Crystallization and Structural Determination of an Enzyme:Substrate Complex by Serial Crystallography in a Versatile Microfluidic Chip
Summary
A versatile microfluidic device is described that enables the crystallization of an enzyme using the counter-diffusion method, the introduction of a substrate in the crystals by soaking, and the 3D structure determination of the enzyme:substrate complex by a serial analysis of crystals inside the chip at room temperature.
Abstract
The preparation of well diffracting crystals and their handling before their X-ray analysis are two critical steps of biocrystallographic studies. We describe a versatile microfluidic chip that enables the production of crystals by the efficient method of counter-diffusion. The convection-free environment provided by the microfluidic channels is ideal for crystal growth and useful to diffuse a substrate into the active site of the crystalline enzyme. Here we applied this approach to the CCA-adding enzyme of the psychrophilic bacterium Planococcus halocryophilus in the presented example. After crystallization and substrate diffusion/soaking, the crystal structure of the enzyme:substrate complex was determined at room temperature by serial crystallography and the analysis of multiple crystals directly inside the chip. The whole procedure preserves the genuine diffraction properties of the samples because it requires no crystal handling.
Introduction
Crystallography is a method to decipher the 3D architecture of biological macromolecules. The latter is important to understand how an enzyme selects and processes its substrates. The determination of a crystal structure requires the crystallization of the target macromolecule and the conditioning of the crystals for their analysis by X-ray diffraction1. Both crystal preparation and handling are crucial but delicate steps that can affect crystal quality and diffraction properties, and, thus, the resolution (i.e., the accuracy) of the resulting 3D structure. To facilitate the preparation of high-quality crystals and eliminate unnecessary handling to preserve their diffraction properties, we designed a user-friendly and versatile microfluidic device called ChipX2,3,4.
In this article, we will demonstrate how to load the protein solution into ChipX channels using conventional laboratory material to prepare crystals by counter-diffusion. This crystallization method provides an efficient screening of supersaturation and of potential nucleation conditions along the microfluidic channels containing the enzyme solution due to the concentration gradient generated by the diffusion of the crystallizing agent5,6.
The chip setup is simple, it uses only standard laboratory pipets and does not require any costly equipment. When crystals have grown in ChipX, ligands of the enzyme can be introduced by diffusion. Diffraction data are then collected at room temperature on a series of crystals contained in the channels of the chip using a synchrotron X-ray source. The structural study described here led to the determination of structures of a tRNA maturation enzyme in its apo form and in complex with an analog of its CTP substrate introduced by soaking. This protein called CCA-adding enzyme polymerizes the CCA trinucleotide tail at the 3' end of tRNAs. The comparison of the two 3D images obtained by serial crystallography reveals the local conformational changes related to the binding of the ligand in conditions that are more physiological than those used in cryo-crystallography. The protocol described in this video is generally applicable to any biomolecule, be it a protein, a nucleic acid or a multi-component complex.
Protocol
1. Setting up crystallization assays in ChipX
NOTE: The ChipX microfluidic device can be obtained from the authors. A description of the chip is given in Figure 1. Solutions containing the crystallant (or crystallizing agent) used to trigger crystallization may be of commercial origin or prepared by the experimenter.
- Loading the biomolecule sample
NOTE: The sample volume actually required to perform an individual counter-diffusion assay in the straight section of each channel of ChipX is 300 nL. However for convenience we suggest to load 5 µL to completely fill the eight channels taking into account the variable length of their curved section and inlet dead volumes.- Pipet 5 µL of enzyme solutions using standard 10 µL pipet and tip.
- Introduce the tip vertically in the sample inlet and inject the solution until the eight channels are filled up to their opposite end (entry of the crystallant reservoir).
- Inject 1 µL of paraffin oil in the sample inlet in order to disconnect the channels from each other.
- Recover the extra solution in the crystallant reservoir at the extremity of each channel using a standard 10 µL pipet.
- Seal the sample inlet with a 1 cm x 1 cm piece of tape.
- Loading the crystallization solutions
- Pipet 5 µL of crystallization solution using standard 10 µL pipet and tip. The reservoir volume is 10 µL, but loading only half of it avoids overflow when sealing with tape and facilitates further addition of ligand for soaking experiments. If initial crystallization conditions were obtained by vapor diffusion, increase the crystallant concentration by a factor of 1.5 – 2. Solutions can be different in every reservoir (in the presented case, 1 M diammonium hydrogen phosphate, 100 mM sodium acetate, pH 4.5 was used throughout).
- Orient the pipet tip towards the entry of the channel in the funnel shaped part of the reservoir to avoid the formation of an air bubble upon solution deposition. It would prevent the contact between the two solutions and crystallant diffusion into the channel.
- Inject the crystallant solution into the reservoir.
- Seal the reservoirs with a 2.5 cm x 1 cm piece of tape.
- Incubate the chip at 20 °C (temperature can be adjusted depending on the target, typically between 4 and 37 °C 4).
2. Protein labeling with carboxyrhodamine for fluorescence detection
NOTE: This step is optional. It must be performed prior to sample loading to facilitate the detection of crystals in the chip using fluorescence. The detailed method of trace fluorescent labeling was described by Pusey and coworkers7. All steps are carried out at room temperature.
- Dissolve 5 mg of carboxyrhodamine ester powder in 1 mL anhydrous dimethyl-formamide, split the solution in 0.6 µL aliquots to be stored at -20 °C.
- Prepare a 1 M Na-borate pH 8.75 stock solution.
- Dilute the stock to prepare the reaction buffer at 0.05 M Na-borate pH 8.75.
- Rinse a desalting column (7 kDa MWCO, 0.5 mL) with 800 µL of reaction buffer.
- Centrifuge the column for 1 min at 1400 x g, remove the filtrate.
- Repeat this operation twice (steps 2.4-2.5) to wash the column.
- Deposit 80 µL of protein in its storage buffer on the column (the protein can be diluted down to 1 mg/mL to increase the volume if needed).
- Centrifuge the column for 1 min at 1400 x g. This step is intended to transfer the protein from its storage buffer to the reaction buffer.
- Recover the flow-through (containing the protein in the reaction buffer) and mix it with 0.6 µL of carboxyrhodamine solution.
- Incubate 5 min at room temperature.
- Meanwhile, rinse the column 3 x with the storage buffer, centrifuge the column for 1 min at 1400 x g and discard the filtrate.
- Deposit the reaction solution on the column.
- Centrifuge the column for 1 min at 1400 x g and recover the flow-through (i.e., solution of labeled protein in its storage buffer).
- Supplement the stock protein solution with 0.1-1 % (w/w) of labeled protein.
- Setup the ChipX crystallization assays as described in section 1.
- Check for the presence of protein crystals in the assays by exciting the fluorescent probe with a 520 nm wavelength light source.
3. Crystal observation
NOTE: The ChipX device may be handled without special care, even with crystals inside, except if the temperature needs to be controlled.
- Use any stereomicroscope to check the outcome of crystallization assays in ChipX. Its footprint has the standard dimensions of microscope slides and is compatible with any system and slide holder.
- Check the content of microfluidic channels starting from the reservoir where the crystallant concentration is the highest to the sample inlet where the crystallant concentration is the lowest. The ChipX material is transparent to visible light, compatible with the use of polarizers, as well as with UV illumination for protein crystal identification by intrinsic tryptophane fluorescence8.
- Record crystal positions using the labels embossed along the channels or mark crystal locations with a permanent marker by drawing color dots next to them on the chip surface.
4. Crystal soaking with ligands
NOTE: This procedure is optional. It is used to introduce ligands, enzyme substrates or heavy atoms into the crystals and should be carried out at least 24-48 h before X-ray analysis to allow the compound diffusion along the channels and into the crystals.
- Gently remove the sealing tape from the reservoirs.
- Add up to 5 µL of ligand solution in one or several reservoirs using a 10 µL micropipet (in the example, 3 µL of 10 mM cytidine-5'-[(α,β)-methyleno] triphosphate (CMPcPP) solution was added to achieve a final concentration of 3.75 mM). CMPcPP is a non-hydrolizable analog of CTP, a natural substrate of the enzyme.
- Seal the reservoirs with a 2.5 cm x 1 cm piece of tape.
- Incubate the chip under controlled temperature for 24-48 h to allow ligand diffusion along the channels of the chip.
5. Crystal analysis by serial crystallography
NOTE: This part of the protocol needs to be adapted depending on the beamline setup and the diffraction properties of the crystals. Only general indications are given for the crystallographic analysis based on experiments performed at X06DA beamline (SLS, Villigen, Switzerland).
- ChipX mounting on the beamline goniometer
NOTE: The file for 3D printing the ChipX holder is provided in ref4.- Turn off the cryo-jet of the beamline. The analysis here is carried out at room temperature.
- Mount the ChipX on a dedicated holder with the channel containing the crystals to be analyzed positioned at the center of the holder. The ChipX holder4 does not require any screw or additional part, as it was designed to provide a perfect fit for ChipX.
- Attach the holder to the goniometer.
- Data collection
- Orient the thickest layer (top layer, Figure 1) of ChipX (in this orientation labels along the channels are directly readable using the centering camera of the beamline), towards the direct beam and the thinnest face behind the crystal to minimize the attenuation of the diffracted signal as described in ref3.
- To avoid collision of ChipX with the surrounding material (beamstop, collimator), restrict goniometer movements in the range ±30° (0° corresponding to the channels being perpendicular to the X-ray beam).
- Find crystals position with the help of the labels embossed along the channels.
- Select a crystal position.
- Center the crystal either by standard low dose grid/raster screening or 1-click procedure (the video shows an example of grid screening).
- Collect diffraction data within the range -30° / +30°.
- Restart the procedure at steps 5.2.4-5.2.6 on another crystal in the same channel after translation of the chip.
- Manually realign another ChipX channel at the center of the holder and carry on data collection on crystals present in this channel.
- Use standard crystallographic packages and procedures to process and merge the data, then to solve and refine the structure.
Representative Results
The microfluidic chip described here was designed to enable easy setup of crystallization assays and crystal analysis at room temperature. The procedure described above and in the video was applied in the frame of the structural characterization of the CCA-adding enzyme from the cold-adapted bacterium Planococcus halocryophilus. This enzyme belongs to an essential polymerase family that catalyses the sequential addition of the 3' CCA sequence on tRNAs using CTP and ATP9,10.
The chip was first used to prepare crystals of the enzyme for structural analysis by the method of counter-diffusion. To this end, the enzyme solution was loaded in the eight microfluidic channels (crystallization chambers) by a single injection in the sample inlet of the chip (see Figure 1). The enzyme was used at 5.5 mg/mL in its storage buffer containing 20 mM Tris/HCl pH 7.5, 200 mM NaCl and 5 mM MgCl2. This step was performed manually with a standard 10 µL micropipet. Crystallization solutions (100 mM sodium acetate pH 4.5,1 M diammonium hydrogen phosphate) were then deposited in the reservoirs at the other extremity of the channels.
The loading procedure is straightforward and does not take longer than five minutes (Figure 2). The crystallant then diffuses into the channels, creates a gradient of concentration that triggers crystal nucleation and growth. This gradient evolves dynamically and explores a continuum of supersaturation states5,6 until reaching an equilibrium of crystallant concentration between the channels and the reservoir. Crystallization assays are typically checked under the micoscope over a period of 2 – 4 weeks to track the growth of crystals. Bipyramidal crystals of CCA-adding enzyme appeared throughout the channels after a few days of incubation at 20 °C (Figure 3). The optional fluorescent labeling7 of the protein greatly facilitates the identification of protein crystals and their discrimination from salt crystals (Figure 4).
We exploited the diffusive environment in chip channels to deliver a substrate to the enzyme that builds up the crystals. In the present case, CMPcPP, a CTP analog, was added to the reservoir solutions at a final concentration of 3.75 mM (Figure 5). This addition was performed two days before the crystallographic analysis to allow CMPcPP to reach and occupy the catalytic site of the enzyme, as later confirmed by the crystal structure (see below).
We manufactured a chip holder (Figure 6) in polylactic acid using a 3D printer. The holder enables chip mounting on goniometers using standard magnetic heads. Hence the chip can be easily positioned and translated in the X-ray beam to bring the crystals in diffraction position. The data collection strategy needs to be adapted depending on beamline characteristics and on crystal properties. In the case of the CCA-adding enzyme, data were collected at X06DA and X10SA beamlines, Swiss Light Source (SLS), with an X-ray wavelength of 1.0 Å and Pilatus 2M-F and 6M pixel detectors, respectively. 30-60° of rotation were collected on each crystal at room temperature with images of 0.1° or 0.2° and 0.1 s exposure (see Table 1). Partial datasets were processed individually and cut when the resolution of diffraction patterns started to decay due to radiation damage (detected by the decrease of signal-to-noise ratio 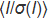 and CC1/2, and an increase of Rmeas in the high resolution shell). Full datasets were reconstituted by merging data from 5 crystals (Table 1). Crystal structures were derived by molecular replacement using standard crystallographic packages and procedures for data processing11 and refinement12. The comparison of the structures of the enzyme and of its complex with CMPcPP reveals the local conformational adaptation that accompanies substrate binding in the active site of the CCA-adding enzyme (Figure 7).
and CC1/2, and an increase of Rmeas in the high resolution shell). Full datasets were reconstituted by merging data from 5 crystals (Table 1). Crystal structures were derived by molecular replacement using standard crystallographic packages and procedures for data processing11 and refinement12. The comparison of the structures of the enzyme and of its complex with CMPcPP reveals the local conformational adaptation that accompanies substrate binding in the active site of the CCA-adding enzyme (Figure 7).
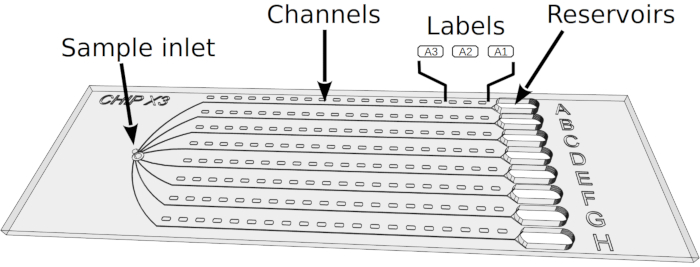
Figure 1: ChipX design. The chip consists of a top layer made of COC (thickness: 1 mm) in which eight microfluidic channels and reservoirs are imprinted. The entire chip is sealed with a layer of COC (thickness: 0.1 mm). All channels are connected to a single inlet on the left hand side for simultaneous sample injection and to individual reservoirs on the right hand side in which crystallization solutions are deposited. The channels, which constitute the actual crystallization chambers of the chip, are 4 cm long and have a cross section of 80 µm x 80 µm. Labels (A1, A2, A3, etc.) embossed along the channels facilitate crystal positioning under the microscope and the preparation of a sample list for data collection. ChipX has the size of a standard microscope slide (7.5 cm x 2.5 cm). Please click here to view a larger version of this figure.
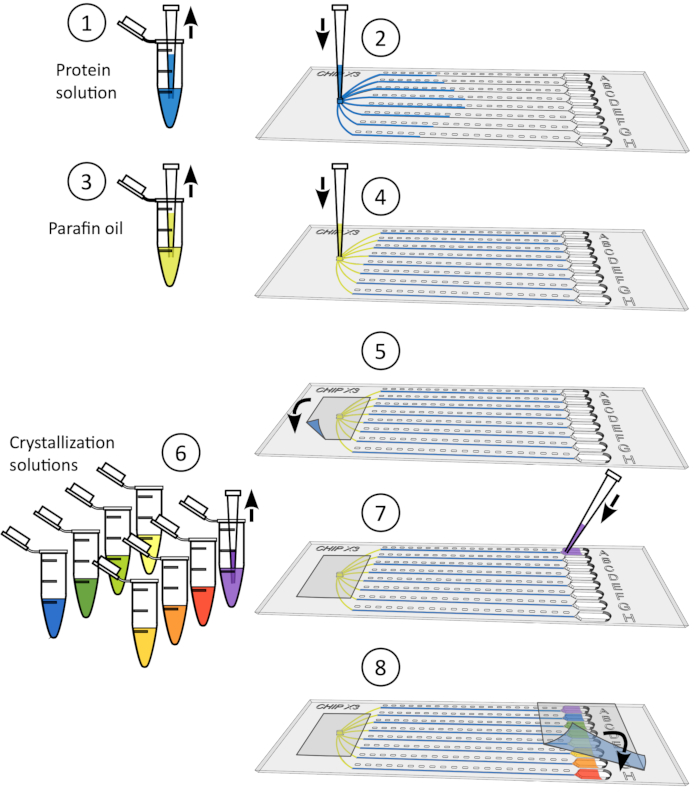
Figure 2: Setting up crystallization assays in ChipX. 1) Deposit 5-6 µL of enzyme solutions using standard 10 µL pipet and tip. 2) Introduce the tip vertically in the sample inlet and inject the solution in the eight channels. 3) Pipet 1 µL of paraffin oil. 4) Introduce the tip vertically in the sample inlet and inject the oil in order to disconnect the channels from each other. 5) Seal the inlet with a piece of tape. 6) Pipet 5 µL of crystallization solution using standard 10 µL pipet and tip. Solutions can be different in every reservoir (e.g., from a screening kit). 7) Orient the pipet tip towards the entry of the channel in the funnel shaped part of the reservoir (to avoid the formation of an air bubble upon solution deposition) and inject the crystallant solution in the reservoir. 8) Seal the reservoirs with a piece of tape and incubate the chip at controlled temperature. Please click here to view a larger version of this figure.
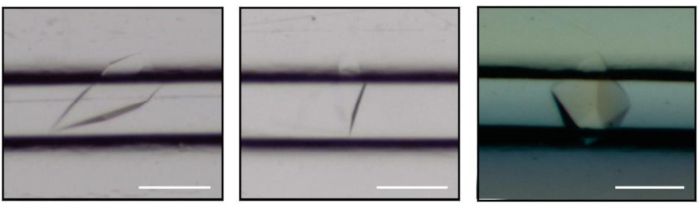
Figure 3: Crystals of CCA-adding enzyme grown by counter-diffusion in the microfluidic channels of ChipX. Scale bar is 0.1 mm. Please click here to view a larger version of this figure.
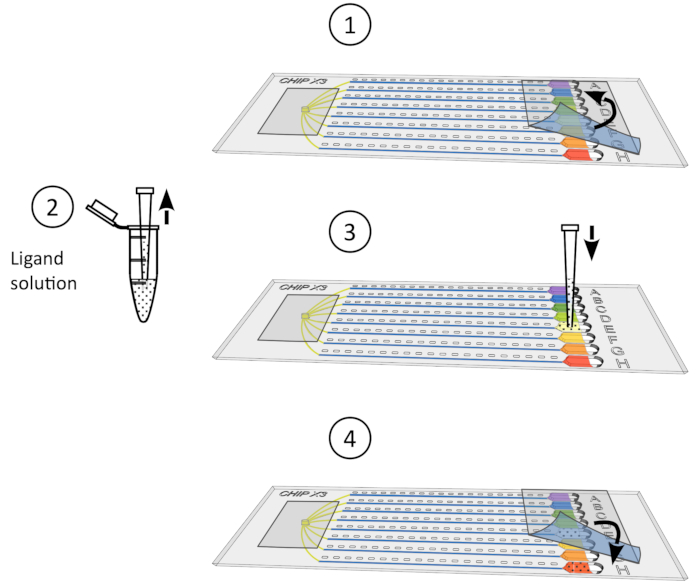
Figure 4: Crystal soaking procedure. 1) Gently remove the tape from the reservoirs. 2) Deposit up to 5 µL of ligand solution using a 10 µL micropipet. 3) Add the ligand to one or several reservoirs. 4) Seal again the reservoirs with a piece of tape and incubate the chip under controlled temperature for 24-48 h before data collection. Please click here to view a larger version of this figure.
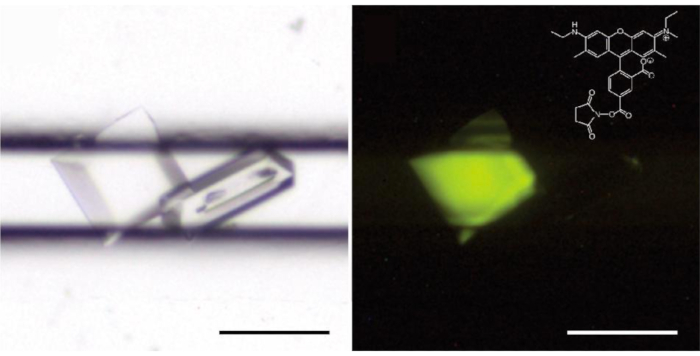
Figure 5: Trace fluorescent labeling discriminates protein (left) from salt (right) crystals. The CCA-adding enzyme solution contained 0.4 % (w/w) of protein labeled with carboxyrhodamine. On the right, crystals are illuminated with a 520 nm wavelength light source and the image is taken with a low pass filter at 550 nm (LP550); (inset) structure of carboxyrhodamine-succinimidyl ester. Please click here to view a larger version of this figure.
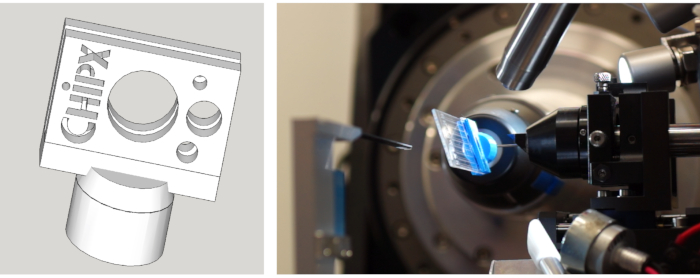
Figure 6: (Left) Drawing of the ChipX holder and (Right) ChipX mounted on the goniometer of beamline X06DA at SLS (Villigen, Switzerland) for serial crystal analysis. Please click here to view a larger version of this figure.
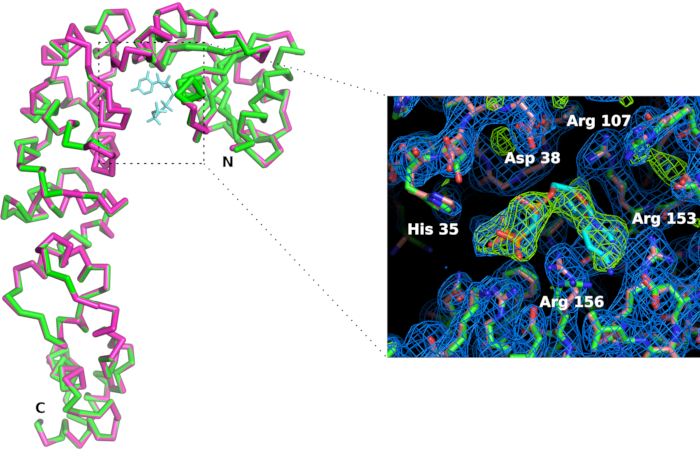
Figure 7: Comparison of CCA-adding enzyme active site in the apo form (in pink) and in the complex with a CTP analog (in green). Although the overall conformation of the enzyme is not affected, the binding of the CMPcPP ligand is accompanied by a slight reorganization of side chains in the active site. The 2Fo-Fc electron density map (in blue) is contoured at 1.2 sigma. The difference electron density map contoured at 4 sigma (in green) confirms the presence of the ligand in the active site. Please click here to view a larger version of this figure.
| Crystallized sample | CCA-adding enzyme | CCA-adding enzyme + CMPcPP |
| Crystal analysis | ||
| X-ray beamline | SLS – X06DA | SLS – X10SA |
| Wavelength (Å) | 1.000 | 1.000 |
| Temperature (K) | 293 | 293 |
| Detector | Pilatus 2M-F | Pilatus 6M |
| Crystal-detector distance (mm) | 300 | 400 |
| Crystals collected | 6 | 14 |
| Crystals selected | 5 | 5 |
| Rotation range per image (°) | 0.1 | 0.2 |
| Exposure time per image (s) | 0.1 | 0.1 |
| No. of images selected | 1000 | 540 |
| Total rotation range (°) | 100 | 108 |
| Space group | P43212 | P43212 |
| a, c (Å) | 71.5, 293.8 | 71.4, 293.6 |
| Mean mosaicity (°) | 0.04 | 0.04 |
| Resolution range (Å) | 46 – 2.54 (2.6 – 2.54) | 48 – 2.3 (2.4 – 2.3) |
| Total No. of reflections | 176105 (9374) | 232642 (32937) |
| No. of unique reflections | 23922 (1598) | 34862 (4066) |
| Completeness (%) | 90.6 (84.6) | 99.5 (100.0) |
| Redundancy | 7.5 (6.0) | 6.7 (8.1) |
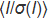 |
8.1 (1.3) | 6.9 (0.7) |
| Rmeas (%) § | 18.6 (126.0) | 18.0 (231.2) |
| CC1/2 (%) £ | 98.7 (55.0) | 98.7 (46.9) |
| Overall B factor from Wilson plot (Å2) | 57.4 | 60.6 |
| Crystallographic refinement | ||
| No. of reflections, working set / test set | 23583 / 1180 | 34840 / 3405 |
| Final Rcryst (%) / Rfree (%) | 18.8 / 21.4 | 20.0 / 22.9 |
| No. of non-H atoms: overall / protein / ligand / solvent | 2998 / 2989 / 0 / 9 | 3057 / 2989 / 29 / 10 |
| R.m.s. deviations for bonds (Å) / angles (°) | 0.009 / 1.23 | 0.010 / 1.22 |
| Average B factors (Å2): overall / protein / ligand / solvent | 60.1 / 60.1 / 0 / 52.7 | 62.5 / 62.6 / 60.1 / 55.5 |
| Ramachandran plot: most favored (%) / allowed (%) | 98.1 / 1.9 | 97.2 / 2.8 |
| PDB id | 6IBP | 6Q52 |
Table 1: Data collection and refinement statistics
§ Redundancy-independent Rmeas = Σhkl(N/N-1)1/2Σi | Ii(hkl)– <I(hkl)>| / ΣhklΣi Ii(hkl), where N is the data multiplicity 17.
£ Data with low <I/σ(I)> in outer shell (<2.0) were included based on CC1/2 criterion (correlation between two random halves of the dataset > 50%) as proposed by Karplus & Diederichs 18.
Discussion
Current protocols in biocrystallography involve the preparation of crystals using methods such as vapor diffusion or batch13, 14, and their transfer into a microloop for cryo-cooling15, 16 before performing the diffraction analysis in a nitrogen jet at cryogenic conditions. In contrast, direct crystal cryo-cooling is not possible in ChipX3 and crystals cannot be extracted from their microfluidic channel, which can be seen as limitations of this setup. However, the protocol described in the article provides a fully integrated pipeline for the determination of crystal structures at room temperature (i.e., in more physiological conditions). Even though data collection at room-temperature causes an increase of radiation damage19, this effect is counterbalanced by fast data acquisition time (a maximum of 60° rotation is collected on each crystal) and by merging of several partial datasets. Both ChipX design and material were optimized to reduce background scattering and diffraction signal attenuation3, and data collection can be performed on crystals with dimensions equivalent to half the size of the channels (40 µm)4.
To summarize, the main advantages of the protocol are the following. The crystals are produced in a convection-free environment (microfluidic channels), which is very favorable to the growth of high-quality crystals. The counter-diffusion method implemented in ChipX is very efficient at screening the supersaturation landscape; the diffusion of crystallants into the chip channel creates a concentration and supersaturation wave that helps determine appropriate nucleation and growth conditions5. Crystals are never directly handled, but are analyzed in situ, inside the chip, which preserves their genuine diffraction properties (i.e., does not alter crystal mosaicity by physical interaction or cryocooling)20. The diffraction analysis is performed on a series of crystals distributed along the chip channels with low dose exposure to minimize radiation damage, and a full dataset is assembled by merging partial data from the series. The standard footprint and simple design of ChipX will allow in the future a complete automation of in situ data collection using synchrotron or XFEL facilities. All steps of the protocol are carried out in ChipX. From the experimenter point-of-view, chip setup is simple and easy to perform with standard pipets and does not require any extra equipment. The tree-like channel connection at the sample inlet minimizes dead volumes in the system, which is important when working with samples that are difficult to purify or that are only available in limited quantity.
In conclusion, the lab-on-a-chip approach implemented in ChipX simplifies and efficiently miniaturizes the process of crystallization by counter-diffusion and crystal structure determination, allowing to go from the sample to its 3D structure in a single device. It is widely applicable and offers a user-friendly, cost-effective solution for routine serial biocrystallography investigations at room temperature.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge the Swiss Light Source (Villigen, Switzerland) for beamtime allocation to the project on beamlines X10SA (PXII) and X06DA (PXIII), Alexandra Bluhm for her contribution to structure refinement, Clarissa Worsdale for the recording of the voiceover and François Schnell (Université de Strasbourg) for his assistance in video editing and SFX. This work was supported by the French Centre National de la Recherche Scientifique (CNRS), the University of Strasbourg, the LabEx consortium "NetRNA" (ANR-10-LABX-0036_NETRNA), a PhD funding to R.dW from the Excellence initiative (IdEx) of the University of Strasbourg in the frame of the French National Program "Investissements d'Avenir", a PhD funding to K.R. from the French-German University (UFA-DFH, grant no. CT-30-19), the Deutsche Forschungsgemeinschaft (grant no. Mo 634/10-1). The authors benefitted from the PROCOPE Hubert Curien cooperation program (French Ministry of Foreign Affair and Deutscher Akademischer Austauschdienst).
Materials
| Axioscope A1 stereomicroscope | Zeiss | Crystal observation (step 3) | |
| Carboxyrhodamine succinimidyl ester | Invitrogen | C-6157 | Protein labeling (step 2) |
| CMPcPP | Jena Bioscience | NU-438 | Crystal soaking (step 4) |
| Crystal clear sealing tape | Hampton research | HR3-511 | ChipX sealing (step 1) |
| Parafin oil | Hampton research | HR3-411 | ChipX loading (step 1) |
| Ultimaker 2 extended+ | Ultimaker | 3D printer – Representative results | |
| UV light source | Xtal Concepts Gmbh | XtalLight100c | Crystal observation (step 3) |
| Zeba spin desalting column 7K MWCO | ThermoFisher Scientific | 89882 | Protein labeling (step 2) |
References
- Giegé, R., Sauter, C. Biocrystallography: past, present, future. HFSP Journal. 4 (3-4), 109-121 (2010).
- Dhouib, K., et al. Microfluidic chips for the crystallization of biomacromolecules by counter-diffusion and on-chip crystal X-ray analysis. Lab on a Chip. 9 (10), 1412-1421 (2009).
- Pinker, F., et al. ChipX: A Novel Microfluidic Chip for Counter-Diffusion Crystallization of Biomolecules and in Situ Crystal Analysis at Room Temperature. Crystal Growth & Design. 13 (8), 3333-3340 (2013).
- de Wijn, R., et al. A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography. IUCrJ. 6 (3), 454-464 (2019).
- García-Ruiz, J. M. A supersaturation wave of protein crystallization. Journal of Crystal Growth. 232 (1-4), 149-155 (2001).
- Otálora, F., Gavira, J. A., Ng, J. D., García-Ruiz, J. M. Counterdiffusion methods applied to protein crystallization. Progress in Biophysics and Molecular Biology. 101 (1-3), 26-37 (2009).
- Pusey, M., Barcena, J., Morris, M., Singhal, A., Yuan, Q., Ng, J. Trace fluorescent labeling for protein crystallization. Acta Crystallographica Section F Structural Biology Communications. 71 (7), 806-814 (2015).
- Meyer, A., Betzel, C., Pusey, M. Latest methods of fluorescence-based protein crystal identification. Acta Crystallographica Section F Structural Biology Communications. 71 (2), 121-131 (2015).
- Betat, H., Rammelt, C., Mörl, M. tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization. Cellular and Molecular Life Sciences. 67 (9), 1447-1463 (2010).
- Ernst, F. G. M., Erber, L., Sammler, J., Jühling, F., Betat, H., Mörl, M. Cold adaptation of tRNA nucleotidyltransferases: A tradeoff in activity, stability and fidelity. RNA Biology. 15 (1), 144-155 (2018).
- Kabsch, W. XDS. Acta Crystallographica. Section D, Biological Crystallography. 66 (2), 125-132 (2010).
- Adams, P. D., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 66 (2), 213-221 (2010).
- Dessau, M. A., Modis, Y. Protein Crystallization for X-ray Crystallography. Journal of Visualized Experiments. (47), e2285 (2011).
- Sauter, C., Lorber, B., McPherson, A., Giegé, R., Arnold, E., Himmel, D. M., Rossmann, M. G. Crystallization – General Methods. International Tables of Crystallography, Vol. F, Crystallography of Biological Macromolecules. , 99-120 (2012).
- Garman, E. “Cool” crystals: macromolecular cryocrystallography and radiation damage. Current Opinion in Structural Biology. 13 (5), 545-551 (2003).
- Li, D., Boland, C., Aragao, D., Walsh, K., Caffrey, M. Harvesting and Cryo-cooling Crystals of Membrane Proteins Grown in Lipidic Mesophases for Structure Determination by Macromolecular Crystallography. Journal of Visualized Experiments. (67), e4001 (2012).
- Diederichs, K., Karplus, P. A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature Structural Biology. 4 (4), 269-275 (1997).
- Karplus, P. A., Diederichs, K. Linking Crystallographic Model and Data Quality. Science. 336 (6084), 1030-1033 (2012).
- de la Mora, E., Coquelle, N., Bury, C. S., Rosenthal, M., Holton, J. M., Carmichael, I., Garman, E. F., Burghammer, M., Colletier, J. -. P., Weik, M. Radiation damage and dose limits in serial synchrotron crystallography at cryo- and room temperatures. Proceedings of the National Academy of Sciences of the United States of America. 117 (8), 4142-4151 (2020).
- Nave, C. A. Description of Imperfections in Protein Crystals. Acta Crystallographica. Section D, Biological Crystallography. 54 (5), 848-853 (1998).

