Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data
Summary
LEfSe (LDA Effect Size) is a tool for high-dimensional biomarker mining to identify genomic features (such as genes, pathways, and taxonomies) that significantly characterize two or more groups in microbiome data.
Abstract
There is growing attention toward closed biological genomes in the environment and in health. To explore and reveal the intergroup differences among different samples or environments, it is crucial to discover biomarkers with statistical differences among groups. The application of Linear discriminant analysis Effect Size (LEfSe) can help find good biomarkers. Based on the original genome data, quality control, and quantification of different sequences based on taxa or genes are carried out. First, the Kruskal-Wallis rank test was used to distinguish between specific differences among statistical and biological groups. Then, the Wilcoxon rank test was performed between the two groups obtained in the previous step to assess whether the differences were consistent. Finally, a linear discriminant analysis (LDA) was conducted to evaluate the influence of biomarkers on significantly different groups based on LDA scores. To sum up, LEfSe provided the convenience for identifying genomic biomarkers that characterize statistical differences among biological groups.
Introduction
Biomarkers are biological characteristics that can be measured and can indicate some phenomena such as infection, disease, or environment. Among them, functional biomarkers may be specific biological functions of single species or common to some species, such as gene, protein, metabolite and pathways. Besides, taxonomic biomarkers indicate an unusual species, a group of organisms (kingdom, phylum, class, order, family, genus, species), the Amplicon Sequence Varient (ASV)1, or the Operational Taxonomic Unit (OTU)2. In order to find biomarkers more quickly and accurately, a tool for analyzing the biological data is necessary. The differences between classes can be explained by LEfSe coupled with standard tests for statistical significance and additional tests encoding biological consistency and effect relevance3. LEfSe is available as a galaxy module, a conda formula, a docker image, and included in bioBakery (VM and cloud)4. Generally, the analysis of microbial diversity often uses a non-parametric test for the uncertain distribution of a sample community. The rank sum test is a non-parametric test method, which uses the rank of samples to replace the value of samples. According to the difference of sample groups, it can be divided into two samples with the Wilcoxon rank sum test and into multiple samples with the Kruskal-Wallis test5,6. Notably, when there are significant differences among multiple groups of samples, a rank-sum test of pairwise comparison of multiple samples should be performed. LDA (which stands for Linear Discriminant Analysis) invented by Ronald Fisher in 1936, is a type of supervised learning, also known as Fisher’s Linear Discriminant7. It is a classic and popular algorithm in the current field of machine learning data mining.
Here, the LEfSe assay has been optimized by Conda and Galaxy servers. Three groups of 16S rRNA gene sequences are analyzed to demonstrate the significant differences between different groups with LDA scores of microbial communities and visualization results.
Protocol
NOTE: The protocol was sourced and modified from the research of Segata et al.3. The method is provided at https://bitbucket.org/biobakery/biobakery/wiki/lefse.
1. Preparation of input file for analysis
- Prepare the input file (Table 1) of LEfSe, which could be easily generated by many workflows8 or previous protocols9 with the original files (sample file and corresponding species annotation file).
2. LEfSe native analysis (limited to the Linux server)
- LEfSe Installation
NOTE: The LEfSe pipeline is recommended to be installed with Conda10.- Run the following commands to exclude the possibility of dependencies conflict. Create a conda environment for LEfSe (This step is recommended but not required.). -n stands for the environment name.
$ conda create -n LEfSe-env - To activate the LEfSe environment that was created, run:
$ source activate LEfSe-env - To install LEfSe with channel bioBakery where -c stands for channel name, run:
$ conda install -c biobakery lefse
- Run the following commands to exclude the possibility of dependencies conflict. Create a conda environment for LEfSe (This step is recommended but not required.). -n stands for the environment name.
- Format data for LEfSe
- Run the following command to format the original file to the internal format for LEfSe. Table.txt is the input file and Table-reformat.in is the output file. -c is used to set the feature, which is used as class (default 1) and -o is used to set the normalization value (default -1.0 meaning no normalization).
$ format_input.py Table.txt Table-reformat.in -c 1 -o 1000000
- Run the following command to format the original file to the internal format for LEfSe. Table.txt is the input file and Table-reformat.in is the output file. -c is used to set the feature, which is used as class (default 1) and -o is used to set the normalization value (default -1.0 meaning no normalization).
- Calculation of linear discriminant analysis (LDA) effect size
- Run the following command. The purpose of this step is to perform LDA of the previous result and generate the result file for the visualization. Table-reformat.in is generated using the previous step and is used as the input file in this step. Table-reformat.res is the result file.
$ run_lefse.py Table-reformat.in Table-reformat.res
- Run the following command. The purpose of this step is to perform LDA of the previous result and generate the result file for the visualization. Table-reformat.in is generated using the previous step and is used as the input file in this step. Table-reformat.res is the result file.
- Visualization by plots
- Plot the LEfSe results. To plot the effect size of the biomarkers in a pdf file,.Table-reformat.res is generated using the previous step and the LDA.pdf is the plot file. –format is used to set the output file format.
$ plot_res.py Table-reformat.res LDA.pdf –format pdf - Plot the cladogram. To draw the species tree and display the biomarkers in a cladogram. cladogram.pdf is the output file.
$ plot_cladogram.py Table-reformat.res cladogram.pdf –format pdf - Plot one feature (optional) To plot the differences of a single biomarker among different groups. -f is used to set the features of plot. If one was set, the –feature_name must be given.
$ plot_features.py -f one –feature_name "k__Bacteria.p__Firmicutes.c__Bacilli.o__Bacillales" –format pdf Table-reformat.in Table-reformat.res Bacillales.pdf - Plot the differential features (optional) to draw all the features, but there is too much to be done with caution. –archive is used to choose whether to compress the results. ./ means the path of the results.
$ plot_features.py -f diff –archive none –format pdf Table-reformat.in Table-reformat.res ./
- Plot the LEfSe results. To plot the effect size of the biomarkers in a pdf file,.Table-reformat.res is generated using the previous step and the LDA.pdf is the plot file. –format is used to set the output file format.
3. LEfSe online analysis (galaxy)
- Go to the huttenhower galaxy server11: http://huttenhower.sph.harvard.edu/galaxy.
- Upload the files. Press the Up arrow button on the left pane and upload the file. Click on Choose local file to select the input file and select the format tabular, and then click on the Start button.
NOTE: Referred to the webpage (https://bitbucket.org/biobakery/biobakery/wiki/lefse), use the script (taxonomy_summary.R) to generate the input file of LEfSe, and the format (each column with a group name, each line with a different level of annotation separated by “|”) is required as shown in Table 1. A schematic overview of the uploading process is shown in Figure 1. - Format the data for LEfSe. Click on the LEfSe | Format Data for LEfSe link on the left pane, and select the specific rows for class in the file, and click on the Execute button. A schematic overview of the operational process and the parameters used are shown in Figure 2.
- Calculate the LDA effect size. Click on the LEfSe | LDA Effect Size (LEfSe) link on the left pane, and select parameter values according to the analysis requirements. Click on Execute. A schematic overview of the operational process and the parameters used are shown in Figure 3.
- Plot the LEfSe results. Click on the LEfSe | Plot LEfSe Results link on the left pane, and click on the Execute button. A schematic overview of the operational process and the parameters used are shown in Figure 4.
- Plot the cladogram. Click on Plot Cladogram on the left pane, and click on the Execute button after selecting the parameter values. A schematic overview of the operational process and the parameters used are shown in Figure 5.
- Plot one feature by clicking on Plot One Feature on the left pane, and clicking on the Execute button after selecting parameter values. A schematic overview of the operational process and the parameters used are shown in Figure 6.
- Plot differential features by clicking on Plot Differential Features on the left pane, and clcking on the Execute button after selecting parameter values. A schematic overview of the operational process and the parameters used are shown in Figure 7.
NOTE: These generated figures can be visualized and downloaded against the resulting output in the right pane.
Representative Results
The LDA scores of microbial communities with significant differences in each group by analyzing the 16S rRNA gene sequences of three samples is shown in Figure 8. The color of the histogram represents different groups, while the length represents the LDA score, which is the influence of the species with significant differences between different groups. The histogram shows the species with significant differences whose LDA score is greater than the preset value. The default preset value is 2.0, so only absolute values of LDA score (abscissa) greater than 2.0 are shown in the plot.
The biomarkers with significant difference and species tree between different classification levels are shown in Figure 9. The circles radiating from the inside to the outside represent the classification levels from phylum to genus (the innermost yellow circle is the kingdom). The diameter of each small circle at the different classification levels represents the size of relative abundance. The species with no significant difference are uniformly colored yellow, and the significantly different species biomarkers are colored with the corresponding groups. The classes A, B, and C are the group names of microbial samples collected. Red nodes represent the microbial groups that play an important role in the red group (A); green nodes represent the microbial groups that play an important role in the green group (B); and blue nodes represent the microbial groups that play an important role in the blue group (C). The corresponding species name of the biomarkers not shown in the plot are shown on the right side, and the letter numbers correspond to that in the plot (only showing differential species from the phylum to the family by default for aesthetic purposes).
The abundance of one biomarker that has differences among different groups according to the LEfSe results is shown in Figure 10. In the relative abundance barplot, the solid line represents the average relative abundance, the dotted line represents the median relative abundance, and each column represents the relative abundance of each sample in different groups.
Table 1: The example file for LEfSe analysis online. Please click here to download this table.
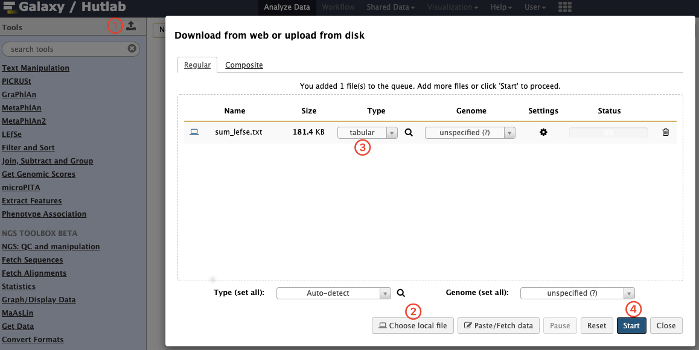
Figure 1: Schematic overview of the uploading process. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
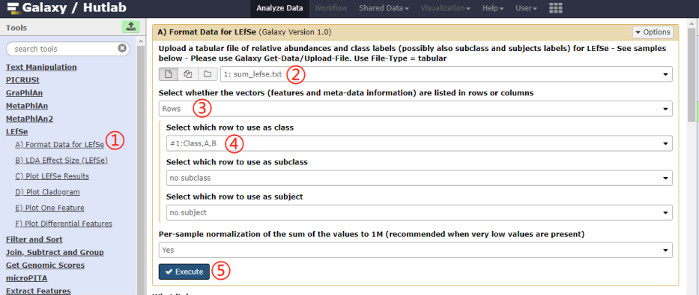
Figure 2: Schematic overview of the operational process for changing data format. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
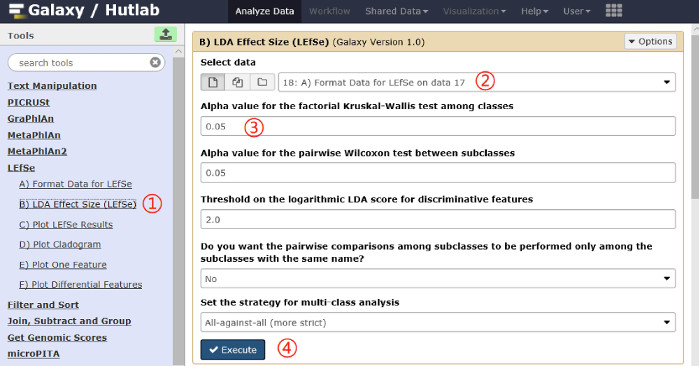
Figure 3: Schematic overview of the operational process for calculating LDA effect size. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
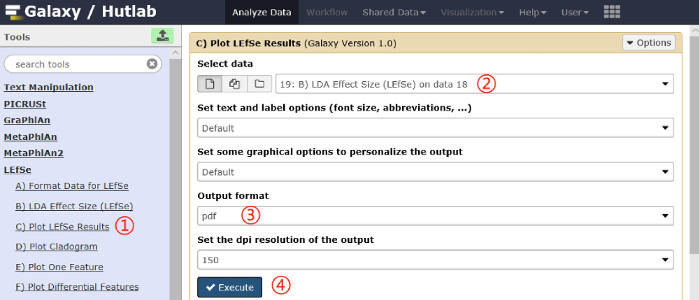
Figure 4: Schematic overview of the operational process for plotting LEfSe results. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
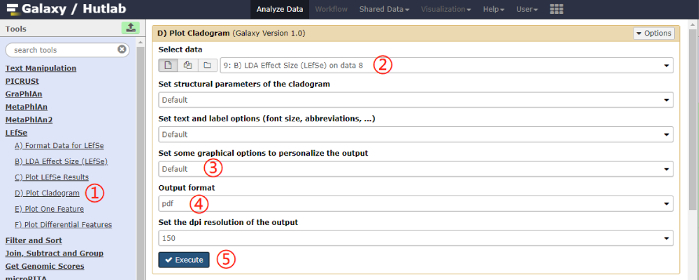
Figure 5: Schematic overview of the operational process for plotting cladogram. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
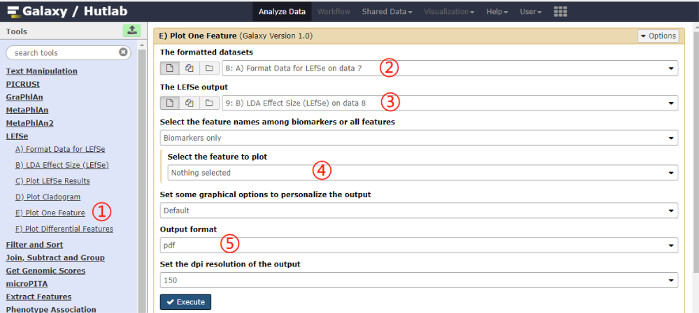
Figure 6: Schematic overview of the operational process for plotting one feature. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
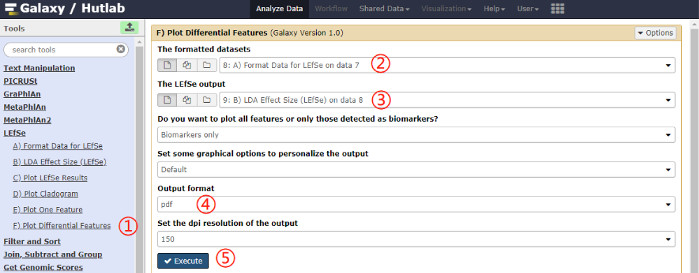
Figure 7: Schematic overview of the operational process for plotting differential features. Click on the red numbers in sequential order on the figure. Please click here to view a larger version of this figure.
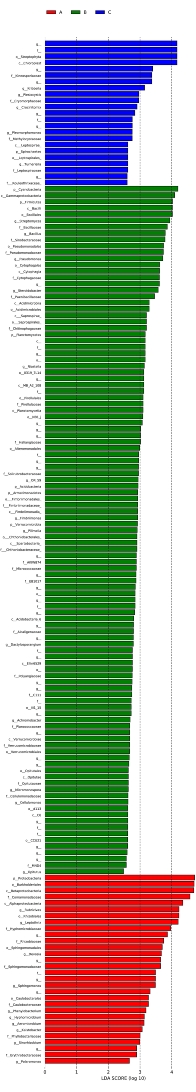
Figure 8: Histogram of the distribution of LDA values. LDA scores of microbial communities with significant differences in each group were analyzed by LDA Effect Size according to their influences and correlations. Please click here to download this figure.
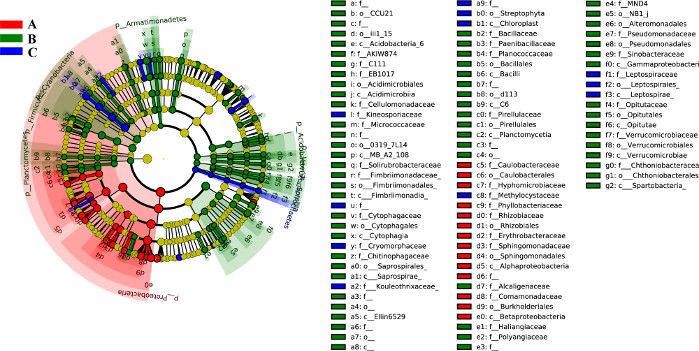
Figure 9: Cladogram. The typical plot of the cladogram obtained by the protocol, which enables the representation of the difference between different classification levels of three groups. Please click here to view a larger version of this figure.
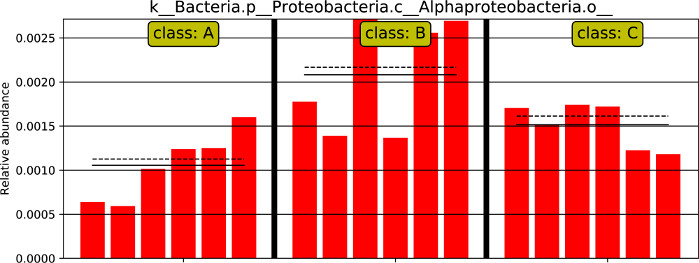
Figure 10: One feature plot. The abundance barplot of one biomarker that has differences among different groups according to the LEfSe results.is shown. Please click here to view a larger version of this figure.
Discussion
Here, the protocol for the identification and characterization of biomarkers within different groups is described. This protocol can easily be adapted for other sample types, such as OTUs of microorganisms. The statistical method by LEfSe can find the characteristic microorganisms in each group (default is LDA >2), that is, the microorganisms that are more abundant in this group relative to the others12. LEfSe is available in both native and web Linux versions where users can also perform LEfSe analysis on web pages. LEfSe is based on the LDA algorithm and needs a species level to draw a species tree. By application of the tool, the relative abundance between groups can be compared. All differential biomarkers could be plotted in a single graph. Also, a single biomarker or all biomarkers can be plotted in batches.
Whether LEfSe is performed through the native server or an online site, there are a lot of tunable parameters to draw the required pictures. Due to the complex structure of input files and the need to convert them to preferred data formats for further analyses of LEfSe, some one-stop services have also been developed. Therefore, the optimization of easier operations can be challenging. On the other hand, there are a few limitations when analyzing complex data using LEfSe. LDA projects a feature that is one dimension less than the category, and if more features are needed, other methods are introduced. The variants of LDA may solve some difficulties. For example, Kernel LDA is a solution if the original data cannot be separated well after projection. Because the amount of computation of LDA is related to the dimension of data, 2DLDA can greatly reduce the amount of computation of LDA. Both LDA and PCA are commonly used dimensionality reduction techniques. PCA (Principal Component Analysisis) dimensionality reduction is directly related to the data dimension, and the projected coordinate system is orthogonal. However, LDA focuses on the ability of classification according to the labeling of categories, so the projected coordinate system is generally not orthogonal.
LEfSe provides assistance for the selection of biomarkers. With many advantages (e.g., adjustable parameters, the detailed results of various parts, application between two or more groups), it has been widely used13. With increasing demand for high-dimensional data analysis, the application of this method will become more and more extensive to explore the biomarkers of the features (organisms, clades, operational taxonomic units, genes, or functions) impacting on human health and disease.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant from Fundamental Research Funds for the Central Public Welfare Research Institutes (TKS170205) and Foundation for Development of Science and Technology, and Tianjin Research Institute for Water Transport Engineering (TIWTE), M.O.T. (KJFZJJ170201).
References
- Bolyen, E., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 37 (8), 852-857 (2019).
- Knight, R., et al. Best practices for analysing microbiomes. Nature Reviews. Microbiology. 16 (7), 410-422 (2018).
- Segata, N., et al. Metagenomic biomarker discovery and explanation. Genome Biology. 12 (6), 60 (2011).
- . Biobakery / lefse [tool] Available from: https://bitbucket.org/biobakery/biobakery/wiki/lefse (2019)
- Kruskal, W. H. A nonparametric test for the several sample problem. The Annals of Mathematical Statistics. 23 (4), 525-540 (1952).
- Wilcoxon, F. Individual comparisons by ranking methods. Biometrics Bulletin. 1 (6), 80-83 (1945).
- Fisher, R. A. The use of multiple measurements in taxonomic problems. Annals of Eugenics. 7 (1), 179-188 (1936).
- Liu, Y. X., et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein and Cell. 41 (7), 1-16 (2020).
- Shahi, S. K., Zarei, K., Guseva, N. V., Mangalam, A. K. Microbiota analysis using two-step PCR and next-generation 16S rRNA gene sequencing. Journal of Visualized Experiments: JoVE. (152), e59980 (2019).
- Grüning, B., et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nature Methods. 15 (7), 475-476 (2018).
- Blankenberg, D., Chilton, J., Coraor, N. Galaxy external display applications: closing a dataflow interoperability loop. Nature Methods. 17 (2), 123-124 (2020).
- Langille, M. G. I., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 31 (9), 814-821 (2013).
- Shilei, Z., et al. Reservoir water stratification and mixing affects microbial community structure and functional community composition in a stratified drinking reservoir. Journal of Environmental Management. 267, 110456 (2020).

