Super-Resolution Imaging to Study Co-Localization of Proteins and Synaptic Markers in Primary Neurons
Summary
This protocol shows how to employ super-resolution microscopy to study protein co-localization in primary neuronal cultures.
Abstract
Synapses are the functional elements of neurons and their defects or losses are at the basis of several neurodegenerative and neurological disorders. Imaging studies are widely used to investigate their function and plasticity in physiological and pathological conditions. Because of their size and structure, localization studies of proteins require high-resolution imaging techniques. In this protocol, we describe a procedure to study in primary neurons the co-localization of target proteins with synaptic markers at a super-resolution level using structured illumination microscopy (SIM). SIM is a patterned-light illumination technique that doubles the spatial resolution of wide-field microscopy, reaching a detail of around 100 nm. The protocol indicates the required controls and settings for robust co-localization studies and an overview of the statistical methods to analyze the imaging data properly.
Introduction
The understanding and view of the synapse has changed enormously since its first description by Foster and Sherrington in 18971. Since then, our knowledge of neuronal communication and the molecular processes behind it has grown exponentially2. It has become clear that synapses can be thought of as a two-compartment system: a pre-synaptic compartment containing vesicles for the release of neurotransmitters and a post-synaptic compartment with receptors3. This simplistic view, in the past twenty years, has evolved into a complex network of the proteins required to transduce transmitter binding into signaling4.
The gains in the understanding are partially due to super-resolution techniques that overcame the diffraction limit of conventional light microscopy to suit the dimension of synapses better5,6,7,8,9,10. Due to the diffraction limit, an optical microscope cannot reach a resolution above 200 nm laterally11,12. To bypass this limit, super-resolution techniques were created, using different approaches and reaching different sub-diffraction limit resolutions: SIM, STED (Stimulated Emission Depletion Microscopy), PALM (PhotoActivated Localization Microscopy) and STORM (Stochastic Optical Reconstruction Microscopy)13,14. SIM doubles the spatial resolution of laser-based wide-field microscopy systems by inserting a diffraction grating into the excitation beam path15. The movable grating diffracts the laser beams to create a known illumination pattern, usually stripes. This purposely structured light pattern is superimposed to the unknown spatial distribution of the fluorescent dye (of the sample). The interference fringes formed by the two patterns encode for otherwise indistinguishable fine details with normal wide-field microscopy. The final super-resolved image is obtained by combining and decoding with mathematical methods several raw images of the same sample obtained by the translations and rotations of the diffraction grating. The resolution of the super-resolved images reaches 100 nm in the lateral and 500 nm in the axial directions for 2D-SIM15 or 100 nm in the lateral and 250 nm in the axial directions for 3D-SIM16.
The new understanding of the synapse is even more important in the light of the many neurological disorders where synaptic dysfunction plays a major role in onset and progression17,18. Alzheimer’s disease, Down syndrome, Parkinson’s disease, prion diseases, epilepsy, autism spectrum disorders and fragile X syndrome among others have been linked to abnormalities in synaptic composition, morphology and function19,20,21,22.
Recently, using a set of SUMO-specific antibodies, we used SIM to show co-localization in primary hippocampal neurons of the SUMO proteins with the pre- and post-synaptic markers synaptophysin and PSD95 at super-resolution level23. This enabled us to confirm biochemical and confocal microscopy evidence of SUMO localization in neurons.
Here, we describe a protocol to study the localization of proteins in mouse hippocampal primary neurons. At the same time, this protocol may be adapted to different types of primary neuronal cultures.
Protocol
1. Primary cultures
- Culture mouse hippocampal primary neurons in chambered coverslips (such as Ibidi µ-Slide 8 Well or Nunc Lab-Tek Chambered Coverglass) that match the objective requirement for #1.5 (0.17 mm) coverslip thickness.
- Coat chambered coverslips with 100 µL of poly-L-lysine (100 µg/mL).
- The next day, wash the chambered coverslips twice with sterile phosphate-buffered saline (PBS).
- To obtain mouse primary neurons, isolate hippocampi from P1-P4 pups23.
- Place dissected hippocampi in 10 mL of Dissection Media (Table 1) and let them deposit at the bottom of the tube.
- Using a sterile pipette, carefully remove the Dissection Media, leaving the hippocampi undisturbed at the bottom of the tube.
- Add 10 mL of Media 1 (Table 1) to the hippocampi and incubate for 30 minutes at 37 °C.
- Using a sterile pipette, carefully remove Media 1, leaving the hippocampi undisturbed at the bottom of the tube.
- Add 10 mL of Media 2 (Table 1) and leave the (capped) centrifuge tube under the hood horizontally for 45 minutes.
- Let the centrifuge tube stand vertically to allow the tissue to settle at the bottom of the tube.
- Using a sterile pipette, carefully remove Media 2, leaving the hippocampi undisturbed at the bottom of the centrifuge tube.
- Add 2 mL of Media 3 (Table 1).
- Using a p1000 pipette with a filtered tip, mechanically dissociate cells from the tissue.
- Transfer the supernatant, in which are located the isolated neurons, to a 15 mL centrifuge tube.
- Centrifuge the cell suspension for 2 min at 300 x g at room temperature (RT).
- After centrifugation, cells are located at the bottom of the centrifuge tube. Using a sterile pipette discard the supernatant.
- Resuspend cells in 1 mL of Media 4.
- Use a 70 µm filter to eliminate undissociated cells.
- Count viable cells in a Bürker chamber by adding 1 µL of 0.4 % Trypan blue solution to 19 µL of the cell suspension.
- Plate cells at 70,000 cells/well in a volume of 200 µL per well.
- Allow the cells to attach for 2 h in a humidified incubator at 37 °C and 5% CO2.
- Take out the chambered coverslips from the incubator and carefully replace the medium with 200 µL of Culture Media.
- Leave the chambered coverslips in a humidified incubator at 37 °C and 5% CO2.
- Replace one third of the medium with fresh culture media every 5-7 days.
- Wait until hippocampal primary neurons are fully matured (12-14 days after plating) to perform co-localization studies.
2. Immunofluorescence staining
- Take the chambered coverslips from the incubator.
- Remove the medium.
- Quickly wash the wells with 200 µL of PBS.
- Add 4% paraformaldehyde (PFA) in PBS (200 µL/well) to neurons to fix them quickly.
- Incubate the cells for 15 min at RT.
- Remove the PFA solution.
- Permeabilize the cells by adding PBS with 0.2% Triton X-100 (200 µL/well).
- Incubate for 1 min at RT.
- Remove the solution and incubate the samples with 1% bovine serum albumin (BSA) in PBS (200 µL/well) for 1 h at RT to passively cover all free binding surfaces of the plate with an irrelevant protein for the analysis. A BSA-based blocking buffer without Triton X-100 reduces the antibody background more efficiently than the same buffer with 0.2% Triton X-100.
- Remove the solution.
- Add the primary antibody of choice diluted in a PBS solution containing 1% BSA and 0.2% Triton X-100 (120-200 µL/well, depending on the antibody dilution and availability). Incubate the samples for 2 h.
- As a negative control, do not add any primary antibody to one of the wells. Multiple antibodies of different species against different targets may be used at the same time. Use an antibody against MAP2 (a neuronal marker) raised in chicken, an antibody against either PSD95 or synaptophysin raised in mouse, and an antibody against a target protein raised in rabbit. This allows three-color SIM analyses.
- Quickly wash the wells three times with PBS (200 µL/well).
- Add secondary antibodies (dyLight and Alexa secondary antibodies can both be used) diluted in a PBS solution containing 1% BSA and 0.2% Triton X-100 (200 µL/well). Incubate the samples for 1 h at RT.
- Quickly wash the wells three times with PBS (200 µL/well).
- Add Hoechst dye at a concentration of 1 μg/mL diluted in PBS (200 µL/well) to stain nuclei. Incubate the samples for 10 minutes at RT.
- Quickly wash the wells twice with PBS.
- Mount cells using a SIM-compatible mountant. Use 10 µL/well of ProLong Glass Antifade Mountant.
- Cover and protect the cells with a coverglass (e.g., a round coverglass with a diameter of 8 mm). Square ones can also be used.
- Store the chambered coverslips at RT and wait at least 48 h before acquiring images. Diamond Glass requires at least two days of curing before super-resolution acquisitions.
3. Antibody specificity control
NOTE: Use two strategies to assure antibody specificity. The first strategy is to use at least two different antibodies targeting the same substrate. The second strategy is antibody neutralization by incubation with the purified protein target or the epitope used to raise the antibody.
- Incubate the antibody of choice with five times excess of the recombinant target or epitope for 1 h at RT in 1% BSA in PBS.
- After the incubation, use the neutralized antibody at the usual concentration for staining as described above from 2.11.
4. Microscope calibration
NOTE: We routinely use an N-SIM Super-Resolution Microscope System manufactured by Nikon for the super-resolution studies. However, several other companies also offer super-resolution microscopes in their catalogues. Although specific indications for Nikon’s N-SIM system are described, the instructions that follow can be generalized to other systems. Before the acquisition of SIM images, the system requires a proper calibration with specific sub-resolution size fluorescent beads. An example is the TetraSpeck microspheres. These beads are stained with different fluorescent dyes to allow the calibration of different lasers with one sample.
- In a water bath sonicate around 1.8 x 108 fluorescent microspheres for 10 minutes. Nikon’s N-SIM system requires a sparsely populated multicolor beads sample for the calibration. This could differ for other systems that require a dense single layer of sub-resolution size fluorescent beads. Adjust the number of fluorescent particles accordingly.
- Dilute the fluorescent microspheres 1:500 in double distilled water.
- Sonicate a second time for an additional 10 minutes.
- Pipet 15 µL of the diluted beads into a well of a chambered coverslip.
- Let the solution dry for 5 minutes at RT.
- Add 10 µL of the mounting solution and place an 8 mm coverslip on top.
- Wait at least 48 hours to allow proper curing.
- Turn on the microscope and lasers.
- Let the system warm up to reach the thermal equilibrium of all microscope components. N-SIM Super-Resolution Microscope System requires at least 3 hours.
- Select the 100x objective.
- Start the calibration by aligning the lasers to the center of the diffraction grating block. In the N-SIM system, a micrometer knob and a dedicated camera allow centering of the light beams to the target.
- Insert the chambered coverslip in the microscope for viewing. Set the system to the chambered coverslip thickness by adjusting the objective correction collar. NIS software, the proprietary software provided with N-SIM Super-Resolution Microscope systems, has an automatic function to regulate correction collars.
- Adjust grating block focus for each channel to ensure focused structured pattern illumination on the sample. NIS software provides an automatic function for this task.
- Next, acquire raw 3D-SIM images of the multicolor microspheres. Reconstruct the raw images to obtain a super-resolved image using the microscope software or the open-source software platform for the analysis of biological images ImageJ24 and the plugin fairSIM25.
- Calculate, for each separated wavelength, the Fourier transform of the super-resolved image obtained in 4.14. If the transformed image fails to obtain a correct flower-like pattern, restart calibration from 4.11 since super-resolution has not been achieved.
- In the super-resolved image, select a single microsphere and calculate its intensity profile for each channel to measure the resolution achieved. It should now be close to 100 nm laterally.
- Next, perform channel registration by overlaying a multichannel acquisition of the microspheres. The goal is to collimate all channel signals laterally and axially. This will eliminate chromatic aberrations due to the misalignment of the different channels and help the co-localization analysis.
- Confirm the quality of calibration by using the functions “Illumination Phase Steps” and “Illumination Pattern Focus” of SIMcheck26, a suite of plugins for the open-source application ImageJ. To this end, prepare a chambered coverslip to obtain a dense single layer of TetraSpeck microspheres and acquire a 3D-SIM image of the sample. Analyze the image in ImageJ and, if aberrations are detected, restart microscope calibration from step 4.11.
5. Acquisition
- Start analyzing the sample using a 40x objective in confocal or widefield mode. This allows navigation to the sample, maintaining good details and a large field of view.
- Use MAP2 antibody signal to identify an area representing neuronal processes.
- Acquire images of the sample in confocal mode to determine the quality of the staining. Poor confocal quality will reflect in poor SIM quality, therefore requiring the samples to be discarded.
- If the area and the quality of the images are satisfactory, switch the objective to 100x.
- Apply oil to the 100x objective.
- Acquire a widefield or confocal image that will be used later to assess the quality of the super-resolved image (Figure 1A,B).
- Switch to 3D-SIM mode.
- Using dialog windows to set parameters for acquisition, select the highest bit-depth setting available to maximize color information. Typically, 16-bit is the standard choice. Moreover, to improve signal-to-noise ratio, select a low frequency value for acquisition, such as 1 MHz.
- Using histogram windows, set lasers power to obtain a linear response of signal. To avoid loss of information, limit saturated pixels in the images. The N-SIM system uses an Andor iXon3 camera. When working at 16-bit, choose a target intensity of 16,000 to ensure the linear response of the camera. Alternatively, choose a range between 30,000-45,000 to maximize the dynamic range of the acquisition.
- Set laser power between 0.1% and 50% when imaging the samples and exposure times between 50 ms and 2 s. Laser powers above 50% may cause rapid photobleaching of the fluorophores in use.
- Start acquiring the images in 3D-SIM mode.
- Use SIMcheck, a suite of free plugins for ImageJ, to assess the quality of acquisition of the raw images.
- If SIMCheck does not detect any artifacts or quality issues, acquire a minimum of 10 images from 4 technical replicates to allow statistical analysis.
6. Post-production: image reconstruction
NOTE: 3D-SIM acquired images are raw images that need to be processed to obtain reconstructed super-resolved images. Incorrect reconstruction of raw images can lead to artifacts that would affect the analysis of the samples. Great attention should therefore be paid to properly choosing reconstruction parameters.
- Process the raw images using the microscope reconstruction analysis software to obtain a super-resolved image (Figure 1C). Alternatively, use the freely available ImageJ plugin fairSIM to reconstruct raw images.
- Calculate the Fourier transform of the super-resolved images using the microscope reconstruction software or ImageJ plugin SIMCheck. A good reconstructed image should return, for each channel, a flower-like image. If the reconstructed images fail to recreate a flower-like shape, restart from the raw images and reconstruct them by modifying the reconstruction parameters such as Wiener filtering, apodization and zero-order suppression27. In NIS software, using the preview to monitor how changing the parameters affects the final resolved image, modify the parameters i) Illumination Modulation Contrast, ii) High Resolution Noise Suppression and iii) Out of Focus Suppression.
- Next, analyze the reconstructed image to unbiasedly detect artefacts by using NanoJ-SQUIRREL28, an ImageJ-based plugin to assess the quality of super-resolved images.
- If NanoJ-SQUIRREL detects artifacts, restart from the raw images and reconstruct them by modifying the reconstruction parameters such as Wiener filtering, apodization and zero-order suppression. In NIS software, using the preview to monitor how changing the parameters affects the final resolved image, modify the parameters Illumination Modulation Contrast, High Resolution Noise Suppression and Out of Focus Suppression.
- Use the super-resolved images to calculate the co-localization profile and/or Pearson’s and Mander’s coefficients.
7. Co-localization with profile analysis
NOTE: As a first step to study co-localization between synaptic markers and a protein of interest, take a super-resolved image and analyze a single locus to determine signal overlap.
- Identify a single locus on the super-resolved image.
- Obtain the intensity profiles of the fluorescent signals of the locus of interest.
- Export the data.
- Use GraphPad Prism, or a similar analysis software, to normalize all signal peaks and obtain comparable signal intensities for each channel with the final goal of determining locus specific co-localization.
8. Quantification of Pearson’s and Mander’s coefficients
NOTE: If profile analysis has suggested single locus co-localization, a more general analysis of the whole image can be carried out by calculating Pearson’s and Mander’s coefficients29,30.
- Use JACoP31, an ImageJ plug-in, to determine the two parameters of co-localization: Pearson’s and Mander’s.
9. Statistical analysis
- Use GraphPad Prism, or a similar analysis and graphing software, to process data collected with JACoP.
- Use at least 40 SIM images for each condition analyzed to obtain graphs and for statistical relevance.
Representative Results
We present here the standard workflow to study neuronal proteins co-localization. We first calibrated the microscope and next we performed SIM analysis of the samples. To calibrate the system, we used fluorescent microspheres of 0.1 μm diameter. Upon obtaining super-resolved 3D-SIM images of the beads, the underlying image data are Fourier-transformed to re-convert them to a spatial frequency representation. In Figure 2A, the distinct flower pattern is presented as an indication of super-resolution detail levels. We next measured the resolution achieved by calculating the full width at half maximum (FWHM) of the peak of a single bead’s intensity profile (Figure 2B,C). Finally, we corrected chromatic aberration by channel registration, again using fluorescent microspheres (Figure 3A,B). Next, we started analyzing the sample with a 100x objective and we acquired 3D-SIM images. We used SIMCheck to assess evenness of field-illumination or movement during acquisition (Figure 4A). We checked differences in intensity between illumination pattern angles (Figure 4B) and we calculated the ratio of the modulation contrast to noise to measure the local stripe contrast (Figure 4C). Finally, we estimated the effective resolution of the reconstruction (Figure 4D).
We next confirmed the quality of the super-resolved images by using NanoJ-SQUIRREL. In the first reconstructed image (Figure 5A), NanoJ-SQUIRREL detected the presence of artefacts (Figure 5C). We changed the reconstruction parameters to obtain a new super-resolved image (Figure 5B) and NanoJ-SQUIRREL confirmed the lack of artifacts (Figure 5D). After having calibrated the system and assessed the quality of the reconstructed images, we next started analyzing the primary neuronal cultures stained with an antibody against MAP2, a neuronal marker, PSD95, a post-synaptic marker and the target protein SUMO1. We first analyzed the sample performing four-channel confocal microscopy with a 40x objective (Figure 6A). Upon selecting an area representing neuronal processes, we switched to a 100x objective. We acquired both confocal and SIM images of the same area to assess quality of reconstruction with NanoJ-SQUIRREL and perform co-localization analysis. In Figure 6B, we show the super-resolved 3D-SIM image of neurons stained for SENP1 and drebrin. Co-localization in super-resolved images can be analyzed with profile analysis (Figure 7A) and quantification of Pearson’s and Mander’s coefficients (Figure 7B).
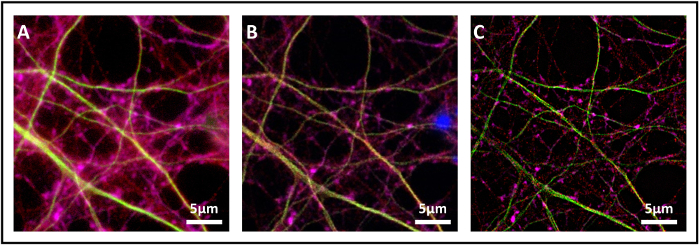
Figure 1: Comparison of widefield, confocal and SIM acquisitions. (A) Widefield image of primary hippocampal neurons immunostained for SENP1 (green), drebrin (red) and MAP2 (mauve). DAPI was used to stain nuclei. Scale bar 5 µm. (B) Confocal image of the same sample of panel A. (C) SIM image of the same sample of panel A and B. Please click here to view a larger version of this figure.
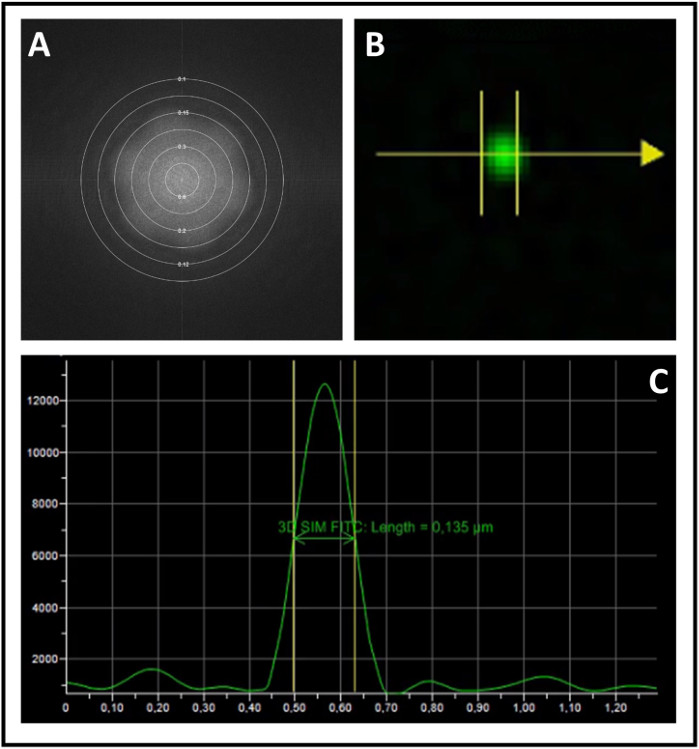
Figure 2: Analysis of a 3D-SIM image of microspheres for microscope calibration. (A) Fast Fourier transform of an acquisition of microspheres with its flower-like shape. (B) Selection of a single microsphere to determine lateral spatial resolution. (C) Intensity profile of the single microsphere in B with the measurement of its FWHM. The values represent the resolution achieved by the instrument. Please click here to view a larger version of this figure.
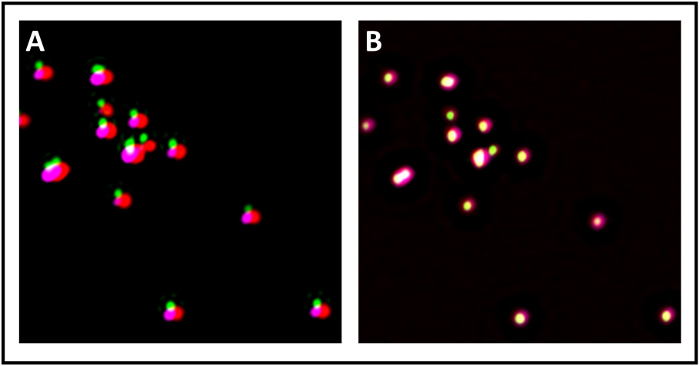
Figure 3: Three channel registration. (A) Acquisition of multicolor (wavelengths 488nm, 555nm and 647nm) TetraSpeck microspheres before registration. (B) Acquisition of the same sample after calibration. Please click here to view a larger version of this figure.
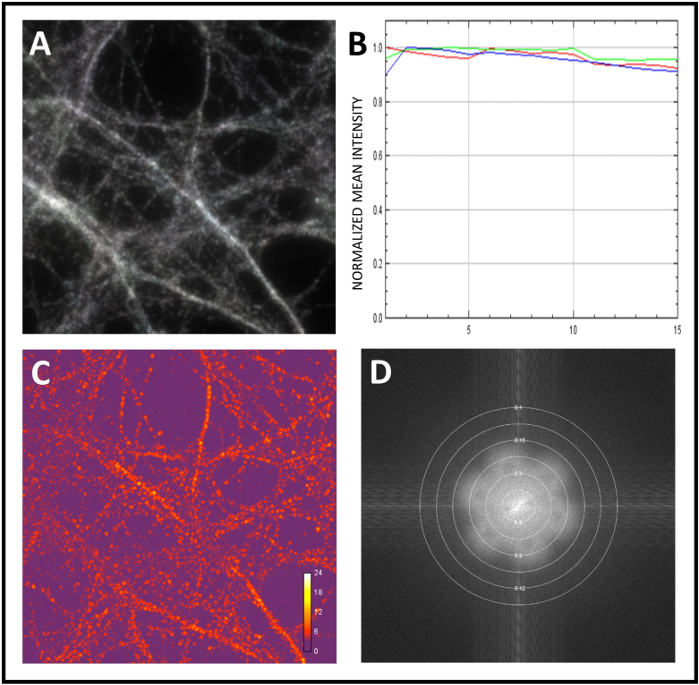
Figure 4: Quality assessment of raw and reconstructed images using SIMcheck. (A) Motion and Illumination Variation analysis using SIMCheck. Signal grey to white represents homogeneous illumination and absence of movement during acquisition. (B) Channel Intensity Profile obtained by analyzing the raw image. In this example there intensity variation is minimal, to suggest lack or bleaching or fluctuations. (C) Raw Modulation Contrast to calculate the ratio of the modulation contrast-to-noise within the image. The heatmap shows modulation contrast variations. (D) Reconstructed Fourier Plot to analyze the amplitude Fourier spectrum to determine the effective resolution of the reconstruction. Please click here to view a larger version of this figure.
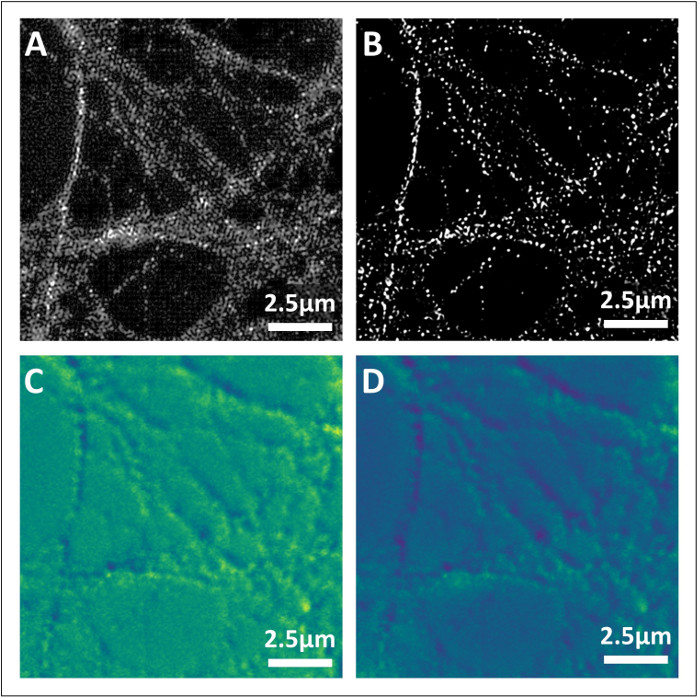
Figure 5: Assessment of super-resolution image quality using NanoJ-SQUIRREL. (A) Reference super-resolution image with artefacts. (B) Reference super-resolution image of good quality. (C) Image representing NanoJ-SQUIRREL error map of A. Lighter areas represent large scale artifacts, while darker ones represent correct reconstruction. (D) NanoJ-SQUIRREL error map of B. Please click here to view a larger version of this figure.
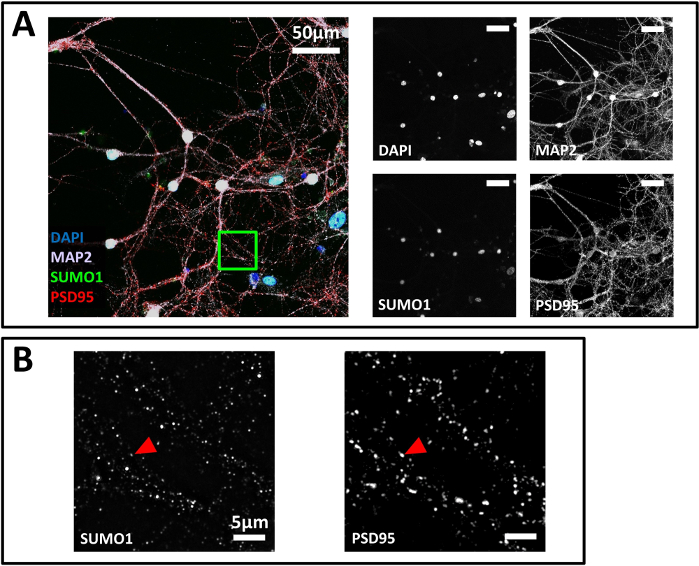
Figure 6: SIM image sample. (A) Confocal microscopy of primary neurons. A 40X-objective was chosen to obtain an overview of the sample while maintaining good resolution. Cells were immunostained for SUMO1 (green), PSD95 (red) and MAP2 (mauve). DAPI was used to stain nuclei. Scale bar 50 µm. Images were displayed as Z projection. (B) SIM images for SUMO1 and PSD95 on the area highlighted in the green box in panel A using a 100X objective. Red arrowheads indicate the position of the inset shown in A used to calculate the intensity profile. Scale bar 5 µm. Please click here to view a larger version of this figure.
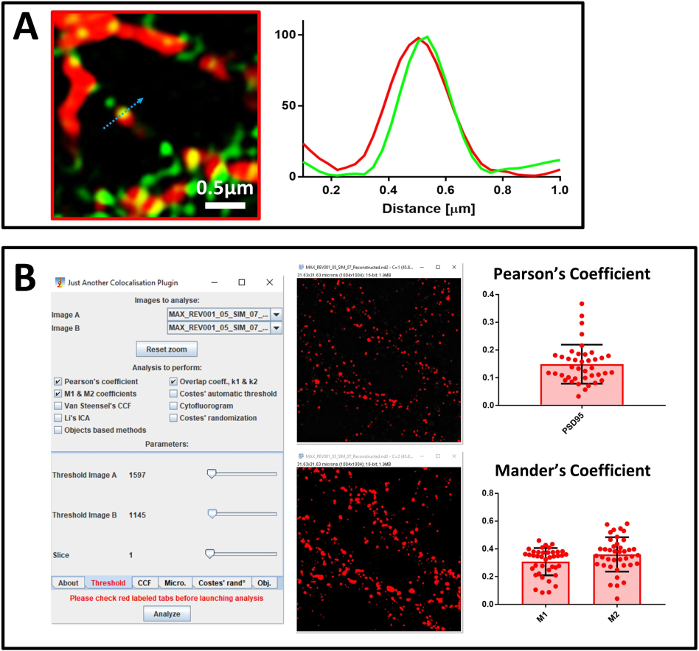
Figure 7: Co-localization analysis. (A) Super-resolved image of primary neurons stained with an antibody against SENP1 (in green) and drebrin (in red), scale bar 0.5 µm, and its intensity profile. The values of the graph were normalized for each channel to 100 (arbitrary unit) and correspond to the pixel intensity shown by the blue arrow. (B) Analysis using JACoP to calculate Pearson’s Correlation Coefficient and Mander’s coefficient between SENP1 and drebrin. Windows of the plug-in set up and visual threshold are shown. Mander’s coefficient is expressed by two values – SENP1 fraction that co-localizes with drebrin (M1) and the drebrin fraction that co-localizes with SENP1 (M2). Please click here to view a larger version of this figure.
Discussion
Elucidating the structure and composition of the synapse is crucial for understanding the physiological and pathological processes that regulate memory and cognition. While in the normal state, synapses are the building blocks of memory, they also underlie complex neurological disorders such as Alzheimer’s disease32. The protocol described here serves to study the co-localization of neuronal proteins with a super-resolution microscopy technique called SIM. Using a particular pattern of illumination, SIM can reach a resolution of about 0.1 μm, which is suited for the study of synapses, which normally measure between 0.03 and 0.15 μm. For even greater detail, other super-resolution techniques such as STED (Stimulated Emission Depletion Microscopy), PALM (PhotoActivated Localization Microscopy) or STORM (Stochastic Optical Reconstruction Microscopy), that can reach a resolution of 10-20 nm, may be applied33.
Here, we describe the analysis of co-localization of target proteins with synaptic markers in primary neurons. The protocol can be applied to any primary culture of neuronal cells, such as hippocampal, cerebellar or cortical neurons and even to cultures of primary neurons that do not belong to the central nervous system, such as enteric nervous system neurons. The key to the analysis at super-resolution level, however, is the reagents used during acquisition, such as chambered coverslips and mounting solutions compatible with the diffraction index of the objective. We used chambered coverslips for their ease of use, but the classical, cheaper method of growing primary neurons on coated coverglass is nevertheless valid, particularly with a high precision #1.5H (0.17 mm) coverglass. In addition, a mounting media that can reach a refraction index as close as possible to the refraction index of glass (1.52) and an immersion oil for the 100x objective with a refraction index of 1.515 should be used. Constant room temperature and stabilized tables are also mandatory to guarantee the accuracy of the acquisitions.
We use both dyLight and Alexa secondary antibodies in the SIM studies. Due to their narrow peaks of excitation and emission and good quantum yield, they are indicated for super-resolution techniques that require the best signal to noise ratio. Dempsey et al. compared Alexa, dyLight and other fluorophores for super-resolution imaging34.
During acquisition, we routinely set the camera at 1 MHz over 10 MHz. 1 MHz, thanks to a slower acquisition speed, gives the images more accuracy and less noise than 10 MHz. 1 MHz read-out mode can also record with a bit depth of 16 bit (compared to the maximum 14 bit of 10 MHz), giving more color information and a more precise color gradient to the images. However, 10 MHz, with its speed, is useful for live images. To avoid bleaching and preserve fluorophore, we also set laser power as low as possible. To improve signal intensity, gain values can be increased. It is worth noting that lower gain guarantees cleaner images without enhancing noise. In general, best results are obtained while imaging within 7 µm from the bottom of the chambered coverslip. This is especially important when using a 100x objective with oil immersion. If deeper acquisition across the cells/tissues is required, a better choice may be the use of a 60x objective with water immersion.
One of the main challenges in performing SIM studies is image reconstruction35. Obtaining super-resolved images without artefacts and aberration requires not only the use of ad-hoc experimental conditions, but also careful calibration of the system and parameter optimization to obtain the final images. In the protocol, we describe how to avoid some of the most common mistakes by assessing calibration of the system and quality analysis of raw and reconstructed images. Specifically, we describe the use of the ImageJ plugins SIMCheck and NanoJ-SQUIRREL to assure correct instrument settings to prevent common artifacts of super-resolved images. The applications allow for an unbiased quality assessment of the final images that is not based on subjective benchmarking the results against prior knowledge of the structures of study.
We suggest using synaptophysin and PSD95 or drebrin as pre- and post-synaptic markers, though other markers are valid as well. A huge body of literature describes proteins such as bassoon as synaptic markers36,37. It is worth noting that pre- and postsynaptic markers are however present throughout the cell, excluding the nucleus. Much of their signal is non-synaptic but represents proteins in transport or degradation, background or other artifacts. It is therefore important to carefully choose the area of the analysis. We use MAP2 antibody signal to choose axon and dendritic terminals.
In the analysis of co-localization we use two approaches. The first is a visual approach, based on profile analysis that shows single events of co-localization and identifies the contribution of each channel. A caveat of this approach, however, is the poor statistical power. For this reason, we decided also to use a second method based on analysis of a larger number of events representative of the entire field of each image. This method is based on calculation of the Pearson’s correlation coefficient and Mander’s M1 and M2 coefficients. We use the Pearson’s correlation coefficient to describe the overlap of signals in the image and Mander’s M1 and M2 to describe reciprocal co-localization between signals of interest38. For the calculation, we employ the ImageJ plugin JACoP, since it has a feature that allows you to set a manual threshold to discard any background contribution to the analysis, especially critical for Mander’s analysis.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Edoardo Micotti for constructive criticism of the manuscript. This study was supported by BrightFocus A2019296F, by Fondo di Beneficenza – Gruppo Intesa Sanpaolo (LC), by Fondazione Regionale per la Ricerca Biomedica (Care4NeuroRare CP_20/2018) (CN), by the Marie Skłodowska-Curie Innovative Training Network (JK) and by Fondazione Telethon TCP15011, Alzheimer's Association AARG-17-505136 (LF).
Materials
| 0.4% Trypan blue solution | Thermo Fisher Scientific | 15250061 | Chemical |
| 70 µm filter | Corning | 352350 | Equiment |
| Alexa | Thermo Fisher Scientific | – | Antibody |
| Antibody SENP1 | Santa Cruz | sc-271360 | Antibody |
| B27 Supplement | Life Technologies | 17504044 | Chemical |
| Bovine serum albumin | Merck | 5470 | Chemical |
| CaCl2 | Merck Life Science | 21115 | Chemical |
| Chambered coverslips | Ibidi | 80826 | Equiment |
| DyLight | Thermo Fisher Scientific | – | Antibody |
| FBS (Hyclone) | GIBCO | SH3007002 (CHA1111L) | Serum |
| FluoSpheres carboxylate-modified microspheres, 0.1 μm, yellow–green fluorescent | Thermo Fisher Scientific | F8803 | Equiment |
| Glucose | Merck Life Science | G8769 | Chemical |
| Glutamax | GIBCO | 35050061 | Chemical |
| HEPES | Merck Life Science | H3537 | Chemical |
| L-Cystein | Merck Life Science | C6852-25g | Chemical |
| MAP2 | Merck | AB15452 | Antibody |
| MEM | Life Technologies | 21575022 | Medium |
| MgCl | Merck Life Science | M8266 | Chemical |
| NaOH | VWR International | 1,091,371,000 | Chemical |
| Neurobasal A | Life Technologies | 10888022 | Medium |
| N-SIM Super Resolution Microscope | Nikon | – | Instrument |
| Papain | Merck Life Science | P-3125 | Chemical |
| paraformaldehyde | Thermo Fisher Scientific | 28908 | Chemical |
| Pen/Strep 10x | Life Technologies | 15140122 | Chemical |
| phosphate-buffered saline | Gibco | 10010023 | Chemical |
| Poly-L lysine | Sigma | P2636 | Chemical |
| ProLong Diamond Glass Antifade Mountant | Thermo Fisher Scientific | P36970 | Chemical |
| PSD95 | NeuroMab | K28/43 | Antibody |
| Round coverglass | Thermo | 12052712 | Equiment |
| SUMO1 | Abcam | ab32058 | Antibody |
| Synaptophysin | Merck | S5768 | Antibody |
| Triton X-100 | Merck | T8787 | Chemical |
| Trypsin inhibitor | Merck Life Science | T9003-500MG | Chemical |
References
- Foster, M., Sherrington, C. S. . A textbook of physiology, part three: The central nervous system (7th ed.). , (1897).
- Choquet, D., Triller, A. The Dynamic Synapse. Neuron. 80 (3), 691-703 (2013).
- McAllister, A. K. Dynamic Aspects of CNS Synapse Formation. Annual Review of Neuroscience. 30 (1), 425-450 (2007).
- Yuzaki, M. Two Classes of Secreted Synaptic Organizers in the Central Nervous System. Annual Review of Physiology. 80 (1), 243-262 (2018).
- Baddeley, D., Bewersdorf, J. Biological Insight from Super-Resolution Microscopy: What We Can Learn from Localization-Based Images. Annual Review of Biochemistry. 87 (1), 965-989 (2018).
- Sigal, Y. M., Zhou, R., Zhuang, X. Visualizing and discovering cellular structures with super-resolution microscopy. Science. 361 (6405), 880-887 (2018).
- Vangindertael, J., et al. An introduction to optical super-resolution microscopy for the adventurous biologist. Methods and Applications in Fluorescence. 6 (2), 022003 (2018).
- Badawi, Y., Nishimune, H. Super-resolution microscopy for analyzing neuromuscular junctions and synapses. Neuroscience Letters. 715, 134644 (2020).
- Scalisi, S., Barberis, A., Petrini, E. M., Zanacchi, F. C., Diaspro, A. Unveiling the Inhibitory Synapse Organization Using Superresolution Microscopy. Biophysical Journal. 116 (3), 133 (2019).
- Yang, X., Specht, C. G. Subsynaptic Domains in Super-Resolution Microscopy: The Treachery of Images. Frontiers in Molecular Neuroscience. 12, (2019).
- Monro, T. Beyond the diffraction limit. Nature Photonics. 3 (7), 361 (2009).
- Won, R. Eyes on super-resolution. Nature Photonics. 3 (7), 368-369 (2009).
- Wegel, E., et al. Imaging cellular structures in super-resolution with SIM, STED and Localisation Microscopy: A practical comparison. Scientific Reports. 6 (1), 27290 (2016).
- Galbraith, C. G., Galbraith, J. A. Super-resolution microscopy at a glance. Journal of Cell Science. 124 (10), 1607-1611 (2011).
- Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Journal of Microscopy. 198 (2), 82-87 (2000).
- Gustafsson, M. G. L., et al. Three-Dimensional Resolution Doubling in Wide-Field Fluorescence Microscopy by Structured Illumination. Biophysical Journal. 94 (12), 4957-4970 (2008).
- Brose, N., O’Connor, V., Skehel, P. Synaptopathy: dysfunction of synaptic function. Biochemical Society Transactions. 38 (2), 443-444 (2010).
- Tyebji, S., Hannan, A. J. Synaptopathic mechanisms of neurodegeneration and dementia: Insights from Huntington’s disease. Progress in Neurobiology. 153, 18-45 (2017).
- Won, H., Mah, W., Kim, E. Autism spectrum disorder causes, mechanisms, and treatments: focus on neuronal synapses. Frontiers in Molecular Neuroscience. 6, (2013).
- Pfeiffer, B. E., Huber, K. M. The State of Synapses in Fragile X Syndrome. The Neuroscientist. 15 (5), 549-567 (2009).
- Pavlowsky, A., Chelly, J., Billuart, P. Emerging major synaptic signaling pathways involved in intellectual disability. Molecular Psychiatry. 17 (7), 682-693 (2012).
- Senatore, A., Restelli, E., Chiesa, R. Synaptic dysfunction in prion diseases: a trafficking problem. International Journal of Cell Biology. 2013, 543803 (2013).
- Colnaghi, L., et al. Super Resolution Microscopy of SUMO Proteins in Neurons. Frontiers in Cellular Neuroscience. 13, (2019).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Müller, M., Mönkemöller, V., Hennig, S., Hübner, W., Huser, T. Open-source image reconstruction of super-resolution structured illumination microscopy data in ImageJ. Nature Communications. 7 (1), 10980 (2016).
- Ball, G., et al. SIMcheck: a Toolbox for Successful Super-resolution Structured Illumination Microscopy. Scientific Reports. 5 (1), 15915 (2015).
- Schaefer, L. H., Schuster, D., Schaffer, J. Structured illumination microscopy: artefact analysis and reduction utilizing a parameter optimization approach. Journal of Microscopy. 216 (2), 165-174 (2004).
- Culley, S., et al. NanoJ-SQUIRREL: quantitative mapping and minimisation of super-resolution optical imaging artefacts. Nature Methods. 15 (4), 263-266 (2018).
- Manders, E. M. M., Verbeek, F. J., Aten, J. A. Measurement of co-localization of objects in dual-colour confocal images. Journal of Microscopy. 169 (3), 375-382 (1993).
- Adler, J., Parmryd, I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 77 (8), 733-742 (2010).
- Bolte, S., Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy. 224, 213-232 (2006).
- Bae, J. R., Kim, S. H. Synapses in neurodegenerative diseases. BMB Reports. 50 (5), 237-246 (2017).
- Godin, A. G., Lounis, B., Cognet, L. Super-resolution Microscopy Approaches for Live Cell Imaging. Biophysical Journal. 107 (8), 1777-1784 (2014).
- Dempsey, G. T., Vaughan, J. C., Chen, K. H., Bates, M., Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nature Methods. 8 (12), 1027-1036 (2011).
- Karras, C., et al. Successful optimization of reconstruction parameters in structured illumination microscopy – A practical guide. Optics Communications. 436, 69-75 (2019).
- Bereczki, E., et al. Synaptic markers of cognitive decline in neurodegenerative diseases: a proteomic approach. Brain: A Journal of Neurology. 141 (2), 582-595 (2018).
- Gilestro, G. F., Tononi, G., Cirelli, C. Widespread Changes in Synaptic Markers as a Function of Sleep and Wakefulness in Drosophila. Science. 324 (5923), 109-112 (2009).
- Adler, J., Parmryd, I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 77 (8), 733-742 (2010).

