 h at RT.
- Dialyze the reaction product by using a dialysis bag (1,000 MWCO) for 3 days as described in step 2.5 to obtain AS1411-g-DOX-g-PEI-g-PEG@AuNPs.
6. Sample characterization
- Dissolve the CT-PEG polymer (step 1.8) and PEI-g-PEG copolymer (step 2.7) in chloroform-d in nuclear magnetic resonance (NMR) tubes, respectively. Analyze the samples using a 600 MHz NMR spectrometer equipped with a 14.09 T superconducting magnet and 5.0 mm 600 MHz broadband Z-gradient high resolution probe to confirm the chemical structure34.
- Disperse AuNPs, DOX, and AS1411, and prepared PEI-g-PEG@AuNPs (step 3.4), DOX-g-PEI-g-PEG@AuNPs (step 4.4), and AS1411-g-DOX-g-PEI-g-PEG@AuNPs (step 5.4), respectively in ultrapure water. Then, transfer to cuvettes and record ultraviolet-visible (UV-vis) spectra using a UV-vis spectrophotometer.
- Attach a double-sided adhesive (~2 mm x 2 mm) to the aluminum foil, and dip the sample solution (PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-g-PEI-g-PEG@AuNPs) on the whole tape uniformly. Analyze the samples using an X-ray photoelectron spectroscopy analyzer.
- Disperse PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-g-PEI-g-PEG@AuNPs solutions, respectively in ultrapure water. Then, transfer to cuvettes and evaluate the size distribution using dynamic light scattering.
- Disperse PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-g-PEI-g-PEG@AuNPs solutions in ultrapure water (one drop of sample per 5 mL of ultrapure water for each sample). Sonicate for 2 h. Dip the copper grid into sample solutions and dry under a infrared lamp. Characterize the morphology using a transmission electron microscope.
- Inject 1 mg of AS1411-g-DOX-g-PEI-g-PEG@AuNPs into a 20 kDa MWCO dialysis cassette, then place in 80 mL of phosphate buffered saline (PBS) with 5% bovine serum albumin (BSA). Stir at 37 °C.
- At the predetermined timepoints, collect 100 µL aliquots and replace with fresh PBS. Use a UV-vis spectrophotometer to measure the DOX fluorescence intensity of the aliquots.
7. CCK-8 assay of AS1411-g-DOX-g-PEI-g-PEG@AuNPs nanoparticles
- Grow A549 cells in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin under a humidified atmosphere of 95% air and 5% CO2 at 37 °C. Replace the culture medium every 2 days. Use cells at passage 5 for the cell proliferation and cytotoxicity assays to quantitatively evaluate the cytotoxicity of prepared AS1411-g-DOX-g-PEI-g-PEG@AuNPs nanoparticles.
- Add 100 μL of nanoparticle solution into each well containg 1 mL of cell medium. After culturing for 24 h and 48 h, remove the culture media from cell culture plates, then add 300 μL of fresh culture media and 30 μL of cell counting kit-8 (CCK-8) kit solutions immediately to each well. Incubate for 4 h in a CO2 incubator at 37 °C.
- Transfer 200 μL of reaction solutions from step 7.2 into a 96 well plate. Read the optical density (OD) of each well at 570 nm with a microplate reader.
- Observe the morphology of cells at 24 h and 48 h under a microscope.
Representative Results
1H NMR spectroscopy was used to confirm the successful synthesis of CT-PEG polymer and PEI-g-PEG copolymers (Figure 2). Figure 2a shows that the methylene proton signal at δ = 3.61 ppm and carboxyl proton signal at δ = 2.57 ppm confirm the successful synthesis of CT-PEG polymers. Figure 2b shows that the methylene proton signal of PEG at δ = 2.6 ppm and proton signal of PEI at δ = 1.66 ppm confirm the synthesis of PEI-g-PEG copolymers.
UV-vis spectroscopy was conducted to determine the successful functionalization of prepared copolymer on AuNPs (Figure 3). In UV-vis spectra, the presence of the bands at ~523 nm, 507 nm, and 260 nm corresponds to the surface plasmon resonance (SPR) peaks of AuNPs, DOX, and AS1411, respectively (Figure 3a). The bands at ~360 nm in UV-vis spectrum of PEI-g-PEG@AuNPs, ~532 nm in UV-vis spectrum of DOX-g-PEI-g-PEG@AuNPs, and ~546 nm in the UV-vis spectrum of AS1411-g-DOX-g-PEI-g-PEG@AuNPs confirm the successful synthesis of PEI-g-PEG copolymers attached to AuNPs. They also confirm that DOX and AS1411 were loaded on functionalized AuNPs gradually (Figure 3b).
X-ray photoelectron spectroscopy (XPS) was used to investigate the chemical bond of copolymer on AuNPs (Figure 4). The XPS spectrum of PEI-g-PEG@AuNPs showed C1s, O1s, N1s, and Au4f peaks indicated the connection between AuNPs and PEI-g-PEG copolymer (Figure 4a). There was a slight change in the XPS spectrum of DOX-g-PEI-g-PEG@AuNPs, as DOX was further grafted on PEI-g-PEG@AuNPs (Figure 4b). Furthermore, the appearance of P2p peak for AS1411-g-DOX-g-PEI-g-PEG@AuNPs was mainly due to the successful graft of AS1411 on DOX-g-PEI-g-PEG@AuNPs (Figure 4c). The size distribution of prepared nanoparticles was analyzed using DLS (Figure 5). Compared to PEI-g-PEG@AuNPs, the average hydration diameter slightly increased in DOX-g-PEI-g-PEG@AuNPs and further increased once the AS1411 was grafted on.
TEM was used to determine the morphology of the nanoparticles, and the images showed that all nanoparticles were uniform without aggregation (Figure 6). Due to the interactions between copolymers on the surface of AuNPs, the distance of AuNPs gradually increased. A cell viability test was used to determine the target property of the prepared DOX delivery system (Figure 7 and Figure 8). CCK-8 results (Figure 7) showed that 1) the A549 cell number decreased after culturing with AS1411-g-DOX-PEI-g-PEG@AuNPs over time and 2) the cell number decreased with increased concentration of nanoparticles. Compared to the free DOX group, the cell number increased, indicating that the toxicity was reduced.
Together with optical microscopy images (Figure 8), the results show that the cell number decreased after culturing with AS1411-g-DOX-g-PEI-g-PEG@AuNPs (Figure 8a−d) compared to the control group without adding nanoparticles (Figure 8e,f). Furthermore, the DOX release profile from prepared AS1411-g-DOX-g-PEI-g-PEG@AuNPs in PBS was investigated (Figure 9). The results show that the sustained release of DOX from functionalized nanoparticles caused the decrease in A549 cells, and the cumulative DOX release was about 63.5% ± 3.2% at 72 h.
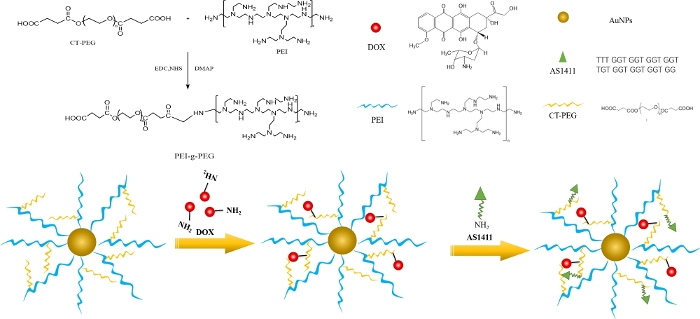
Figure 1: Schematic illustration of synthesis of PEI-g-PEG@AuNP, DOX-g-PEI-g-PEG@AuNP, and AS1411-g-DOX-g-PEI-g-PEG@AuNP. Please click here to view a larger version of this figure.
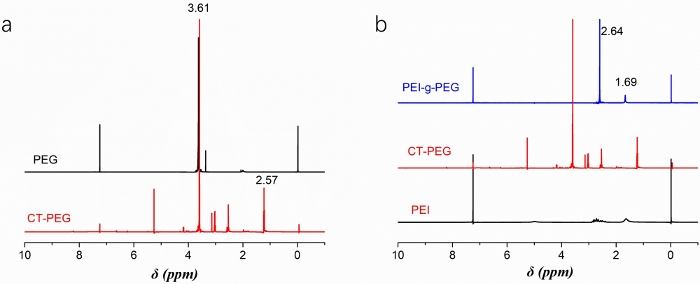
Figure 2: 1H NMR spectra of (a) synthesized CT-PEG polymer and (b) PEI-g-PEG copolymer. Please click here to view a larger version of this figure.
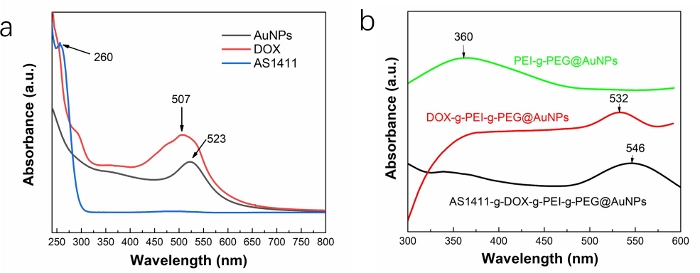
Figure 3: UV-vis spectra of (a) AuNPs, DOX, and AS1411, and (b) PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-g-PEI-g-PEG@AuNPs. Please click here to view a larger version of this figure.
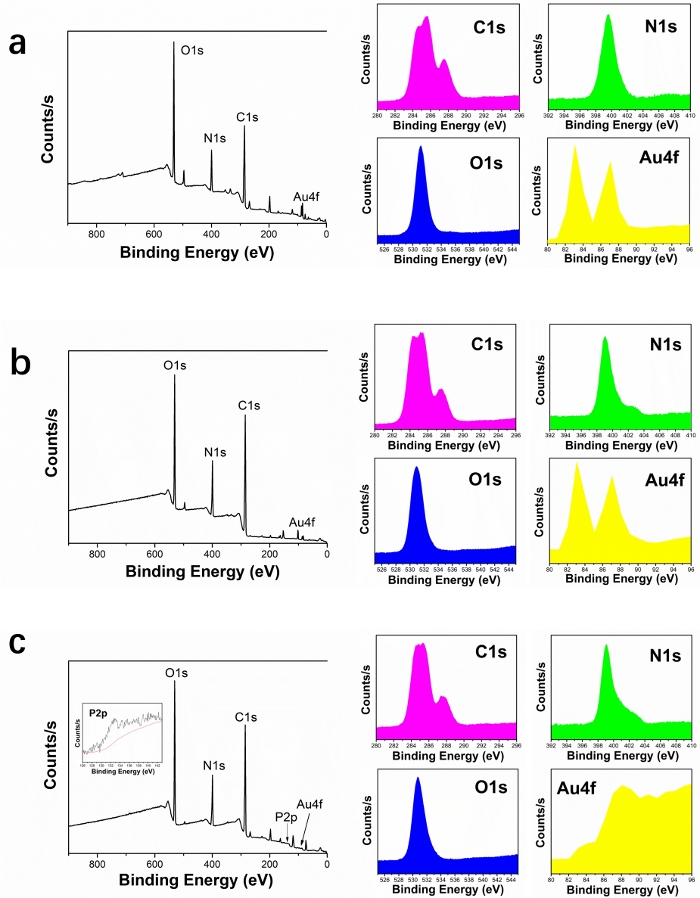
Figure 4: XPS spectra of (a) PEI-g-PEG@AuNPs, (b) DOX-g-PEI-g-PEG@AuNPs, and (c) AS1411-g-DOX-g-PEI-g-PEG@AuNPs. Please click here to view a larger version of this figure.
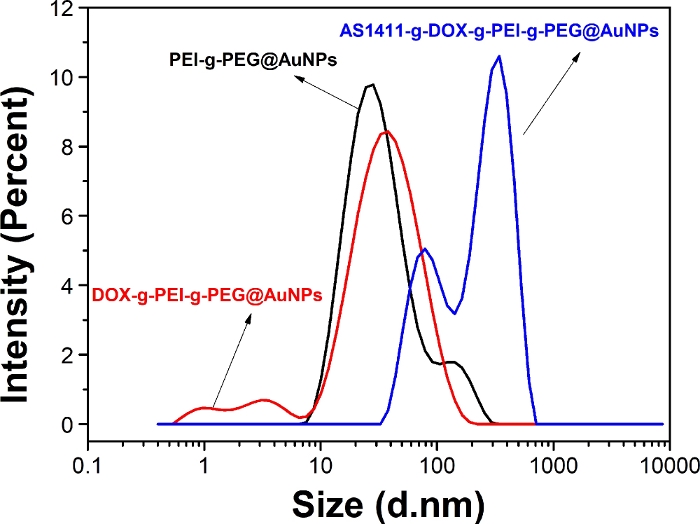
Figure 5: Size distribution of PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-g-PEI-g-PEG@AuNPs.
d.nm = average diameter of the nanoparticle. Please click here to view a larger version of this figure.
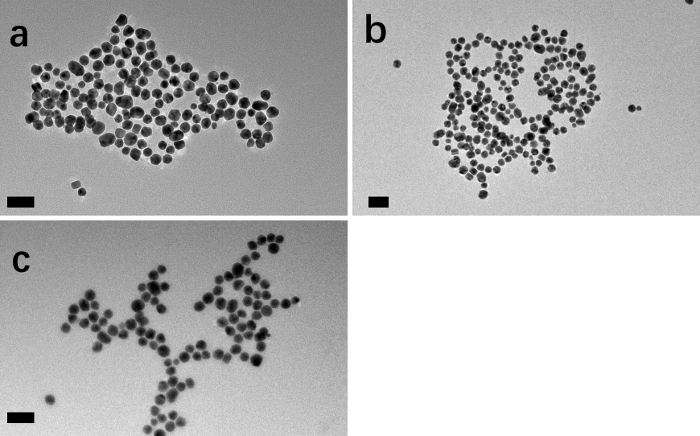
Figure 6: TEM images of (a) PEI-g-PEG@AuNPs, (b) DOX-g-PEI-g-PEG@AuNPs, and (c) AS1411-g-DOX-g-PEI-g-PEG@AuNPs.
Scale bars = 50 nm. Please click here to view a larger version of this figure.
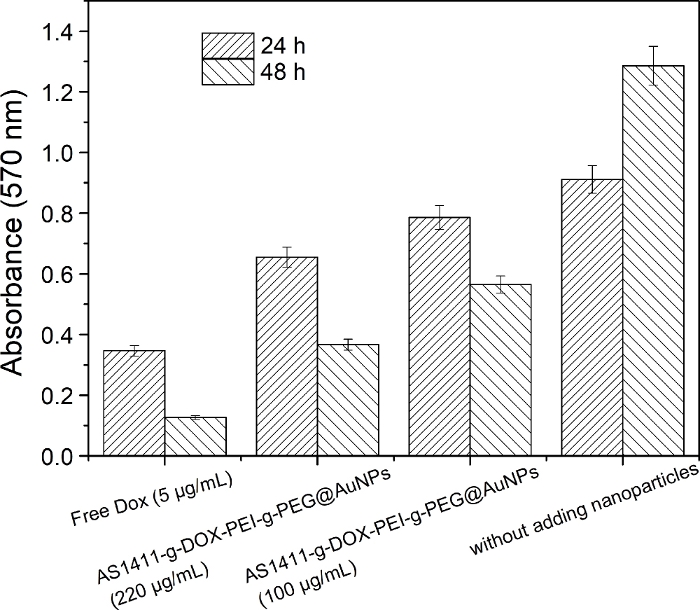
Figure 7: Optical density values at 570 nm (OD570) of A549 cells after culturing with AS1411-g-DOX-g-PEI-g-PEG@AuNPs (220 μg/mL and 110 μg/mL) for 24 h and 48 h, respectively.
Cells with free DOX and cells without adding nanoparticles are included as control groups. Please click here to view a larger version of this figure.
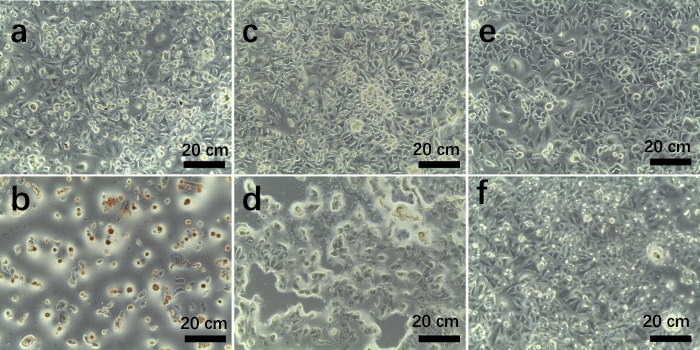
Figure 8: Optical microscopic images of A549 cells after culturing with AS1411-g-DOX-g-PEI-g-PEG@AuNPs at 220 μg/mL (a,b) and 110 μg/ml (c,d), or culturing without adding nanoparticles as control group (e,f) at 24 h (top panels) and 48 h (bottom panels). Please click here to view a larger version of this figure.
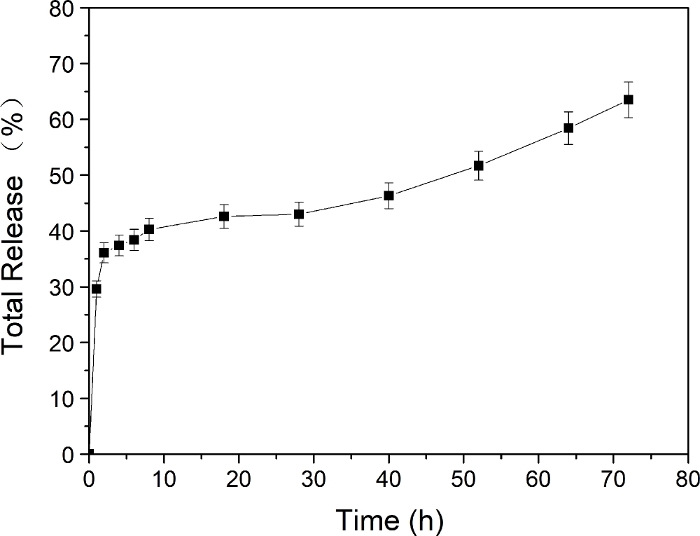
Figure 9: Release profile of DOX from AS1411-g-DOX-g-PEI-g-PEG@AuNPs in PBS for 72 h. Please click here to view a larger version of this figure.
Discussion
The 1H NMR spectrum (Figure 2) confirm the successful synthesis of CT-PEG copolymer and PEI-g-PEG copolymer. The molecular weights of PEG and PEI were 1,000 and 1,200, respectively. Additionally, the EDC/NHS catalytic system was used to synthesize PEI-g-PEG copolymer via amide reactions. It should be noted that if the molecular weights of PEG and PEI changed for synthesizing PEI-g-PEG copolymer, then the reaction time and catalytic system needs to be reevaluated. Also, the reaction condition for PEI-g-PEG copolymer coating on AuNPs needs to be further adjusted, mainly because the molecule weight and structure of PEG-g-PEI copolymer can influence the coating efficiency and diameter of AuNPs. Afterwards, the morphology of copolymer functionalized AuNPs can be changed, as well. The number of amino groups from PEI polymer can influence the structure of final PEI-g-PEG copolymer synthesis, and the crosslink action between PEI and CT-PEG will inevitably occur. Thus, step 2.4 needs to be performed carefully, and the PEI solution needs to be slowly added drop-by-drop. After the synthesis reaction, dialysis (steps 2.5 and 2.6) needs to be operated to remove the crosslinked copolymer and unreacted polymers.
Furthermore, DOX and AS1411 are sequentially functionalized on PEI-g-PEG@AuNPs via amide reactions, and the EDC/NHS catalytic system is used. It requires 3 days for each reaction (step 4.3 and step 5.3) here; however, if the reaction time requires less than 3 days, the functionalization efficiency will decrease. When requiring more than 3 days, the same result has been obtained. It should be noted that chemical EDC, NHS, and unconnected DOX or AS1411 can be removed through dialysis treatment (step 4.4 and step 5.4). UV-vis spectra and XPS are effective methods to investigate the successful functionalization of copolymer on nanoparticles, and consistent results have been obtained (Figure 3 and Figure 4).
Different from the characteristic UV-vis bands of AuNPs, DOX, and AS1411, unique peaks of PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-g-PEI-g-PEG@AuNPs are challenging to observe, due to the overlapping of each peak. Furthermore, we have performed a different method to fabricate DOX-g-PEI-g-PEG@AuNPs (synthesize DOX-g-PEI-g-PEG first, and accomplishment of functionalization on AuNPs); however, gold nanoparticles using such an approach has led to low DOX loading efficiency35,36. Thus, it should be noted that the method for synthesis of DOX-g-PEI-g-PEG@AuNPs in this work will ensure a sufficient DOX loading efficiency as well as further release profile. If an experiment does not take DOX loading efficiency of gold nanoparticles into account, there are other methods to obtain AS1411-g-DOX-g-PEI-g-PEG@AuNPs. These include synthesis of DOX-g-PEI-g-PEG or AS1411-g-DOX-g-PEI-g-PEG via an amide reaction first, then functionalization on gold nanoparticles. Thus, the method used here as well as the obtained copolymers can be applied to diverse medical applications, such as tissue engineering.
The size distribution and morphology of prepared nanoparticles can be investigated by DLS and TEM. DLS data (Figure 5) show that the hydration diameter nanoparticles vary from different coatings, and more than one peak appears for each sample. Regarding the structure of PEI-g-PEG (Figure 1), the normal distribution curve of DLS is not observed. It should be noted that the nanoparticles are dispersed in ultrapure water during the DLS test, different volume ratios are used, and multi-peaks still exist, due to interactions between copolymers on the surface of nanoparticles. Thus, TEM images are used to confirm the morphology of nanoparticles. TEM images of PEI-g-PEG@AuNPs, DOX-g-PEI-g-PEG@AuNPs, and AS1411-g-DOX-PEI-g-PEG@AuNPs are shown in Figure 6.
Based on different components of the surfaces of gold nanoparticles, distances between nanoparticles change. Besides, the prepared nanoparticles dispersed in water are stable according to zeta potential tests (-29 to 50 mV for different nanoparticles after time tests). The further functionalization of DOX and AS1411 (sections 4 and 5 of the protocol) do not influence the diameter of gold nanoparticles. It can be concluded that the UV-vis is an effective method to confirm DOX and AS1411 loaded on nanoparticles without using all testing methods.
The targeted property on cancer cells was investigated using A549 cells cultured with different concentrations of prepared AS1411 and DOX loaded AuNPs and without adding nanoparticles as control group. At the same time, the effects of free DOX on A549 cell viability was also tested (Figure 7 and Figure 8). Compared to the group without adding nanoparticles, the prepared AS1411-g-DOX-PEI-g-PEG@AuNPs lead to a decrease in A549 cells. However, while the concentration of nanoparticles decrease (100 µg/mL), the cells show better activity at 24 h compared to the free DOX group. This is mainly because the PEI-g-PEG copolymer has excellent cytocompatibility37 and the nonspecific toxicity of branched PEI polymer is ameliorated.
Finally, due to the targeted property of aptamer AS1411, the obtained nanoparticles are accumulated in cancer cells instead of healthy cells. Once the aptamer is recognized, the DOX is released to kill the cancer cells. The release profile of DOX from AS1411-g-DOX-PEI-g-PEG@AuNPs in PBS was recorded (Figure 9). This protocol demonstrates an approach for preparing aptamers and DOX grafted on copolymer modified AuNPs via a multi-step amide reaction. The synthesized nanoparticles have potential for cancer therapy applications.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (31700840); the Key Scientific Research Project of Henan Province (18B430013, 18A150049). This research was supported by the Nanhu Scholars Program for Young Scholars of XYNU. The authors would like to thank bachelor student Zebo Qu from the College of Life Sciences in XYNU for his helpful works. The authors would like to acknowledge the Analysis & Testing Center of XYNU for the use of their equipment.
Materials
| 4-Dimethylaminopyridine | Macklin | D807273 | |
| A549 cell | ATCC CCL-185TM | ||
| AS1411 | BBI Life Sciences Corporation | 5'-d (TTTGGTGGTGGTGGTTGTGGTGGTGGTGG) FL-AS1411 (fluorophore-labeled AS1411) | |
| Anhydrous Tetrahydrofuran (THF) | SinoPharm Chemical Reagent Co., Ltd | ||
| Cell counting kit-8 (CCK-8) | Sigma Aldrich | 96992-500TESTS-F | |
| Dichloromethane | Traditional Chinese medicine | 80047318 | |
| Diethyl ether (Et2O) | SinoPharm Chemical Reagent Co., Ltd | ||
| Dimethyl sulfoxide | Macklin | D806645 | |
| Dulbecco's modified Eagle's medium (DMEM) | Sigma Aldrich | ||
| Doxorubicin hydrochloride | Rhawn | R017518 | |
| Ether absolute | Traditional Chinese medicine | 80059618 | |
| Field Emission Transmission Electron Microscope | FEI Company | Tecnai G2 F 20 | |
| Gold(III) chloride trihydrate | Rhawn | R016035 | |
| Laser Particle-size Instrument | Malvern Instruments Ltd | ZetasizerNanoZS/Masterszer3000E | |
| Microplate Reader | Molecular Devices | SpectraMax 190 | |
| N-(3-Dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride | Macklin | N808856 | |
| N-Hydroxysuccinimide | Macklin | H6231 | |
| NMR software | Delta 5.2.1 | ||
| Nuclear Magnetic Resonance Spectrometer | JEOL | JNM-ECZ600R/S3 | |
| Origin 8.5 | OriginLab | ||
| Penicillin | Sigma Aldrich | V900929-100ML | |
| Phosphate-buffered saline | Sigma Aldrich | P4417-100TAB | |
| Poly(ethylene glycol) | Sigma Aldrich | 81188 | BioUltra, average Mn ~ 1000 |
| Poly (ethyleneimine) solution | Sigma Aldrich | 482595 | average Mn ~ 1200, 50 wt.% in H2O |
| Sodium borohydride, powder | Acros | C18930 | |
| Streptomycin | Sigma Aldrich | 85886-10ML | |
| Succinic anhydride | Traditional Chinese medicine | 30171826 | |
| Tetrahydrofuran | Traditional Chinese medicine | 40058161 | |
| Triethylamine | Traditional Chinese medicine | 80134318 | |
| UV/VIS/NIR Spectrometer | Lambda950 | Lambda950 | |
| X-ray Photoelectron Spectrometer | Thermo Fisher Scientific | K-ALPHA 0.5EV |
References
- Abad, J. M., Bravo, I., Pariente, F., Lorenzo, E. Multi-tasking base ligand: a new concept of AuNPs synthesis. Analytical and Bioanalytical Chemistry. 408 (9), 2329-2338 (2016).
- Siegel, R. L., Miller, K. D., Jemal, A. Cancer statistics, 2019. CA-A Cancer Journal for Clinicians. 69 (1), 7-34 (2019).
- Jang, B., Kwon, H., Katila, P., Lee, S. J., Lee, H. Dual delivery of biological therapeutics for multimodal and synergistic cancer therapies. Advanced Drug Delivery Reviews. 98, 113-133 (2016).
- Gansler, T., et al. Sixty years of CA: a cancer journal for clinicians. CA-A Cancer Journal for Clinicians. 60 (6), 345-350 (2010).
- Li, J., et al. Molecular Mechanism for Selective Cytotoxicity towards Cancer Cells of Diselenide-Containing Paclitaxel Nanoparticles. International Journal of Biological Sciences. 15 (8), 1755-1770 (2019).
- Zhao, D., et al. Precise ratiometric loading of PTX and DOX based on redox-sensitive mixed micelles for cancer therapy. Colloids Surfaces B: Biointerfaces. 155, 51-60 (2017).
- Blum, R. H., Carter, S. K. Adriamycin. A new anticancer drug with significant clinical activity. Annals of Internal Medicine. 80 (2), 249-259 (1974).
- de Lima, R. D. N., et al. Low-level laser therapy alleviates the deleterious effect of doxorubicin on rat adipose tissue-derived mesenchymal stem cells. Journal of Photochemistry Photobiology B. 196, 111512 (2019).
- Markowska, A., Kaysiewicz, J., Markowska, J., Huczynski, A. Doxycycline, salinomycin, monensin and ivermectin repositioned as cancer drugs. Bioorganic & Medicinal Chemistry Letters. 29 (13), 1549-1554 (2019).
- Songbo, M., et al. Oxidative stress injury in doxorubicin-induced cardiotoxicity. Toxicology Letters. 307, 41-48 (2019).
- Ewer, M. S., Ewer, S. M. Cardiotoxicity of anticancer treatments. Nature Reviews Cardiology. 12 (9), 547-558 (2015).
- Gabizon, A., Shmeeda, H., Barenholz, Y. Pharmacokinetics of pegylated liposomal Doxorubicin: review of animal and human studies. Clinical Pharmacokinetics. 42 (5), 419-436 (2003).
- Xu, X., Ho, W., Zhang, X., Bertrand, N., Farokhzad, O. Cancer nanomedicine: from targeted delivery to combination therapy. Trends in Molecular Medicine. 21 (4), 223-232 (2015).
- Feng, S., Nie, L., Zou, P., Suo, J. Effects of drug and polymer molecular weight on drug release from PLGA-mPEG microspheres. Journal of Applied Polymer Science. 132 (6), 41431 (2015).
- Chen, D., et al. Injectable Temperature-sensitive Hydrogel with VEGF Loaded Microspheres for Vascularization and Bone Regeneration of Femoral Head Necrosis. Materials Letters. 229, 138-141 (2018).
- Abadeer, N. S., Murphy, C. J. Recent Progress in Cancer Thermal Therapy Using Gold Nanoparticles. The Journal of Physical Chemistry C. 120 (9), 4691-4716 (2016).
- Riley, R. S., Day, E. S. Gold nanoparticle-mediated photothermal therapy: applications and opportunities for multimodal cancer treatment. WIREs Nanomedicine and Nanobiotechnology. 9 (4), 1449 (2017).
- Fratoddi, I., et al. Highly Hydrophilic Gold Nanoparticles as Carrier for Anticancer Copper(I) Complexes: Loading and Release Studies for Biomedical Applications. Nanomaterials (Basel). 9 (5), 772 (2019).
- Lee, S. M., et al. Drug-loaded gold plasmonic nanoparticles for treatment of multidrug resistance in cancer. Biomaterials. 35 (7), 2272-2282 (2014).
- Dreaden, E. C., Alkilany, A. M., Huang, X., Murphy, C. J., El-Sayed, M. A. The golden age: gold nanoparticles for biomedicine. Chemical Society Reviews. 41 (7), 2740-2779 (2012).
- Wei, T., et al. Anticancer drug nanomicelles formed by self-assembling amphiphilic dendrimer to combat cancer drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 112 (10), 2978-2983 (2015).
- Galluzzi, L., Buque, A., Kepp, O., Zitvogel, L., Kroemer, G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 28 (6), 690-714 (2015).
- Muddineti, O. S., Ghosh, B., Biswas, S. Current trends in using polymer coated gold nanoparticles for cancer therapy. International Journal of Pharmaceutics. 484 (1-2), 252-267 (2015).
- Hu, W., et al. Methyl Orange removal by a novel PEI-AuNPs-hemin nanocomposite. Journal of Environmental Sciences. 53, 278-283 (2017).
- Gu, F. X., et al. Targeted nanoparticles for cancer therapy. Nano Today. 2 (3), 14-21 (2007).
- Srinivasarao, M., Galliford, C. V., Low, P. S. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nature Reviews Drug Discovery. 14 (3), 203-219 (2015).
- Liu, Z., Shi, Y., Chen, Z., Duan, L., Wang, X. Current progress towards the use of aptamers in targeted cancer therapy. Chinese Science Bulletin (Chinese Version). 59 (14), 1267 (2014).
- Ghosh, P., Han, G., De, M., Kim, C. K., Rotello, V. M. Gold nanoparticles in delivery applications. Advanced Drug Delivery Reviews. 60 (11), 1307-1315 (2008).
- Vandghanooni, S., Eskandani, M., Barar, J., Omidi, Y. Antisense LNA-loaded nanoparticles of star-shaped glucose-core PCL-PEG copolymer for enhanced inhibition of oncomiR-214 and nucleolin-mediated therapy of cisplatin-resistant ovarian cancer cells. International Journal of Pharmaceutics. 573, 118729 (2020).
- Andghanooni, S., Eskandani, M., Barar, J., Omidi, Y. AS1411 aptamer-decorated cisplatin-loaded poly(lactic-co-glycolic acid) nanoparticles for targeted therapy of miR-21-inhibited ovarian cancer cells. Nanomedicine. 13 (21), 2729-2758 (2018).
- Palmieri, D., et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proceedings of the National Academy of Sciences of the United States of America. 112 (30), 9418-9423 (2015).
- Pichiorri, F., et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. Journal of Experimental Medicine. 210 (5), 951-968 (2013).
- Hou, S., McCauley, L. K., Ma, P. X. Synthesis and erosion properties of PEG-containing polyanhydrides. Macromolecular Bioscience. 7 (5), 620-628 (2007).
- Nie, L., et al. Injectable Vaginal Hydrogels as a Multi-Drug Carrier for Contraception. Applied Sciences. 9 (8), 1638 (2019).
- Zou, P., Suo, J., Nie, L., Feng, S. Temperature-responsive biodegradable star-shaped block copolymers for vaginal gels. Journal of Materials Chemistry. 22 (13), 6316-6326 (2012).
- Etrych, T., Šubr, V., Laga, R., Říhová, B., Ulbrich, K. Polymer conjugates of doxorubicin bound through an amide and hydrazone bond: Impact of the carrier structure onto synergistic action in the treatment of solid tumours. European Journal of Pharmaceutical Sciences. 58, 1-12 (2014).
- Safari, F., Tamaddon, A. M., Zarghami, N., Abolmali, S., Akbarzadeh, A. Polyelectrolyte complexes of hTERT siRNA and polyethyleneimine: Effect of degree of PEG grafting on biological and cellular activity. Artificial Cells, Nanomedicine, and Biotechnology. 44 (6), 1561-1568 (2016).

