Cryo-Electron Microscopic Grid Preparation for Time-Resolved Studies using a Novel Robotic System, Spotiton
Summary
The protocol presented here describes the use of Spotiton, a novel robotic system, to deliver two samples of interest onto a self-wicking, nanowire grid that mix for a minimum of 90 ms prior to vitrification in liquid cryogen.
Abstract
The capture of short-lived molecular states triggered by the early encounter of two or more interacting particles continues to be an experimental challenge of great interest to the field of cryo-electron microscopy (cryo-EM). A few methodological strategies have been developed that support these "time-resolved" studies, one of which, Spotiton-a novel robotic system-combines the dispensing of picoliter-sized sample droplets with precise temporal and spatial control. The time-resolved Spotiton workflow offers a uniquely efficient approach to interrogate early structural rearrangements from minimal sample volume. Fired from independently controlled piezoelectric dispensers, two samples land and rapidly mix on a nanowire EM grid as it plunges toward the cryogen. Potentially hundreds of grids can be prepared in rapid succession from only a few microliters of a sample. Here, a detailed step-by-step protocol of the operation of the Spotiton system is presented with a focus on troubleshooting specific problems that arise during grid preparation.
Introduction
The potential for cryo-EM to capture and reveal transient conformational states of proteins on the sub-second timescale (time-resolved cryo-EM) has been realized by several groups, beginning with Berriman and Unwin1 whose technique built upon the standard plunge freezing method developed by Dubochet for preparing cryo-EM grids2. They added an atomizer just above the cryogen cup that used compressed nitrogen gas to spray a fine mist of a second sample onto a plunging EM grid containing a first sample that had been applied and blotted to a thin aqueous layer. Although this system could achieve mixing times as low as 1 ms, it still demanded manual blotting of the first sample by the user-a technically challenging task-and a relatively high volume of the second sample. Moreover, in practice, it was difficult to know where mixing of the two samples had occurred, requiring the use of ferritin nanoparticles as a fiducial marker in the mixed sample. Subsequent efforts by Howard White and co-workers improved control and reproducibility of this spraying-mixing approach by incorporating computer control of the blotting and spraying steps3,4. To address how well and where the samples mix, the same group5 and others6,7,8,9,10 have moved to a mixing-spraying approach11 in which two samples are mixed either in narrow capillary tubes under the pressure of syringe pumps or in microfabricated, microfluidic chips driven by nitrogen gas. These premixing systems not only ensure complete mixing, but also enable the fine-tuning of mixing times to increase the resolution of time-resolved studies.
The introduction of piezoelectric dispensers as an alternate way to apply sample to EM grids in the Spotiton system enabled both the precise targeting of sample deposition and the requirement of a much smaller sample volume to make a grid12. Later, the use of nanowire grids and en route sample application ("on-the-fly" spotting) removed the need for a blotting step and reduced application-to-vitrification times13,14. For the new approach to time-resolved cryo-EM described here, a second dispenser along with the necessary control hardware and software upgrades were added to the Spotiton system to enable the delivery of a second sample onto a moving nanowire grid almost immediately after deposition of the first15. The two overlapping samples mix on the grid as they are wicked by the nanowires into a thin aqueous layer prior to vitrification. Mixing times as low as 90 ms can be achieved. This protocol intends to provide practical information as to how to conduct time-resolved experiments using piezoelectric dispensing and nanowire grids. Additionally, as the hardware and software are being modified to enhance ease of use, consistency, and throughput, this protocol also serves as an up-to-date description of the previously reported method15.
Protocol
1. Set up the Spotiton machine and software
- Prepare the system to dispense (Figure 1).
- Open the main valve on the nitrogen supply tank. Ensure the system reservoir is filled with degassed, ultrapure water. Turn on the computer. Turn on the Spotiton system at the multioutlet powerstrip.
- Click on the desktop icon (Figure 2A) to open the user interface (UI) of the Spotiton software (Figure 2B). In the Tools menu, select Initialize Stages to initialize and home the 3-axis robots (grid stage and pipette stage) and the rotating dispenser head assembly (theta stage). Ensure the dispenser tips are pointing down prior to initialization and homing.
- On the main window, click on Go to SafePosition to send the robots to the SafePosition (Figure 3).
NOTE: Moving to SafePosition can be done with robots in any position without risk of collision. - On the Aspirate tab, select Prime to flush the dispenser heads multiple times with water from the reservoir. Continue until uninterrupted streams of water can be seen emerging from the two tips.
NOTE: This may need to be repeated if a significant volume of air entered the fluid lines when the tips were flushed with methanol at last shutdown.
- Inspect dispenser performance
- On the Inspect tab, send each tip to the inspection camera to test-fire water, and match the pattern of droplet production from the two dispensers (Figure S1).
NOTE: If a tip fails to fire due to a bubble (Figure S2A) or debris, clean the tips at the wash station (Figure S2B) using the Ultrasonic Wash function accessed on the Aspirate tab. - Adjust the firing amplitude (unitless) for each dispenser and sample to achieve a stream of discrete droplets of consistent size.
- Visually confirm equivalent droplet production by the two dispensers.
- Press the Record button in the upper camera monitor to record a video of Tip 1 firing. Play back the video of Tip 1 firing in the right-side monitor at the same time Tip 2 is fired in the upper camera monitor (Figure S1).
NOTE: Videos of each tip firing are recorded and stored.
- Press the Record button in the upper camera monitor to record a video of Tip 1 firing. Play back the video of Tip 1 firing in the right-side monitor at the same time Tip 2 is fired in the upper camera monitor (Figure S1).
- Dispensers are now ready to test-fire on a grid.
NOTE: This protocol assumes that correct alignment of the grid and pipette stages at their respective plunge positions has been verified. Failure to confirm correct alignment could result in a collision, damaging the tips or the robots.
- On the Inspect tab, send each tip to the inspection camera to test-fire water, and match the pattern of droplet production from the two dispensers (Figure S1).
- Test-fire water on a grid.
- Ensure robots are at the SafePosition.
- Remove the tweezers from the mount on the grid robot using the supplied Allen key.
- On a nearby benchtop, position a test grid, nanowire side up, on the edge of the grid block. Carefully grab the rim of the grid, position correctly in the tweezers (Figure S3), and remount the tweezers.
- On the Cryo tab, click on Tip to Camera to move the tips into the field of view of the upper plunge path camera (upper camera). Ensure Live is selected in the upper camera monitor, and the upper camera light is turned on (dial on front of machine cabinet (Figure 1).
- Mount the tweezers on the grid robot. Position Tip 1, visible in the upper camera monitor, by clicking the mouse within the monitor.
NOTE: Only Tip 1 will be visible and will move to the location of the click. - Click Grid to Camera to position the grid in front of the upper camera. Adjust Tip 1 position as before, if needed.
- Choose either Tip1, Tip2, or Tip1 and Tip2 to select either one or both dispensers to test-fire.
NOTE: When test-firing the tips individually, use the mouse to position Tip 1 in the upper camera monitor at different lateral locations on the grid. This will deposit liquid stripes from the two tips in a non-overlapping pattern and allow the user to confirm each tip has fired. - On the Cryo tab, ensure Vitrify Grid is not selected, click on Queue Target, then on Plunge.
NOTE: The pipette stage will rise slightly, followed by the grid stage. The grid robot then plunges the grid past the firing dispensers and upper and lower cameras, coming to rest just above the deck. An image capture from the upper camera appears above an image capture from the lower camera on the left-hand side of the UI. - Evaluate the upper and lower images: Did each tip fire when selected? When fired together, did the liquid from the two tips overlap fully, creating a noticeably thicker stripe than when fired individually?
- If either tip failed to fire, perform one or multiple ultrasonic wash cycles until clear firing is observed.
- If the sample stripes do not fully overlap, adjust the lateral alignment of one of the dispensers using an Allen key to turn the lateral adjustment screw on one of the dispensers one quarter turn, and perform a test plunge. Repeat until stripes fully overlap.
NOTE: If both tips fired successfully and as expected, the system is ready to prepare sample grids.
2. Prepare two-sample, mixed grids
- Update the acceleration, deceleration, and velocity values in the Machine Parameters XML file to attain the mixing time of interest.
NOTE: Tables correlating these values to mixing times have been previously reported15. - Ready the sample and grids.
NOTE: From this point in the protocol, 20-30 min will elapse before the first grid is prepared, and samples should be kept at an appropriate temperature during this time interval.- Dilute two samples (i.e., protein and ligand or other interacting partner) to the desired concentrations with an appropriate buffer, ideally, the same for both.
NOTE: Spotiton grids require 1.5-2 times higher concentrations of protein than typically used for automatic plunge freezers. This may be due to the minimal time the protein spends in a thin aqueous layer on the grid surface during a plunge, giving particles less opportunity to concentrate at the air-water interface. - Fill the cryogen bowl with liquid nitrogen.
- Plasma clean 3-4 nanowire grids. Use 5 W, hydrogen and oxygen, 1.5 min as a starting point.
NOTE: The most effective plasma cleaning duration and recipe may change from day-to-day based on ambient temperature and humidity and the batch of nanowire grids used. Alternative gas mixtures or a glow discharger may also be effective, but have not been tested for this purpose. As water and protein in buffer solution are often wicked at different speeds by nanowires, it is best practice to evaluate the wicking of the grids to be used that day on a plunge in which the actual sample is dispensed.
- Dilute two samples (i.e., protein and ligand or other interacting partner) to the desired concentrations with an appropriate buffer, ideally, the same for both.
- Set the humidity level in the system.
NOTE: In addition to the conditions used to both make and plasma clean the grids, humidity is another primary factor that impacts the wicking speed of nanowire grids. Although the target percent humidity for a particular session is determined empirically for each day and batch of grids, 90-95% is a good starting point.- Ensure the nebulizer is filled sufficiently with ultrapure water. Plug in the nebulizer, and observe the vapor exit the central port on the nebulizer cap.
- Observe the live humidity monitor in the main window, or open the ambient humidity tracker under Reports | Ambient (Figure S4). Check humidity levels in the two zones: Chamber and Shroud.
NOTE: Chamber includes all areas within the Spotiton enclosure. Shroud is the area immediately encompassing the "at camera" positions of the grid and dispenser tips. The black resin shroud maintains the set humidity level when the enclosure doors are opened to load a grid. - Once the target humidity values are reached, maintain these values either automatically or manually from the Humidity tab. Select Automatic control, and choose a setpoint and threshold to maintain the setpoint within the threshold limit chosen. Alternatively, select Manual control, and adjust the humidity level using the two fan controls: chamber fan and shroud fan.
NOTE: Turning on the chamber fan pulls the vapor through a filter in the left port and prevents larger water droplets from entering the chamber, preventing further increase in ambient humidity. As long as the chamber doors remain closed, the humidity level will stabilize at the desired percentage. Turning on the shroud fan pulls the vapor through the right port into the shroud. This both reduces vapor release into the chamber and increases the humidity level within the shroud.
- Load the sample into the dispensers.
- Add 5 µL of each sample into the sample cups.
NOTE: To avoid introducing bubbles, dispense the volume very carefully onto the inner sidewall of the cup, then shake down to force the sample to the bottom of the cup. - Load the sample cups into the holding tray, the sample for Tip 1 on the left, for Tip 2 on the right. Push the tray back into the machine until it seats.
- On the Aspirate tab, select 3 µL for the volume to be aspirated by each tip. Ensure the sample tray has been seated securely, then click on Aspirate and observe how the pipette stage moves the dispenser heads into the sample cups.
- Verify successful aspiration of both samples by removing the sample cups and observing a drop in the liquid level.
- On the Inspect tab, send each tip to the inspection camera to confirm unobstructed dispensing. Adjust the amplitude as needed to match droplet formation from each tip (see section 1.2).
NOTE: Amplitude will likely need to be increased for the tip that dispenses the protein sample. - The system is now ready to prepare a sample grid.
- Add 5 µL of each sample into the sample cups.
- Freeze sample grids
- Load a freshly plasma cleaned grid into the tweezers, but do not mount the tweezers yet. Ensure humidity level is elevated, ~90-95%. Fill the ethane cup.
- Perform a final test-fire of both tips in front of the inspection camera, confirming no obstruction. On the Cryo tab, click on Tip to camera.
- Fill ethane cup. If ethane ice forms, melt as needed with additional ethane gas.
NOTE: Steps 2.5.4 and 2.5.5 should be completed relatively quickly (<20 s) to minimize (i) saturation of the nanowires in the high humidity of the chamber and (ii) the likelihood that the tip firing the protein sample will clog. - Mount the tweezers with the grid onto the grid stage. On the Cryo tab, click on Grid to camera. Ensure Tip 1 is positioned correctly in the upper camera monitor (see steps 1.3.4-1.3.5).
- Click on Vitrify Grid, Queue Target, then Plunge. Click on OK when prompted to command the grid robot to hop the grid from ethane into liquid nitrogen and release it onto the submerged shelf.
NOTE: The tweezers then rise back into the chamber. After the hop to liquid nitrogen, a prompt appears asking if the grid dropped from the tweezers. If it did not drop, click on No, and the tweezers will open and close several times to detach the grid. - Examine images of the grid (Figure S5) to decide if it should be kept or discarded.
- If keeping the grid, transfer it to the pre-cooled grid box. Alternatively, spot subsequent grids and then transfer all grids at once to the grid box, taking care that each grid can be identified by its position in the resting area.
- To transfer the grid to a grid box, pre-cool fine-tipped forceps, gently grasp the grid by the edge, and place it into a grid box slot, starting with the first slot left of the notch going clockwise.
- When all the grids are loaded, attach the grid box lid to the lid tool, and precool in liquid nitrogen. Screw the lid onto the grid box, and tighten with a lid-tool or a precooled screwdriver.
- Use large forceps to quickly transfer the closed grid box to a small dewar for imaging or long-term storage.
- Evaluate grid suitability for downstream EM imaging.
- Observe the images from the upper and lower plunge path cameras that are displayed on the left side of the UI immediately after a grid is plunged. Use these images to evaluate wicking speed, successful firing of both tips, and the extent of overlap of the two samples stripes.
- Review and compare the grid images from the upper and lower cameras along with the machine settings and humidity measurements at the time of the plunge using the experiment viewer: Reports | Experiment.
- Select grids for imaging in the electron microscope that demonstrate evidence of good wicking.
Representative Results
Figure 4 shows images of grids prepared during a single time-resolved Spotiton session by mixing RNA polymerase and a 105 bp DNA oligomer carrying a promoter sequence for 150 ms prior to vitrification15. Seen in the figure are images taken by the two high-speed cameras of six grids at two timepoints following sample application. These cameras, unique among plunge-style freezers, allow the user to immediately decide whether to keep or discard a grid based on the observed extent of wicking by the nanowires and the likelihood the liquid samples were drawn into an aqueous layer thin enough for EM imaging. Although a single grid can provide sufficient ice for a complete dataset, once optimal conditions have been achieved, several grids are prepared to keep on hand in the event one or more is lost or contaminated. Of the six grids prepared in this session, the image captures show only one with suboptimal wicking (Figure 4F).
The grids shown were prepared (steps 2.5.1 through 2.5.8) in succession over a period of 40 min after an hour to ready the samples and the machine as described in the protocol (steps 1.1.1 through 2.4.6). Of the six grids, two were used for data collection, and the rest were saved for later analysis if needed. The pattern of ice on a vitrified grid (Figure 5C) matches closely the pattern of deposited liquid seen in the upper camera image (Figure 5B). Effective wicking of the mixed samples, made apparent by the lack of a visible liquid stripe in the lower camera image (Figure 5A), occurs along the nanowire-covered grid bars, and sample rarely overflows into squares adjacent to those in which it landed. Within ice-filled squares, the ice is typically thickest within holes at the center of the square and becomes thinner in holes closer to the grid bars (Figure 5E). Often holes immediately adjacent to the grid bars are empty due to proximity to the nanowires (Figure 5F).
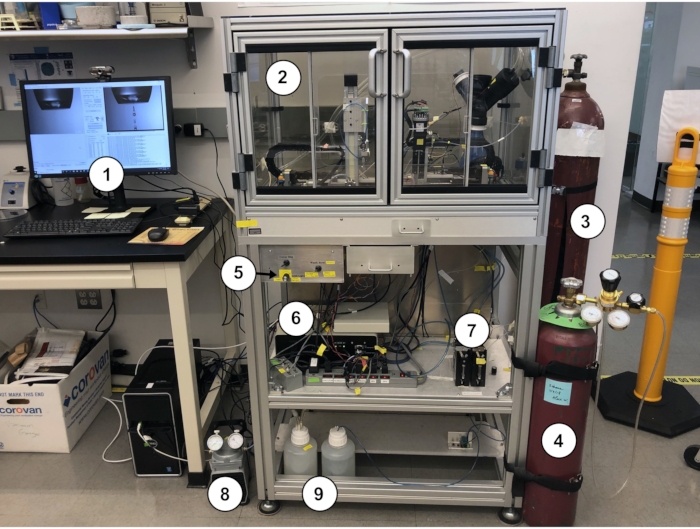
Figure 1: The time-resolved Spotiton system. 1. Operator's workstation; 2. Environmental chamber; 3. Nitrogen supply; 4. Ethane supply 5. Backlight control for upper plunge path camera 6. Piezoelectric dispenser controllers; 7. Syringe pumps; 8. Vacuum pump; 9. Wash water supply and waste bottles. Please click here to view a larger version of this figure.
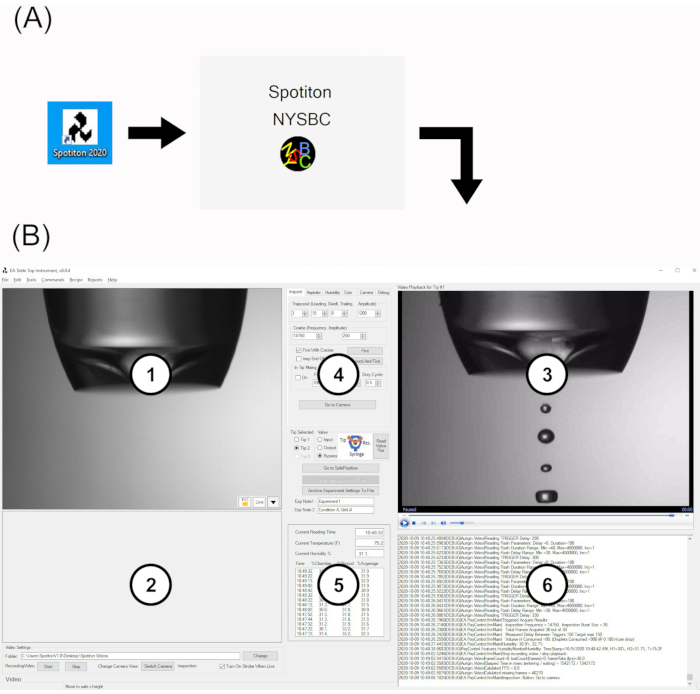
Figure 2: Spotiton software user interface. (A) Desktop icon and splash screen. (B) The main UI is composed of six areas: 1. display area for upper plunge path ("upper") and tip inspection cameras; 2. display area for lower plunge path ("lower") camera; 3. Playback area for tip inspection videos; 4. Multifunction tab area; 5. Live humidity monitor; 6. Live system logfile. Abbreviation: UI = user interface. Please click here to view a larger version of this figure.
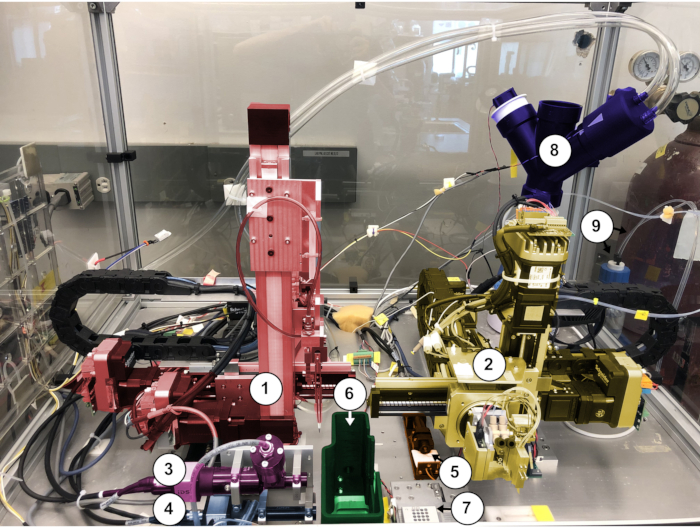
Figure 3: Interior view of Spotiton chamber (robots in SafePosition). 1. Grid robot (red); 2. Dispenser robot (yellow); 3. Upper plunge path camera (pink); 4. Lower plunge path camera (light blue); 5. Tip inspection camera (orange); 6. Humidity shroud (green); 7. sample tray; 8. Nebulizer assembly (dark blue); 9. System reservoir and water lines. Please click here to view a larger version of this figure.
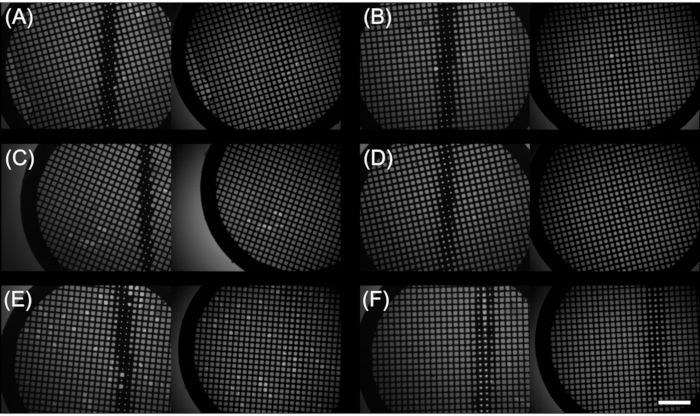
Figure 4: Representative system camera images from a time-resolved Spotiton grid making session. (A–F) Upper (left) and lower (right) plunge path camera images of six grids onto which RNA polymerase and a promoter DNA sequence were applied using Spotiton. Scale bar = 500 µm. Please click here to view a larger version of this figure.
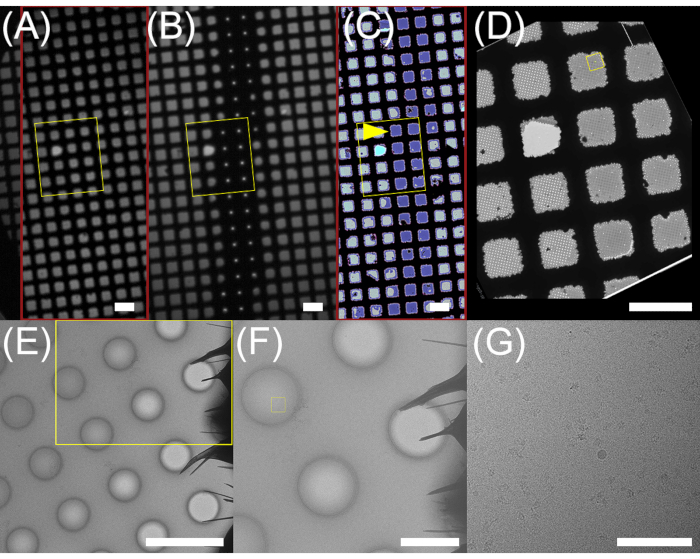
Figure 5: Pattern of ice deposition on a Spotiton-prepared grid. Portions of the (A) lower and (B) upper camera images and (C) atlas of the grid shown in Figure 4F. Approximate locations of ice resulting from sample deposition and mixing are colored lavender. Areas within the square marked with a yellow arrowhead are imaged in (E–G). The region shown in (E) is boxed in yellow in (D). Representative square (E), hole (F), and (G) exposure images collected from the grid shown in (A–D). The boxed areas in the square and hole images correspond to the hole and exposure images, respectively. Scale bars = 100 µm (A–D), 5 µm (E), 2 µm (F), 100 nm (G). Please click here to view a larger version of this figure.
Figure S1: Inspection of tip firing. A live view of Tip 2 firing is seen in the upper camera monitor on the left of the User Interface, while a recording made previously of Tip 1 firing plays on a loop for comparison in the video playback area on the right. Please click here download this File.
Figure S2: Ultrasonic cleaning of dispenser tips. An air bubble in the (A) tip will disrupt droplet formation and prevent tip firing. (B) Tips immersed in water at the ultrasonic cleaning station to remove an air bubble or clear dried protein blocking the tip orifice. Please click here download this File.
Figure S3: Loading grid in tweezers. (A) A grid placed nanowire side up on the edge of the grid block. (B) A grid positioned correctly in the self-closing tweezers. Please click here download this File.
Figure S4: Humidity tracker. The percent relative humidity in the chamber (dark blue) and shroud (light blue) as recorded during a typical grid-making session. The times of grid plunges (green squares) are plotted on the graph. Please click here download this File.
Figure S5: Wicking on nanowire grids during a plunge. Representative upper (A, C, E) and lower (B, D, F) plunge path camera images of wicking on nanowire grids that is too slow (A, B), ideal (C, D), and too fast (E, F). A slight thickening (white arrowheads) indicates grid bars with nanowires that have been saturated by sample. Squares in these regions typically contain ice of appropriate thickness for electron microscopic imaging. Scale bar = 500 µm. Please click here download this File.
Discussion
This protocol outlines the use of the Spotiton robotic system to prepare grids for cryo-EM imaging that carry two samples, generally a protein of interest and an activating ligand, that have been mixed for 90-500 ms. Although the workflow is straightforward, there are a few considerations the user must keep in mind to ensure a productive grid-making session. First, it is not uncommon for one of the piezoelectric dispenser tips to become clogged or blocked preventing it from firing. Such a failure will result in a liquid stripe, seen on image captures from the upper or lower camera, which is visibly narrower than that seen after both tips fired. A blockage could result from an air bubble lodging in the tip, disrupting droplet formation, or from protein sample that has dried out and sealed the narrow tip orifice. Although the aspirated sample is lost in the process, both problems can be resolved by an ultrasonic wash of the tips and re-aspiration of the sample. To prevent subsequent clogging and sample waste, it is crucial to thoroughly prime (flush) the fluid lines prior to sample aspiration and to maintain a high and consistent humidity level within the chamber and the shroud. Additionally, a protein sample with a particularly high concentration can affect tip firing despite well-maintained humidity. Although increasing the firing amplitude on the Inspect tab can partially compensate for weak firing due to high protein concentration, diluting the sample at least 1:2 will improve tip firing and avoid clogging.
Second, it can be difficult to achieve the ideal wicking speed that is needed to generate ice of optimal thickness for the targeted mixing duration. Generally, faster mixing times will require faster wicking, slower mixing times require slower wicking. For the ideally wicked grid, a liquid stripe is clearly discernable in the upper camera image, while in the lower camera, only a very slight thickening of the grid bars in the location of the stripe remains visible. Slow wicking, indicated by a dark stripe on the lower camera image, generally leaves ice that is too thick for imaging. Absence of a stripe on either image indicates fast wicking that may have left no water in the holes (Figure S5). Several factors such as nanowire density, plasma cleaning settings and duration, and time of exposure to and set level of chamber humidity can affect wicking speed. Poor (slow) wicking may be the result of a sparse coating of nanowires on the grid. By slightly diluting and increasing the time of exposure to the nanowire solution16, the density and coverage of nanowires on the grid bars will increase, facilitating faster wicking. If the nanowire density is sufficient, increasing the wattage setting or duration of plasma cleaning will also improve wicking. The settings recommended here are relatively low power and long duration, but may be changed if needed.
However, if both the specific batch of grids and plasma cleaning settings have worked well in a prior session, slow wicking performance may arise from excessive exposure of the nanowires to high humidity within the chamber, leading to their saturation with moisture and reduced liquid-holding capacity. The grid-saturating effect of chamber humidity can be reduced by either minimizing the elapsed time between tweezer mounting and grid plunging or decreasing the system humidity level prior to a plunge. It should be noted, however, that the latter brings the associated risk that the tip loaded with protein sample will clog. To offset this risk, holding the tips in the shroud where a high humidity level is maintained, can increase the allowable amount of time to mount tweezers holding a new grid. Finally, it should be noted that a reduced plunge time (achieved by increasing grid acceleration and/or maximum velocity) can result in grids with thinner ice without actually changing the wicking characteristics of the grid. However, as the mixing time of the two samples will also be reduced, plunge time is not a factor that is typically changed to address slow wicking. To address wicking that is too fast, resulting in ice that is either too thin or absent in the grid holes, the opposite of the measures outlined above can be taken.
Spotiton presents certain advantages and disadvantages when compared to other techniques developed for subsecond time-resolved studies. As the mixed sample stripe contains only 2-4 nL of liquid from each dispenser, a single 3 µL aliquot of each sample is sufficient to prepare many grids-a key advantage when sample is limited. Additionally, observation of sample deposition using integrated cameras, although not entirely unique to Spotiton17, is not a feature of other mixing devices and allows plunged grids to be subjected to a crude pass/fail evaluation, greatly reducing screening time. One key disadvantage of the system is a minimum mixing time of 90 ms, restricted by the physical limitations of the mechanical components, that puts interrogation of faster biological reactions out of reach. By comparison, times less than 10 ms are routinely achieved on existing microfluidic systems. On the Spotiton-based, commercially available chameleon system, design and construction improvements have reduced the minimum plunge time to 54 ms and raise the possibility that addition of a second dispenser could allow faster mixing times than Spotiton can currently offer.
To date, a range of experiments has been conducted to investigate early, short-lived molecular states using Spotiton, including assembly of the 70S ribosome, calcium-triggered conformational changes in a transmembrane ion channel, and constriction of dynamin in response to GTP hydrolysis15. Since the publication of these results, several changes have been incorporated to the system to enhance the throughput, reproducibility, and reporting of Spotiton grid-making sessions. These include, among others, the dual-zone, automatic humidity-monitoring and control system, the experiment viewer feature, the side-by-side tip inspection feature, and several minor upgrades to the UI. The upgraded system will better support future two-sample mixing experiments similar to those previously reported as well as rapid binding assays such as between a small-molecule therapeutic and its protein target or even antibody-antigen complex formation. While current and future time-resolved experiments involving two interacting partners will certainly continue, the addition of a third piezoelectric dispenser and associated hardware could further broaden the range of possible experiments. For example, initial deposition of a detergent followed by the protein of interest, followed by the interacting or activating ligand could remove any potential negative impact of extended exposure to the detergent, often necessary to prevent common suboptimal imaging outcomes such as preferred orientation. In light of both the already published work and potential future applications, Spotiton represents an important tool for the cryo-EM community to facilitate the conduct of subsecond time-resolved studies.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank Peter A. Kahn and Terry Rohde at Engineering Arts LLC (Arizona, USA) for initial design and subsequent development of the Spotiton system. We thank the staff of the Simons Electron Microscopy Center at the New York Structural Biology Center for help and technical support. The work presented here was conducted at the National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the NIH (GM103310) and the Simons Foundation (SF349247).
Materials
| Buffer | N/A | N/A | |
| cryogenic grid storage boxes | EMS (Hatfield, PA; USA) | N/A | |
| Cu/Rh 300 mesh EM grids | EMS (Hatfield, PA; USA) | EMS300-Cu-Rh | treated to coat with nanowires as previously described (Wei et al, 2018) |
| ethane gas | TW Smith Corp. (Brooklyn, NY; USA) | N/A | |
| Grid-handling forceps | EMS (Hatfield, PA; USA) | 78320-5B | |
| liquid nitrogen | Airgas, Inc. (Radnor, PA; USA) | N/A | |
| liquid nitrogen reservoir with brass ethane cup (from FEI Vitrobot) | ThermoFisher Scientific (Waltham, MA; USA) | ||
| Picosystem | Hydro System and Supplies (Durham, NC; USA) | N/A | water purification system |
| Protein/other sample | N/A | N/A | |
| Solarus 950 plasma cleaner | Gatan, Inc. (Pleasanton, CA; USA) | N/A |
References
- Berriman, J., Unwin, N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy. 56 (4), 241-252 (1994).
- Dubochet, J., McDowall, A. W. Vitrification of pure water for electron microscopy. Journal of microscopy. 124 (3), 3-4 (1981).
- White, H. D., Walker, M. L., Trinick, J. A computer-controlled spraying-freezing apparatus for millisecond time-resolution electron cryomicroscopy. Journal of Structural Biology. 121 (3), 306-313 (1998).
- White, H. D., Thirumurugan, K., Walker, M. L., Trinick, J. A second generation apparatus for time-resolved electron cryo-microscopy using stepper motors and electrospray. Journal of Structural Biology. 144 (1-2), 246-252 (2003).
- Kontziampasis, D., et al. A cryo-EM grid preparation device for time-resolved structural studies. IUCrJ. 6, 1024-1031 (2019).
- Lu, Z., et al. Gas-assisted annular microsprayer for sample preparation for time-resolved cryo-electron microscopy. Journal of Micromechanics and Microengineering: Structures, Devices, and Systems. 24 (11), 115001 (2014).
- Chen, B., et al. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure. 23 (6), 1097-1105 (2015).
- Kaledhonkar, S., et al. Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature. 570 (7761), 400-404 (2019).
- Fu, Z., et al. The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Nature Communications. 10 (1), 2579 (2019).
- Mäeots, M. -. E., et al. Modular microfluidics enables kinetic insight from time-resolved cryo-EM. Nature Communications. 11 (1), 3465 (2020).
- Kaledhonkar, S., Fu, Z., White, H., Frank, J. Time-resolved cryo-electron microscopy using a microfluidic chip. Methods in Molecular Biology. 1764, 59-71 (2018).
- Jain, T., Sheehan, P., Crum, J., Carragher, B., Potter, C. S. Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. Journal of Structural Biology. 179 (1), 68-75 (2012).
- Razinkov, I., et al. A new method for vitrifying samples for cryoEM. Journal of Structural Biology. 195 (2), 190-198 (2016).
- Dandey, V. P., et al. Spotiton: New features and applications. Journal of Structural Biology. 202 (2), 161-169 (2018).
- Dandey, V. P., et al. Time-resolved cryo-EM using Spotiton. Nature Methods. 17, 897-900 (2020).
- Wei, H., et al. Optimizing “self-wicking” nanowire grids. Journal of Structural Biology. 202 (2), 170-174 (2018).
- Ravelli, R. B. G., et al. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nature Communications. 11 (1), 2563 (2020).

