Modeling Hepatitis B Virus Infection in Non-Hepatic 293T-NE-3NRs Cells
Summary
This manuscript describes a detailed protocol for Hepatitis B virus (HBV) infection in novel engineered 293T cells (293T-NE-3NRs, expressing human NTCP, HNF4α, RXRα and PPARα) and traditional hepatic cells (HepG2-NE, expressing human NTCP).
Abstract
HBV mainly infects human hepatocytes, but it has also been found to infect extrahepatic tissues such as kidney and testis. Nonetheless, cell-based HBV models are limited to hepatoma cell lines (such as HepG2 and Huh7) overexpressing a functional HBV receptor, sodium taurocholate co-transporting polypeptide (NTCP). Here, we used 293T-NE-3NRs (293T overexpressing human NTCP, HNF4α, RXRα and PPARα) and HepG2-NE (HepG2 overexpressing NTCP) as model cell lines. HBV infection in these cell lines was performed either by using concentrated HBV virus particles from HepG2.2.15 or co-culturing HepG2.2.15 with the target cell lines. HBcAg immunofluorescence for HBcAg was performed to confirm HBV infection. The two methods presented here will help us study HBV infection in non-hepatic cell lines.
Introduction
Hepatitis B affects the lives of more than 2 billion people and is one of the major threats to public health. Approximately 257 million people are chronically infected with hepatitis B virus (HBV) worldwide, causing a big burden to the society1. Hepatocytes are not the only cells infected by HBV and other cells in non-hepatic tissue, such as kidney and testis, are also infected by this virus2,3. Currently, HBV infection cell models are limited to human hepatocytes with only few non-hepatic cell line models. This hampers the study of HBV infection and HBV-related pathology of non-hepatic tissues. Here we present protocols to study HBV infection in non-hepatic 293T cells as well as in hepatoma-based cells.
Sodium taurocholate co-transporting polypeptide (NTCP) is a functional receptor for human hepatitis B and hepatitis D virus4. Hepatocyte nuclear factor 4α (HNF4α), retinoid X receptor α (RXRα) and peroxisome proliferator-activated receptor α (PPARα) are liver-enriched transcription factors restricting viral tropism of HBV. They have been verified to promote HBV pregenomic RNA synthesis and support HBV replication in a nonhepatoma cell line5. We constructed three different cell lines, HepG2 cell lines expressing NTCP (HepG2-NE), 293T cell line expressing NTCP (293T-NE) and 293T cell line expressing four host genes; NTCP, HNF4α, RXRα and PPARα (293T-NE-3NRs). Two methods for HBV infection were developed based on 293T-NE-3NRs (Figure 1). The first method uses inoculation in 293T-NE-3NRs with high viral genome equivalence (high GEq (150), DMSO and PEG8000) for 24 h. The second method employs co-culturing 293T-NE-3NRs with HepG2.2.15, which can produce HBV particles (low GEq (about 1.83) without DMSO and PEG8000), thus closely emulating natural conditions.
HepG2.2.15 cells are derived from the HepG2 line and chronically secrete infectious HBV, as well as subviral particles into the culture medium6,7. Cyclosporin A (CsA) is an immunosuppressant clinically used for the suppression of xenograft rejection. Studies have shown that CsA inhibits HBV entry into cultured hepatocytes by inhibiting the transporter activity of NTCP and blocking the binding of NTCP to large envelope proteins8.
HepG2-NE cells were used as positive control whereas CsA treated cells were used as negative control. By comparing with the positive and negative control groups, we can find out which host genes play a critical role in HBV infection. Additionally, through this mode of HBV infection, we can also find other novel mechanisms or host genes involved in HBV infection.
Protocol
Culture, collection of supernatants from HepG2.2.15 cells and HBV infection should be performed in biosafety level II (P2) or biosafety level III (P3) laboratory according to the biosafety guidance in the country. Laboratory safety practices should be followed to ensure the safety of laboratory personnel, and all should be vaccinated and detected HBsAb positive before performing HBV experiments. Observe the state of the cells at every step before proceeding to the next step. Human serum samples were used in accordance with the approval obtained by the Institutional Review Board of Shantou University Medical College.
1. HepG2.2.15 cell culture
NOTE: HepG2.2.15 cell supernatant, HepG2.2.15 cell supernatant concentrate and all the tips, flasks, plates, and tubes that come in contact with HepG2.2.15 should be disposed after being soaked in 2% virucide overnight.
- Remove the vial containing HepG2.2.15 cells from liquid nitrogen and thaw by gently swirling the vial in a 37 °C water bath.
NOTE: To reduce the possibility of contamination, keep the cap out of the water. Thawing should be rapid (approximately 2 min). - Remove the vial from the water bath as soon as the cells are thawed and disinfect it with 70% ethanol.
NOTE: From this point perform all steps under strict aseptic conditions. - Transfer the cells to a 25 cm2 tissue culture flask and add 4 mL of complete culture medium (DMEM containing 10% FBS, penicillin 100 U/mL, streptomycin 0.1 mg/mL). Incubate the cells at 37 °C in a humidified incubator with 5% CO2.
2. Collection of HepG2.2.15 cell culture supernatant
- Culture HepG2.2.15 cell in T25 flasks at 37 °C, 5% CO2 in a humidified incubator. Change the medium every 3 days.
- Collect the cell supernatant in a 50 mL centrifuge tube. Close the lid firmly and wrap it with a paraffin film.
- Store the HepG2.2.15 supernatant at -20 °C to keep HBV virus active.
3. Concentrating HepG2.2.15 supernatant
- Remove HepG2.2.15 supernatant from -20 °C and thaw at 4 °C.
- To remove the cell fragments, filter the HepG2.2.15 supernatant with a 0.45 µm membrane to a new 50 mL centrifuge tube. First, block the filter by filtering 10 mL of DMEM with 10% FBS before filtering the virus, then filter the cell culture supernatant.
- Add 14 mL of filtered supernatant to a virus concentrator column and close the lid. Centrifuge the column at 3,200 x g for 35 min in a horizontal spinning centrifuge. Collect the HepG2.2.15 supernatant concentrate (about 200 µL) into a 1.5 mL tube.
- Wash the column with DMEM once by adding 200 µL of DMEM and collect it into a 1.5 mL tube.
4. Detection of HBV DNA in supernatant concentrate
- Turn on the dry block heater and set the temperature to 100 °C.
- To ensure that the concentration of HBV DNA in the HepG2.2.15 supernatant concentrate is within the standard curve range, dilute the concentrate 20-fold by adding 10 µL of HepG2.2.15 supernatant concentrate to 190 µL of DMEM in a 1.5 mL microcentrifuge tube. Add 200 µL of HepG2.2.15 supernatant, negative control (HBV-negative serum from healthy donor), HBV positive control (HBV-positive serum from HBV patient) and four dilutions of the quantitative references provided in the kit (2.0 x 106, 2.0 x 105, 2.0 x 104, 2.0 x 103 GEq /mL), respectively to separate 1.5 mL microcentrifuge tubes.
- Add 450 µL of HBV DNA extraction buffer from the kit to all the above eight 1.5 mL tubes, vortex for 15 s and spin down for 10 s. Incubate at 100 °C for 10 min in dry block heater. Centrifuge at 12, 000 x g for 5 min at room temperature.
- Add 27 µL of PCR master mix and 3 µL of Taq polymerase enzyme to the PCR tubes placed on ice.
NOTE: PCR master mix provided in the kit contain the primers against the conserved region of HBV DNA. - Add 20 µL of centrifuged supernatant liquid to the PCR tubes placed on ice. Cover tightly and spin down for 10 s.
- Use the following conditions on a real-time PCR machine: 93 °C for 2 min, 10 cycles of 93 °C for 45 s followed by 55 °C for 60 s; then 30 cycles of 93 °C for 30 s followed by 55 °C for 45 s to carry out real-time PCR.
NOTE: The master mix provided in the commercial kit has primers against the conserved region of HBV DNA. - Export the Ct values obtained and generate a standard curve as shown in Table 1 and Figure 2.
- Based on the standard curve, calculate the HBV DNA concentration of HepG2.2.15 supernatant concentrate diluted 20x as shown in Table 2. Calculate the HBV DNA concentration of HepG2.2.15 supernatant concentrate (Table 2).
- Divide the HepG2.2.15 supernatant concentrate in aliquots and store at -80 °C until use.
5. In vitro infection of HepG2-NE and 293T-NE-3NRs cells
- Prepare the following infection medium in DMEM/F-12: 10% FBS, 4 mM glutamine supplement, 0.1 mM NEAA, 1 mM Sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, 1 μg/mL puromycin, 40 ng/mL dexamethasone, 20 μg/mL hydrocortisone and 5 μg/mL insulin. Store at 4 °C.
NOTE: These NTCP positive cells are puromycin-resistant9. - Add 0.1% gelatin to 5 wells of 48 well-plate, 200 μL to each well. Incubate the plate at 37 °C for 2 h, discard the supernatant. Incubate the plate at 37 °C for another 2 h.
- Seed HepG2-NE at a density of 1 x 104 cells in 2 wells each. Seed 293T-NE-3NRs cells at a density of 1 x 104 cells in 2 wells each (all-trans retinoic acid at 1 μmol/L and clofibric acid at 1 mmol/L final concentrations were added to the medium to activate the RXRα and PPARα nuclear hormone receptors, respectively). Seed 293T-NE in 1 well at a density of 1 x 104 cells.
NOTE: Passage the cells at sub-confluence using 0.25% trypsin. Count the cell number and then seed the cells to an appropriate density. - Incubate the cell at 37 °C, 5% CO2 in a humidified incubator for 24 h. Observe the cells after 24 h and proceed with infection if the cells are healthy.
- Prepare the infection complex as mentioned in Table 3, add HBV (HepG2.2.15 supernatant concentrate), DMSO, PEG8000 and infection medium to 1.5 mL microcentrifuge tube. Prepare CsA stock by dissolving it in DMSO to a concentration of 5 mM and keep the stock at -20 ºC. The working solution of CsA is 5 μM. Seed 1×104 cells per well of the 48-well plate. Add 100 μL of HepG2.2.15 supernatant concentrate (containing HBV 1.5063 ×107 GEq/mL) to infect cells at 150 GEq/cell.
- Add 500 μL of the infection complex to each well and incubate the cells at 37 °C, 5% CO2 in humidified incubator for 24 h.
- Wash the cells twice with 500 μL of PBS before changing the medium. Add 1 mL of the medium in each well. If the cell state is poor and some cells are floating, change the medium directly. This is day 1 of infection. Change the medium gently every 2 days.
- On day 11, wash the cells twice with 500 μL of PBS and fix the cells with ice cold methanol for immunofluorescence.
6. HepG2.2.15 co-culture with HepG2-NE / 293T-NE-3NRs / 293T-NE
- Add 1 mL/well of 0.1% gelatin to 5 wells of 6 well-plate. Incubate the plate at 37 °C for 2 h and discard the supernatant. Incubate the plate again for another 2 h at 37 °C to completely dry the well.
- Seed 1 x 105 cells/well of HepG-NE into 2 wells, 293T-NE-3NRs into 2 wells, and 293T-NE into 1 well of the plate. Seed 1 x 105 cells/well of HepG2.2.15 onto five membrane inserts (24 mm, 0.4 μm pore diameter, uncoated) in a separate 6 well plate. Incubate the cells at 37 °C, 5% CO2 in a humidified incubator for 24 h.
- After 24 h, if the cells are all adherent and in good state, prepare for co-culture. Discard the medium in the 6-well-plate and cell membrane inserts. Place the membrane inserts with HepG2.2.15 into the 6 well plate seeded with HepG-NE, 293T-NE-3NRs and 293T-NE by tweezers. Incubate the cell at 37 °C, 5% CO2 in a humidified incubator, change the medium every 3-4 days.
NOTE: Set groups as following: Well 1: 293T-NE co-culture with HepG 2.2.15; Well 2: HepG2-NE co-culture with HepG 2.2.15; Well 3: 293T-NE-3NRs co-culture with HepG2.2.15; Well 4: HepG2-NE co-culture with HepG 2.2.15 (add CsA); Well 5: 293T-NE-3NRs co-culture with HepG2.2.15 (add CsA); Add the complete medium to well number 1, 2 and 3, add the medium with 5 µM CsA to well number 4 and 5 well, about 9 mL for each well, make sure the media are in contact in order for virus particles to migrate from transwells to infect cells. - After 10 days, remove the membrane inserts, add PBS to the 6-well-plate and wash gently twice, fix the cells with ice cold methanol for immunofluorescence.
7. Perform immunofluorescence for HBcAg
- Fix the cells with ice cold methanol at -20 °C for more than 3 h. Discard the methanol. Wash the cells with PBS for 10 min at room temperature on shaker, repeat for 3 times.
- Add 5% BSA (100 μL/48-well-plate, 500 μL/6-well-plate), shake on horizontal shaker for 1 h at 40 rpm and room temperature.
- Add the HBcAg primary antibody solution (primary antibody: 5% BSA solution =1:200, 100 μL / 48-well-plate, 500 μL / 6-well-plate), shake overnight at 4 °C.
- Wash wells with PBST (PBS with 0.1% tween 20), for 10 min at room temperature on shaker, repeat for 3 times.
- Add the second antibody solution (594 labeled antibody raised in goat against rabbit IgG: PBS = 1:1,000, 100 μL /48-well-plate, 500 μL / 6-well-plate), cover the plate with foil and shake for 2 h at room temperature.
- Wash again with PBST for three times, shake for 10 min each.
- Add 1 μg/mL DAPI, shake for 2 min. Wash twice with PBST on a shaker for 5 min each. Discard PBST. Add PBS and observe under immunofluorescence microscope.
Representative Results
We constructed pSIN-NTCP-EGFP plasmid expressing NTCP and EGFP fusion and with puromycin resistance. The plasmid was transfected into HepG2 and 293T cells to construct stable cell lines HepG2-NE and 293T-NE expressing NTCP and EGFP. Plasmids (pSIN-HNF4α, pSIN-RXRα, pLV-PPARα-puro-flag) with puromycin resistance and expression were transfected into 293T-NE cells to construct a stable cell line expressing 4 host genes9. The expression of NTCP-EGFP can be observed by green fluorescence, and verified by qPCR and western blot (data not shown, but see previous work 9).
Primary antibody of HBcAg and DAPI incubated with fixed cells showed a distinct staining when observed using 10x objective on a fluorescent inverted microscope. Nuclear localization was confirmed by staining with DAPI (Figure 3 and Figure 4-the third column). NTCP-EGFP is expressed on the cell membrane and can be located by the green fluorescence (Figure 3 and Figure 4-the second column). The expression sites of HBcAg can be located by red fluorescence (Figure 3 and Figure 4-the first column). When DAPI, NTCP and HBcAg are in the same cell, it means that the cell has been successfully infected with HBV (Figure 3 and Figure 4-the last column).
The CsA group was the negative control. CsA prevented HBV from entering cells by blocking NTCP. As a positive control, infected HepG2-NTCP-EGFP cells can express HBcAg. HBcAg was detected in 293T-NE-3NRs cells but not in 293T-NE cells, indicating NTCP is not the only factor essential for HBV infection in 293T. The immunofluorescence of cells infected with HepG2.2.15 supernatant concentrate was the same as that of cells co-cultured with HepG2.2.15 cell. It showed that HBcAg was expressed in 293T-NE-3NRs and HepG2-NE cells, this suggests that the four host genes (NTCP, HNF4α, RXRα and PPARα) facilitates HBV infection. But signals for HBcAg in 293T-NE-3NRs were lower compared to HepG2-NE, indicating HBV infection may be also affected by other host factors.
| Quantitative reference | Quantitative reference | Quantitative reference | Quantitative reference | Negative control | HBV positive control | |
| 1 | 2 | 3 | 4 | |||
| Ct | 20.1527 | 17.6647 | 14.5816 | 12.0409 | none | 12.3865 |
| GEq/mL | 2*10^3 | 2*10^4 | 2*10^5 | 2*10^6 | —— | —— |
Table 1: Ct values for the Quantitative references.
| HepG 2.2.15 supernatant |
1/20 HepG 2.2.15 concentrate |
HepG 2.2.15 concentrate |
|
| Ct | 17.716 | 13.1556 | —— |
| GEq/mL | 1.65*10^4 | 7.53*10^5 | 1.5*10^7 |
Table 2: Ct values for HBV DNA obtained from HepG2.2.15 supernatant. Values represent Ct values for HBV DNA obtained from original supernatant, 1/20 dilution or its concentrate.
| 293T-NE | HepG2-NE | 293T-NE-3NRs | |||
| HBV | HBV | HBV+CsA | HBV | HBV+CsA | |
| HBV GEq/cell | 150 | 150 | 150 | 150 | 150 |
| HBV µL | 100 | 100 | 100 | 100 | 100 |
| CsA µL (5 µM) | 0 | 0 | 0.5 | 0 | 0.5 |
| DMSO µL | 10 | 10 | 9.5 | 10 | 9.5 |
| 40 % PEG8000 µL | 50 | 50 | 50 | 50 | 50 |
| Infection medium µL | 340 | 340 | 340 | 340 | 340 |
| Total µL | 500 | 500 | 500 | 500 | 500 |
Table 3: Infection complex. The total volume used was 500 µL. Cyclosporine A was used as a negative control.
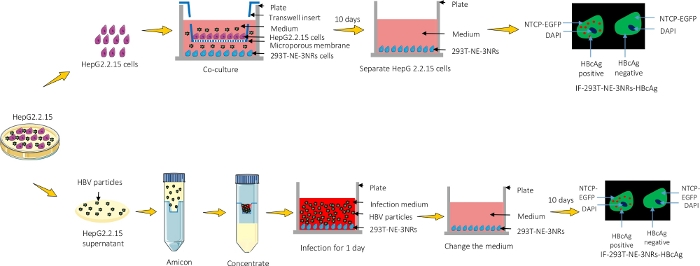
Figure 1: A graphical representation of the protocol. HepG2.2.15 were either co-cultured with 293T-NE-3NRs and infection was observed using immunofluorescence microscopy or the supernatant containing viral particles was concentrated and introduced into 293T-NE-3NRs cultured cells. Please click here to view a larger version of this figure.
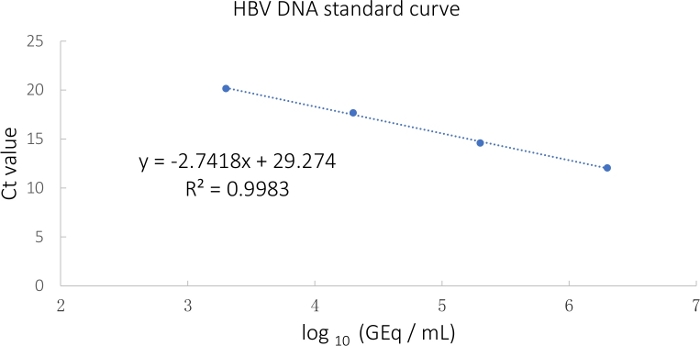
Figure 2: HBV standard curve calculated from the Ct values. Please click here to view a larger version of this figure.
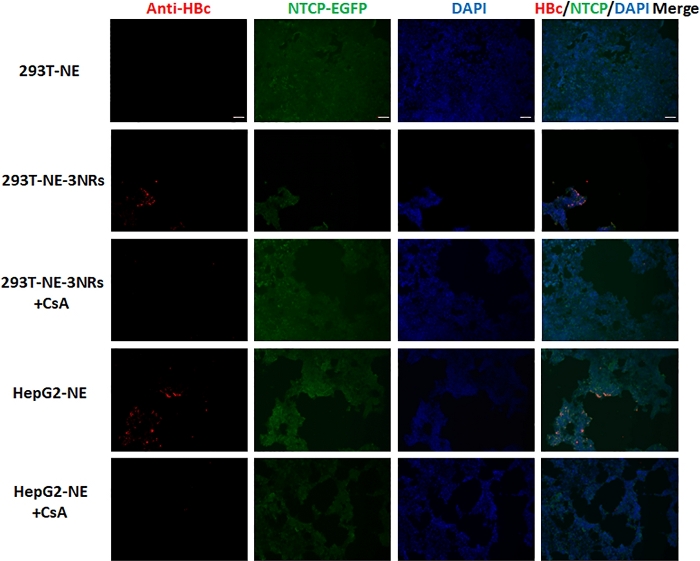
Figure 3: HepG2.2.15 supernatant concentrate was used to infect 293T-NE, 293T-NE-3NRs and HepG2-NE cells with 150 GEq per cell. HBcAg expression was identified by immunofluorescence assay. HBcAg was expressed in 293T-NE-3NRs and HepG2-NE cells, whereas, no expression was observed in these cells treated with CsA (HBV entry inhibitor) and 293T-NE cells. Scale bar = 100 µm. Please click here to view a larger version of this figure.
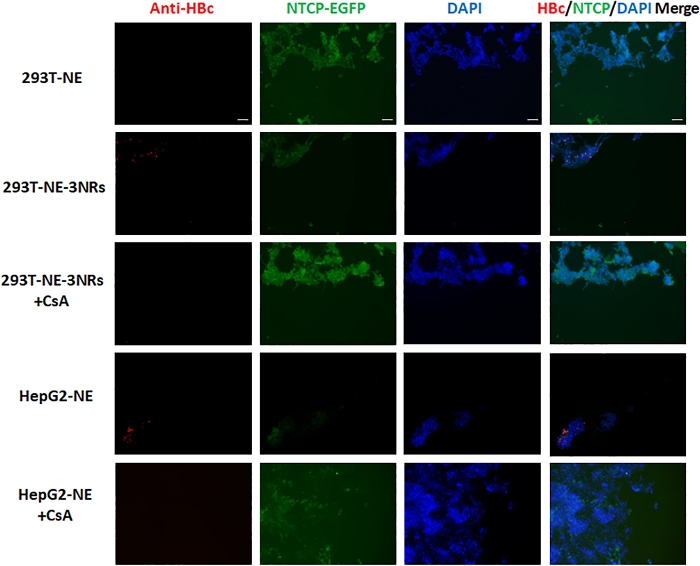
Figure 4: 293T-NE, 293T-NE-3NRs and HepG2-NE co-cultured with HepG2.2.15 cells. HBcAg expression was identified by immunofluorescence assay. HBcAg was expressed in 293T-NE-3NRs and HepG2-NE cells, whereas, no expression was observed in these cells treated with CsA (HBV entry inhibitor) and 293T-NE cells. Scale bar = 100 µm. Please click here to view a larger version of this figure.
Discussion
Here, we introduce protocols for HBV infection in non-hepatic 293T-NE-3NRs and hepatoma-based HepG2-NE cells. 293T-NE-3NRs were suitable for HBV infection at both high and low GEq. The following critical steps need to be taken into consideration while using our protocol. The cell status is an important factor for a successful infection. The infection medium must be changed timely after the initial period of HBV infection. 293T-NE-3NRs cells are typically fragile following infection with high viral titers. Therefore, these cells must be washed gently to avoid detaching the cells after 24 h of infection. When using membrane inserts for culture, we must make sure the initial seeding density is appropriate, and the chamber is sufficiently submerged in media to ensure efficient attachment of viral particles to the 293T-NE-3NRs cells. To detect HBcAg, we must wash the cells gently before fixation to avoid false positive signals. Care must be taken during washing step as the 293T cells tend to detach from the bottom.
Infecting 293T-NE-3NRs by HBV is not as easy as NTCP-expressing HepG2. Unlike HepG2, these cells are easy to detach from the bottom of the plate during culture. And these cells are more fragile than HepG2 after infection using DMSO and PEG8000 or co-culture with HepG2.2.15 for 10 days without passaging. Therefore, we need to coat the plate by 0.1% gelatin and seed appropriate cell number into the plate. More care should be taken to prevent the cells detaching during changing the medium, washing or fixing the cells.
Compared to traditional HBV infection methods, we adopted an engineered 293T cell line, 293T-NE-3NRs, as infection model and co-cultured it with HepG2.2.15. Interestingly, 293T-NE-3NRs cells were successfully infected with HBV even at low GEq which mimics natural physiological conditions more accurately. Modeling HBV infection in 293T-NE-3NRs is a useful complement to the traditional one due to the nonhepatic background of these cells. Application of the method may help us to identify the novel host factors although the infection efficiency of this method is not as high as the traditional one. We believe that the in vitro models presented in this manuscript will help facilitate new discoveries in the field.
A limitation to this method is the infection efficiency. The in vitro HBV infection is not as easy as that in vivo. Current in vitro models for HBV are limited due to their poor susceptibility to HBV infection, where extremely high inocula are required to initiate infection and there is no obvious viral spread10. This is in stark contrast to in vivo observations where a single HBV virion can infect a large portion of hepatocytes in mere weeks11,12. Thus, the identification of novel host factors which affect HBV replication is greatly beneficial to furthering our understanding of HBV and host interactions. Moreover, by mimicking the in vivo liver or kidney development, functional organoids can be generated that would more closely recapitulate the physiological functioning of a human liver or kidney13,14. This would greatly enhance our understanding of HBV infection.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81870432 and 81570567 to X.L.Z.), (No. 81571994 to P.N.S.), Research Fund for International Young Scientists (No. 81950410640 to W.I.); The Li Ka Shing Shantou University Foundation (No. L1111 2008 to P.N.S.). We would like to thank Prof. Stanley Lin from Shantou University Medical College for useful advice.
Materials
| 0.45μm membrane filter | Millex-HV | SLHU033RB | Filter for HepG 2.2.15 supernatant |
| 293T-NE | Laboratory construction | —— | Cell model for HBV infection |
| 293T-NE-3NRs | Laboratory construction | —— | Cell model for HBV infection |
| 594 labeled goat against rabbit IgG | ZSGB-BIO | ZF-0516 | For immunofluorescence assay,second antibody |
| 6-well plate | BIOFIL | TCP010006 | For co-culture |
| Amicon Ultra 15 ml | Millipore | UFC910008 | For concentration of HepG 2.2.15 supernatant |
| BSA | Beyotime | ST023 | For immunofluorescence assay |
| Cyclosporin A | Sangon biotech | 59865-13-3 | inhibitor of HBV infection |
| DAPI | Beyotime | C1006 | For nuclear staining |
| Diagnostic kit for Quantification of Hepatitis B Virus DNA(PCR-Fluorescence Probing) | DAAN GENE | 7265-2013 | For HBV DNA detection |
| DMEM | HyClong | SH30243.01 | For culture medium |
| DMSO | Sigma-Aldrich | D5879 | For improvement of infection efficiency |
| Fetal bovine serum(FBS) | CLARK Bioscience | FB25015 | For culture medium |
| Fluorescence microscope | ZEISS | Axio observer Z1 | For immunofluorescence assay |
| HepG2-NE | Laboratory construction | —— | Cell model for HBV infection |
| HBcAg antibody | ZSGB-BIO | ZA-0121 | For immunofluorescence assay, primary antibody |
| PBS | ZSGB-BIO | ZLI-9062 | For cell wash |
| PEG8000 | Merck | P8260 | For infection medium |
| Penicillin-Streptomycin-Glutamine | Thermo Fisher | 10378016 | For culture medium |
| Transwell | CORNING | 3412 | For co-culture |
| Tween 20 | sigma-Aldrich | WXBB7485V | For PBST |
| Virkon | Douban | 6971728840012 | Viruside |
References
- World Health Organization. Global hepatitis report, 2017. World Health Organization. , (2017).
- Yoffe, B., Burns, D. K., Bhatt, H. S., Combes, B. Extrahepatic hepatitis B virus DNA sequences in patients with acute hepatitis B infection. Hepatology. 12, 187-192 (1990).
- Mason, A., Wick, M., White, H., Perrillo, R. Hepatitis B virus replication in diverse cell types during chronic hepatitis B virus infection. Hepatology. 18, 781-789 (1993).
- Yan, H., et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 1, 00049 (2012).
- Tang, H., McLachlan, A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proceedings of the National Academy of Sciences of the United States of America. 98, 1841-1846 (2001).
- Sells, M. A., Chen, M. L., Acs, G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proceedings of the National Academy of Sciences of the United States of America. 84, 1005-1009 (1987).
- Sells, M. A., Zelent, A. Z., Shvartsman, M., Acs, G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. Journal of Virology. 62, 2836-2844 (1988).
- Watashi, K., et al. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology. 59, 1726-1737 (2014).
- Yang, X., et al. Defined host factors support HBV infection in non-hepatic 293T cells. Journal of Cell and Molecular Medicine. 24, 2507-2518 (2020).
- Xia, Y., et al. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. Journal of Hepatology. 66, 494-503 (2017).
- Ortega-Prieto, A. M., Cherry, C., Gunn, H., Dorner, M. In Vivo Model Systems for Hepatitis B Virus Research. ACS Infectious Diseases. 5, 688-702 (2019).
- Liang, T. J. Hepatitis B: the virus and disease. Hepatology. 49, 13-21 (2009).
- Nie, Y. Z., et al. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 35, 114-123 (2018).
- Nishinakamura, R. Human kidney organoids: progress and remaining challenges. Nature Reviews Nephrology. 15, 613-624 (2019).

