Loss-of-Function Approach in the Embryonic Chick Retina by Using Tol2 Transposon-Mediated Transgenic Expression of Artificial microRNAs
Summary
We have developed a novel loss-of-function approach that involves the introduction and genomic integration of artificial micro-RNA sequences into chick embryos by using in ovo electroporation and the Tol2 transposon system. This technique provides a robust and stable gene knockdown methodology for studies of gene function during development.
Abstract
The chick retina has long been an important model system in developmental neurobiology, with advantages including its large size, rapid development, and accessibility for visualization and experimental manipulations. However, its major technical limitation had been the lack of robust loss-of-function approaches for gene function analyses. This protocol describes a methodology of gene silencing in the developing chick retina that involves transgenic expression of artificial microRNAs (miRNAs) by using the Tol2 transposon system. In this approach, a Tol2 transposon plasmid that contains an expression cassette for the EmGFP (emerald green fluorescent protein) marker and artificial pre-miRNA sequences against a target gene is introduced into the embryonic chick retina with a Tol2 transposase expression construct by in ovo electroporation. In the transfected retinal cells, the transposase catalyzes the excision of the expression cassette from the transposon vector and its integration into host chromosomes, leading to the stable expression of miRNAs and the EmGFP protein. In our previous study, we have demonstrated that the expression of Nel, a glycoprotein that exerts multiple functions in neural development, can be significantly suppressed in the developing chick retina by using this technique. Our results indicate that this methodology induces a stable and robust suppression of gene expression and thus provides an efficient loss-of-function approach for studies of retinal development.
Introduction
The vertebrate retina is an important model system for studying neural development. Despite its peripheral location, the retina is anatomically and developmentally an extension of the central nervous system, and the optic nerve, which consists of axons of retinal ganglion cells, represents a tract within the central nervous system. The chick retina has significant advantages as a model system to study the molecular mechanism of neural development: It is large and develops rapidly; it has structural and functional similarities to the human retina; it is highly accessible for visualization and experimental manipulations. Molecular mechanisms of cell proliferation and differentiation, morphogenesis, and axon guidance during neural development have been extensively studied by using the chicken retina.
In ovo electroporation has been successfully used over the last two decades to introduce ectopic genes into cells in the developing chick embryo. This technique allows for labeling of developing cells, cell fate tracing, and tracing of cell migration and axon tracts, as well as ectopic gene expression for in vivo analysis of gene function. The conditions of in ovo electroporation for efficient ectopic gene expression in chick embryos have been well established1,2,3.
Despite these advantages, the lack of a stable loss-of-function technique for studies of gene function had been a major technical limitation of the chick embryo. Whereas chick embryos electroporated with small interfering RNAs (siRNAs)4 or expression vectors for short hairpin RNAs (shRNAs)5 show knockdown of the targeted gene, gene suppression in those approaches is transient because the effects disappear once cells lose the introduced RNAs or DNAs. A more stable gene suppression can be achieved by delivering siRNAs into chick embryos by an RCAS (Replication Competent Avian sarcoma-leukosis virus (ASLV) long terminal repeat (LTR) with a Splice acceptor) retrovirus system6. The viral vector integrates into the host genome, and the ectopic genes are stably expressed. However, the retrovirus can only integrate into the genome of dividing cells during the mitotic (M) phase of the cell cycle, which may impose a limitation on the developmental stages and/or cell types for which this loss-of-function approach can be applied. In addition, expression of transgenes by RCAS appears slower and less robust than that induced by in ovo electroporation7.
Transposons are genetic elements that move from one location on the genome to another. The Tol2 element is a member of the hAT transposable element family and contains an internal gene encoding a transposase that catalyzes the transposon reaction of the Tol2 element8. When a plasmid vector that carries a gene expression cassette flanked by the sequences of the left and right ends of the Tol2 elements (200 bp and 150 bp, respectively) is introduced into vertebrate cells with a Tol2 transposase expression construct, the expression cassette is excised from the plasmid and integrated into the host genome, which supports a stable expression of the ectopic gene (Figure 1). It has been shown that the Tol2 transposable element can induce gene transposition very efficiently in different vertebrate species, including zebrafish9,10, frogs11, chicks12, and mice13, and thus is a useful method of transgenesis and insertional mutagenesis. The Tol2 transposon system has been successfully used for conditional knockdown of a target gene by genomic integration of siRNA that is processed from long double-stranded RNA14.
This protocol describes a loss-of-function approach in the chick embryo that involves the introduction of artificial microRNAs (miRNAs) by the Tol2 transposon system15,16. In this approach, an expression cassette for the EmGFP (emerald green fluorescent protein) marker and artificial miRNAs against a target gene is cloned into a Tol2 transposon vector. The Tol2 transposon construct is then introduced into the embryonic chick retina with a Tol2 transposase expression construct by in ovo electroporation. In the transfected retinal cells, the transposase catalyzes the excision of the expression cassette from the transposon vector and its integration into host chromosomes, leading to the stable expression of miRNAs and the EmGFP protein. In our previous studies, we successfully knocked down the expression of Nel, an extracellular glycoprotein predominantly expressed in the nervous system, in the developing chick retina (see Representative Results). Our results indicate that stable and efficient gene suppression can be achieved in ovo by this technique.
Protocol
1. Construction of miRNA expression vectors
NOTE: The procedures for constructing miRNA expression vectors (steps 1.1-1.3, 1.5-1.6.) are optimized for the miRNA expression kit, Block-iT Pol II miR RNA expression kit with EmGFP, as previously described15,16. The kit provides the expression vector designed to allow miRNA expression (pcDNA6.2-GW/EmGFP-miRNA), a control vector (pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid), accessory reagents, and instructions to produce miRNA expression vectors (see Table of Materials)17.
- Designing single-stranded DNA oligos encoding pre-miRNAs against the target gene: Design single-stranded DNA oligos ("top strand" oligos (target pre-miRNAs) and "bottom strand" oligos (complements of top strand oligos)) using the online tool, RNAi Designer (see Table of Materials). See Figure 2 for the required features of the single-stranded oligos (Figure 2A) and examples of target sequences (Figure 2B).
NOTE: It is recommended that 5-10 pre-miRNA sequences be generated for a given target gene and screened for knockdown activities in vitro (step 1.4). - Annealing of the top- and bottom-strand oligos to generate a double-stranded oligo
- Set up the following annealing reaction (Table 1) in a sterile 0.5 mL microcentrifuge tube.
- Incubate the reaction mixture at 95 °C for 4 min. Anneal the top- and bottom-strand oligos to generate a double-stranded oligo by allowing the reaction mixture to cool to room temperature (RT) for 5-10 min. Centrifuge the sample briefly (~5 s).
NOTE: The annealed oligos can be stored at -20 °C without degradation for at least a year.
- Cloning the double-stranded oligos into the miRNA expression vector (pcDNA6.2-GW/EmGFP-miRNA (Provided in the miRNA expression kit)): Clone individual double-stranded oligos into the linearized miRNA expression vector, according to the manufacturer's manual17.
- Evaluation of knockdown effects
NOTE: It is recommended that individual miRNA sequences be tested for gene suppression efficiency in vitro before they are applied in ovo. Knockdown efficiency can be tested by transfecting miRNA expression plasmids into a cell line that expresses the target gene. Alternatively, individual miRNA expression plasmids can be co-transfected into cell lines with an expression construct for the target gene. For target genes encoding proteins that are not membrane-anchored in their native state (e.g., soluble proteins), an alkaline phosphatase (AP) fusion protein can be used for monitoring the expression of the target protein. A cDNA sequence encoding the target protein can be fused in frame to human placental alkaline phosphatase in AP-tag vectors (APtag-1- APtag-5; see Table of Materials) and introduced into cells18. When expressed in culture cells (e.g., HEK293T cells), the AP-tagged target protein is secreted at high levels into culture media, and thus knockdown effects of miRNA sequences can be evaluated by measuring the decrease in AP activity in the culture media of miRNA-transfected cells (substeps 1.4.1-1.4.4).- Culture HEK293T cells in a 24-well plate (8 x 104 cells/well) overnight. Transiently transfect the cells with individual miRNA expression constructs with an expression plasmid of an AP-tagged target protein. Use the pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid (provided in the miRNA expression kit) as a control. (If cell lines that stably express an AP-tagged target protein are used, transfect the cells only with individual miRNA expression constructs.)
NOTE: A conventional lipofection reagent (see Table of Materials) is used for transfection. - Collect the conditioned medium 48-72 h after the transfection and heat inactivate the endogenous AP activity in a 65 °C water bath for 5 min. Spinout debris in a desktop microcentrifuge at maximum speed for 5 min.
- Buffer the supernatant with 10 mM HEPES, pH 7.0 and pass it through a 0.45 µm filter.
- Take 100 µL (for measurement in a plate reader) or 500 µL (for a spectrophotometer) of the supernatant and add an equal amount of 2x AP substrate buffer (Table 2). Check the AP activity by measuring OD405 in a plate reader or a spectrophotometer.
NOTE: If the AP activity of the conditioned medium is too high for accurate measurement, dilute it with HBAH buffer (Hanks' balanced salt solution (HBSS), 0.5 mg/mL bovine serum albumin, 20 mM HEPES (pH 7.0)) or another buffer that contains a carrier protein. Do not use phosphate-containing buffers (e.g., PBS) because inorganic phosphate acts as a competitive inhibitor of AP.
- Culture HEK293T cells in a 24-well plate (8 x 104 cells/well) overnight. Transiently transfect the cells with individual miRNA expression constructs with an expression plasmid of an AP-tagged target protein. Use the pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid (provided in the miRNA expression kit) as a control. (If cell lines that stably express an AP-tagged target protein are used, transfect the cells only with individual miRNA expression constructs.)
- Chaining of miRNA sequence
NOTE: Knockdown effects can be enhanced by chaining different miRNAs against the same target gene or repeating the same miRNA. The miRNA expression vector supports the chaining of multiple pre-miRNA sequences and their co-cistronic expression17.- Chain two different pre-miRNA sequences (against the same target gene) that show highest knockdown activities in in vitro assays (step 1.4), according to the manufacturer's instructions17.
- Evaluate gene suppression efficiency of the chained constructs using in vitro assays as described in step 1.4. If three or more pre-miRNA sequences show similarly high knockdown activities, test different combinations of two sequences and use the combinations that show the highest knockdown activity (see Representative Results).
- Transferring the EmGFP-pre-miRNA expression cassette to the Tol2 transposon vector
NOTE: The expression cassette containing EmGFP cDNA and two pre-miRNA sequences is transferred into the Tol2 transposon vector (pT2K-CAGGS vector, see Table of Materials). To this end, the expression cassette (encompassing from the 3' end of the CMV promoter to the miRNA reverse sequencing primer site of the miRNA expression vector) is PCR-amplified using primers with an artificial restriction enzyme site, and the PCR product is cloned into the Tol2 transposon vector. The Tol2 transposon vector contains the ubiquitous CAGGS promoter, which drives the expression of the inserted expression cassette. The CAGGS promoter and the expression cassette are flanked by the Tol2 sequences (Figure 1).- PCR amplification of the EmGFP-pre-miRNA expression cassette: Follow the reaction setup and the thermocycling conditions described in Table 3.
- Ligate the gel-purified PCR product (c. 1.3 kb) into the restriction enzyme-digested Tol2 transposon vector. Electroporate the plasmid into competent E. coli cells (see Table of Materials) and select the recombinants (pT2K-CAGGS-EmGFP-2x miRNA constructs).
- Prepare the plasmid by using a conventional maxiprep kit (see Table of Materials). Check the structure and sequence of the plasmid by restriction mapping and by using the primers used for the PCR, respectively.
2. Egg storage and incubation
- Purchase fertilized White Leghorn (Gallus gallus) eggs from local farms or commercial vendors.
NOTE: Eggs may be kept at 12-16 °C or at 4 °C for up to 1 week prior to incubation without significant loss of viability or delay in development during incubation. - Label the eggs with the start date of incubation and mark the top side of the egg (where the embryo will be positioned). Incubate fertilized eggs in a horizontal position at 38 °C until the embryos have reached Hamburger and Hamilton (HH) stages 10 (33-38 h) -11 (40-45 h)19.
3. In ovo electroporation
- Preparation for in ovo electroporation
- Preparation of 0.25% fast green solution: Dissolve 25 mg of Fast Green FCF in 10 mL of PBS. Filter the solution using a 0.2 µm syringe filter. The solution can be stored at RT.
- DNA cocktail: Prepare the injection solution by mixing the individual pT2K-CAGGS-EmGFP-2x miRNA plasmids (substep 1.6.3) with the Tol2 transposase expression plasmid (pCAGGS-T2TP vector; see Table of Materials) (5 µg/µL each) at the ratio of 2:1. Add 1/10 volume of 0.25% fast green solution to visualize the injected area.
NOTE: The optimum DNA concentration may vary depending on the constructs. - Setting up the microinjection apparatus (Figure 3A,B): The microinjection apparatus consists of the followings (see Table of Materials): Hamilton syringe, 18 G needle (Needle length = 2"), PVC (Polyvinyl chloride) tubing (2 cm length), Micropipette needle (Can be made by pulling capillary tubes with omega dot fiber (1 mm O.D. X 0.75 mm I.D) with a micropipette puller).
- Fill a Hamilton syringe with heavy mineral oil. Attach an 18 G needle to the syringe and fill the inner space of the needle with oil by depressing the syringe plunger.
- Attach a piece of 2 cm long PVC tubing to the end of the needle and fill the tubing with the oil.
- Attach a pulled micropipette needle to the tubing. Break off the tip of the micropipette needle to a 10-20 µm diameter by fine forceps to make a small opening. Fill the entire needle with oil.
NOTE: Care should be taken not to trap any air bubbles in the system, as they would inhibit the flow of the DNA solution.
- Put 5 µL of the colored DNA cocktail (substep 3.1.2) onto a sterile Petri dish. Under the dissecting microscope, place the tip of the micropipette needle into the DNA solution on the sterile Petri dish and slowly draw the solution into the needle.
- Wait until the pressure equilibrates inside and outside the needle (to avoid that air goes into the needle) and take the tip of the needle out of the DNA solution. Keep the tip of the needle submerged in sterile PBS in a small beaker until injection.
- Setting up the electroporation apparatus (Figure 3A,C):
- Set a pair of platinum electrodes with an electrode holder on a micromanipulator. Adjust the spacing between the tip of the electrodes to 2 mm (Figure 3C,E).
- Connect the electrodes to a square wave pulse generator with cables (see Table of Materials).
- Microinjection of DNA solution (Figure 3D)
- Remove a chicken egg from the incubator and wipe the surface of the egg with a tissue paper soaked in 70% ethanol.
- Attach an 18 G needle to a 10 mL syringe. Insert the needle through the blunt end of the egg, angled downward (45°) to avoid damaging the yolk.
- Withdraw 2-3 mL of albumin from the egg. Seal the hole with a piece of Scotch tape. Confirm that the embryo and vitelline membrane are detached from the shell by "candling" the egg with light.
- Remove a piece of eggshell (2-3 cm diameter circle) from the top of the egg using scissors and forceps to expose the embryo. Do not window more than five eggs at any one time to prevent drying out of embryos during electroporation.
NOTE: If it is difficult to open a window without cracking the egg, the entire top of the egg can be covered with Sellotape or Scotch tape before removing a piece of eggshell. - Insert the tip of the needle into the optic vesicle from its proximal side at a 45° angle and inject the DNA cocktail by slowly depressing (or tapping on) the plunger until the blue-colored solution fills the lumen (Figure 3D).
NOTE: Alternatively, a pressure microinjection system (see Table of Materials) can be used for the injection of DNA solutions. - Withdraw the needle and place its tip back into PBS.
- Electroporation (Figure 3E)
- Set pulse parameters of the electroporator as follows: Voltage: 15 V, Pulse length: 50 ms, Pulse interval: 950 ms, Pulse number: 5
- Add a few drops of HBSS onto the vitelline membrane over the embryo. Lower the electrodes into the HBSS using the micromanipulator, perpendicular to the anterior-posterior axis of the embryo.
NOTE: Alternatively, this procedure can be done without adding HBSS, as albumin is a good electric conductor that allows efficient electroporation. - Place the electrodes on either side (anterior (nasal) side and posterior (temporal) side) of the optic vesicle (Figure 3E). Ensure that the electrodes do not touch the embryo or blood vessels. Apply pulsed electric fields.
- Remove the electrodes and gently clean the electrodes with a water-soaked sterile cotton bud to avoid the accumulation of albumin.
- Seal the window with Scotch tape and re-incubate the embryo until the desired developmental stage.
Representative Results
Construction of Tol2 transposon constructs for expression of artificial miRNAs against Nel
Nel (Neural Epidermal growth factor (EGF)-Like; also known as Nell2) is an extracellular glycoprotein. It has structural similarities with thrombospondin-1 and is predominantly expressed in the nervous system20,21. We have previously demonstrated that Nel regulates differentiation and survival of retinal ganglion cells15 and acts as an inhibitory guidance cue for retinal axons22,23,24. In the developing chick retina, Nel expression is detected in the presumptive pigment epithelium at HH15 (embryonic day (E) 2.5). At HH20 (E3.5), Nel expression is also observed in newly differentiated retinal ganglion cells. Strong expression of Nel in the pigment epithelium and retinal ganglion cells continues at least until E1815.
To examine the function of Nel in the development of retinal ganglion cells, expression of the Nel gene was knocked down by introducing artificial miRNAs into the developing retina using in ovo electroporation and the Tol2 transposon system15,16. First, pairs of single-stranded DNA oligonucleotides were designed for 10 potential target sequences in the protein-coding region of the chicken Nel cDNA (GenBank accession number: NM_001030740.1) by using the online RNAi designing tool (see Table of Materials). The oligonucleotide pairs were annealed and individually cloned into the miRNA expression vector. Then, individual constructs were transfected into HEK293T cells that stably express Nel-AP, and their knockdown efficiencies were evaluated by measuring AP activity in the culture media. The pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid was used as a control (Figure 4A).
Three Nel pre-miRNA sequences that showed highest knock down effects were selected (against nucleotides 482-502, 910-930, and 2461-2481 of chicken Nel cDNA, respectively), and two pre-miRNA sequences were tandemly cloned into the miRNA expression vector (pcDNA6.2-GW/EmGFP-2x Nel pre-miRNA), according to the manufacturer's instructions17. The expression cassettes encoding the two pre-miRNA and EmGFP were amplified by PCR with an artificial EcoRI site on both ends (5′-GGGAATTCTCTGGCTAACTAGAGAAC-3′ and 5′-CCGAATTCCCTCTAGATCAACCACT-3′) and cloned into the pT2K-CAGGS vector (pT2K-CAGGS-EmGFP-2x Nel pre-miRNA). The sequence of the expression cassette was confirmed by using the primers used for the PCR. pT2K-CAGGS-EmGFP-2x Nel pre-miRNA constructs were individually co-transfected with the Tol2 transposase expression vector (pCAGGS-T2TP) into HEK293T cells that stably express Nel-AP, and inhibitory effects on Nel-AP expression was evaluated by measuring the AP activities in the culture media. As shown in Figure 4B, knockdown efficiency was significantly enhanced by the chaining of two miRNA sequences. Robust suppression of Nel-AP expression (more than 90%) was observed at least until 13 days after transfection (Figure 4B).
Stable suppression of Nel expression in the developing chick retina
A pT2K-CAGGS-EmGFP-2x Nel pre-miRNA construct (containing target sequences for nucleotides 482-502 and 2461-2481 of the chicken Nel cDNA) was co-transfected with the transposase expression vector (pCAGGS-T2TP) into the temporal or nasal side of the chick retina by in ovo electroporation at HH9-11 (Figure 3, Figure 5A). Sections were prepared from E4.5 or E8 retinae, and effects on Nel expression was evaluated by immunohistochemistry using anti-Nel antibody22. At E4.5, the expression of Nel in the retinal pigment epithelium was significantly reduced in EmGFP expressing cells (Figure 5B–D). At E8, decreases in Nel expression were also clearly observed in retinal ganglion cells (Figure 5E–I). Significant suppression of Nel expression continued at least until E18 (data not shown). The introduction of control miRNA did not affect Nel expression in the retina (Figure 5J–O).
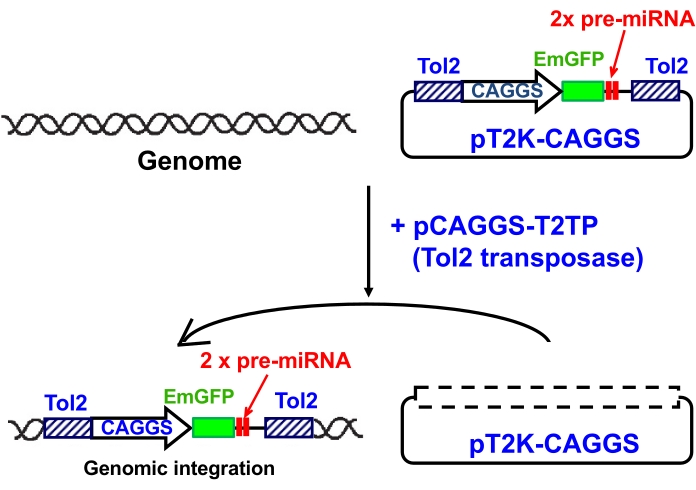
Figure 1: Transposition of Tol2-flanked cDNAs by transposase. A schematic showing transposition of a Tol2-flanked expression cassette for EmGFP and chained two pre-miRNA sequences (2x pre-miRNA) by transposase. When a Tol2 transposon vector containing the expression cassette (pT2K-CAGGS-EmGFP-2x miRNA construct) is introduced into cells with a Tol2 transposase expression construct (pCAGGS-T2TP), the Tol2-flanked expression cassette is excised from the vector and integrated into the host genome by the transposase activity. Expression of EmGFP and miRNAs is driven by the ubiquitous promoter CAGGS. This figure has been modified from Nakamoto et al.15. Please click here to view a larger version of this figure.
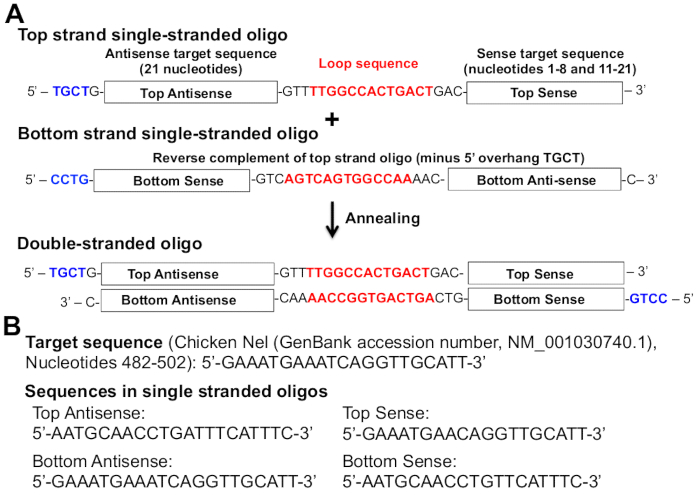
Figure 2: Structure of the engineered pre-miRNA. (A) The designed pre-miRNA sequence is based on the murine miR-155 sequence17. The top strand oligo contains (from the 5' end to the 3' end) a four-nucleotide overhang (TGCT (blue)), antisense target sequence (21 nucleotides), terminal loop sequence (red), and sense target sequence (19 nucleotides). The antisense target sequence represents the mature miRNA sequence. The bottom strand oligo contains a four-nucleotide 5' overhang (CCTG (blue)) and the reverse complement sequence of the top strand oligo but lacks the four-nucleotide overhang at the 3' end (AGCA-3': reverse complement of 5'-TGCT of the top-strand oligo). The sense target sequence lacks nucleotides 9 and 10 of the target sequence. The "extra" two nucleotides in the antisense target sequence form an internal loop in the stem-loop structure of the mature miRNA molecule, which results in a higher knockdown rate17. (B) An example of target sequence against chicken Nel. Sense and antisense sequences in the top and bottom strands for a target sequence of the chicken Nel gene (GenBank accession number NM_001030740.1, nucleotides 482-502) are shown. Please click here to view a larger version of this figure.
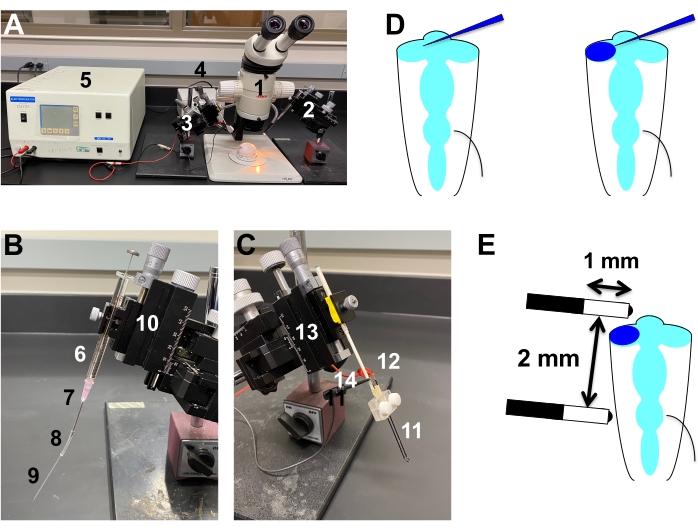
Figure 3: In ovo microinjection and electroporation. (A) Workstation for in ovo microinjection and electroporation. (1) Dissecting microscope. (2) Injection apparatus. (3) Electroporation apparatus. (4) Dual gooseneck microscope illuminator. (5) Electroporator. (B) Injection apparatus. A Hamilton syringe (6) with an 18 G needle (7) is connected to PVC tubing (8) and to a pulled micropipette needle (9) and set on a micromanipulator (10). The inner space of the apparatus is filled with heavy mineral oil. (C) Electroporation apparatus. A set of platinum electrodes (11) with an electrode holder (12) are set on a micromanipulator (13). The electrodes are connected to the electroporator with cables (14). (D) Microinjection of DNA cocktail solution into the optic vesicle at HH 10-11. The pulled micropipette needle is inserted into the optic vesicle from its proximal side.DNA cocktail solution with fast green (blue) is injected into the optic vesicle until the dye fills the entire lumen. (E) Electroporation into the optic vesicle. Electrodes are placed in parallel (The distance between the electrodes is 2 mm) and in a way so that the injected optic vesicle is located between the electrodes: One electrode is located near the anterior (nasal) surface of the optic vesicle, and the other is in the posterior part of the embryo at the level of the hindbrain. Please click here to view a larger version of this figure.
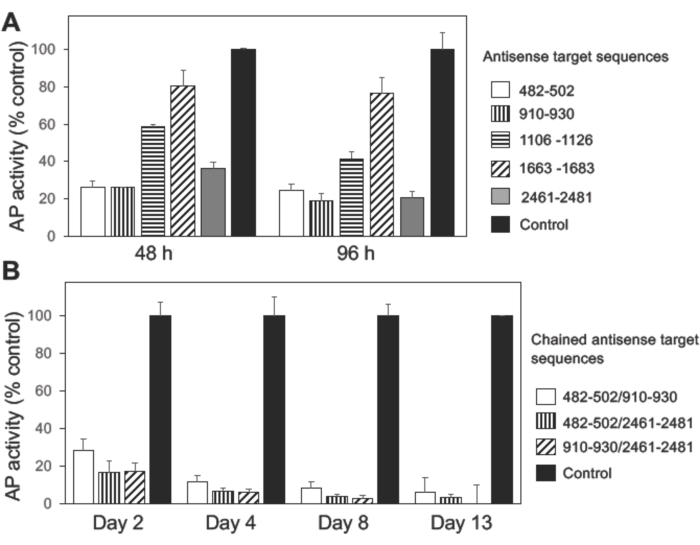
Figure 4: In vitro evaluation of individual or chained pre-miRNA sequences for Nel gene suppression efficiency. (A) Knockdown effects of individual pre-miRNA sequences. Results of five representative constructs (Antisense target sequences corresponding nucleotide numbers 482-502, 910-930, 1106-1126, 1663-1683, and 2461-2481 of chicken Nel cDNA (GenBank accession number: NM_001030740.1)) are shown. Individual miRNA expression constructs (pcDNA6.2-GW/EmGFP-Nel pre-miRNA) were transfected into HEK293T cells that stably express Nel-AP. The pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid was used as a control. Expression of Nel-AP was evaluated by measuring the AP activities in the culture media 48-96 h after transfection. Three Nel pre-miRNA expression constructs (against nucleotides 482-502, 910-930, and 2461-2481, respectively) showed significant suppression of Nel expression. (B) Knockdown effects of chained pre-miRNA sequences. Two of the three most potent pre-miRNA sequences (against nucleotides 482-502, 910-930, and 2461-2481) were tandemly cloned into the pcDNA6.2-GW/EmGFP-miR vector, and the expression cassette (containing the two pre-miRNA sequences and EmGFP cDNA) was transferred to the pT2K-CAGGS vector (pT2K-CAGGS-EmGFP-2x Nel pre-miRNA). The pT2K-CAGGS-EmGFP-2x Nel pre-miRNA constructs were individually co-transfected with expression vectors for Tol2 transposase (pCAGGS-T2TP) into HEK293T cells that stably express Nel-AP, and expression of Nel-AP was evaluated by measuring the AP activities in the culture media from 2 to 13 days after transfection. All three combinations (482-502/910-930, 482-502/2461-2481, 910-930/2461-2481) showed enhanced suppression activities compared to unchained individual pre-miRNA sequences. Significant suppression of Nel-AP expression was observed from day 2 to at least day 13. Please click here to view a larger version of this figure.
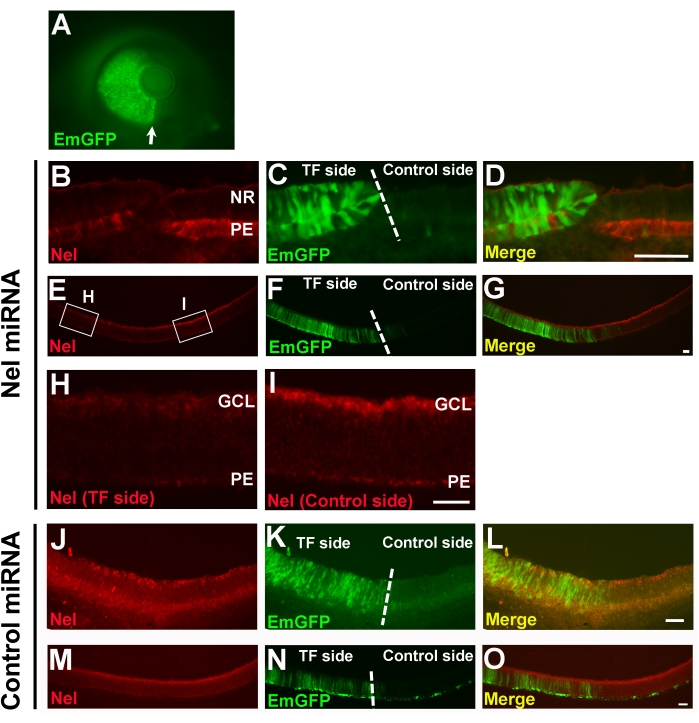
Figure 5: Stable knockdown of Nel expression in the developing chick retina by Tol2 transposon-mediated integration of miRNA transgene. A Tol2 transposon construct containing an expression cassette of two Nel pre-miRNA sequences and EmGFP (pT2K-CAGGS-EmGFP-Nel pre-miRNA (482-502)-Nel pre-miRNA (2461-2481)) (A–I) or a control construct that expresses an unrelated miRNA and EmGFP (J-O) was co-transfected with a transposase expression vector (pCAGGS-T2TP) into the temporal or nasal half of the optic vesicle by in ovo electroporation at HH9-HH11 (E1.5). Retinal sections were prepared at E4.5 (B–D, J–L) or E8 (E–I, M–O), and efficiencies of Nel gene suppression were examined by immunohistochemistry using anti-Nel antibody (red). Since DNA is negatively charged, the transgenes are electroporated into the tissue on the anode side; thus, only half of the retina was labeled with EmGFP (transfection (TF) side, green). (A) Expression of the EmGFP marker gene in the temporal half of the retina at E4.5. The optic fissure (white arrow) represents the boundary between transfected and untransfected (control) sides. (B–I) RNAi knockdown of Nel expression in the developing chick retina. (B–D) At E4.5, Nel expression is significantly reduced (B,D) in retinal pigment epithelium (PE) cells that express EmGFP (C,D). Note the complementary pattern of Nel immunostaining and EmGFP expression in D. NR, neural retina. (E–I) At E8, Nel expression (E,G) in the retinal pigment epithelium and the ganglion cell layer (GCL) is decreased on the transfection side (F,G). (H, I) Higher magnification views of a transfection area and a corresponding control area in E (indicated by small rectangles), respectively. (D,G) Merged images of the left two images. (J–O) Expression of control miRNA. Introduction of the negative control construct does not affect the expression of Nel in E4.5 (J–L) or E8 (M–O) retina. (L,O) Merged images of the left two images. The boundary between transfection and control sides is indicated by a dotted line in C, F, K, and N. Scale bars, 50 µm. (A) and (B–I) have been modified from Nakamoto, M. & Nakamoto, C16 and Nakamoto et al.15, respectively. Please click here to view a larger version of this figure.
| Annealing reaction setup | |
| Top strand oligo (200 μM in TE buffer) | 5 μL (Final concentration 50 μM) |
| Bottom strand oligo (200 μM in TE buffer) | 5 μL (Final concentration 50 μM) |
| 10x Oligo annealing buffer* | 2 μL |
| DNase/RNase-free water* | 8 μL |
| Total | 20 μL |
| * Provided in the miRNA expression kit (see Table of Materials) | |
Table 1: Annealing reaction setup
| 2x AP substrate buffer | |
| 10 M Diethanolamine (pH 9.8) | 4 mL |
| 1 M MgCl2 | 10 μL |
| L-Homoarginine | 45 mg |
| Bovine Serum Albumin (BSA) | 10 mg |
| p-Nitrophenylphosphate | 63 mg |
| H2O | Add to make up a total volume of 20 mL |
Table 2: 2x AP substrate buffer. The solution should be stored in aliquots at -20 °C and be protected from light as p-nitrophenylphosphate is light sensitive.
| Reaction setup | |||
| pcDNA6.2-GW/EmGFP-2x pre-miRNA plasmid (From Section 1.5) or pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid* (0.1 μg/μL) |
1 μL | ||
| 10x Pfu buffer | 5 μL | ||
| dNTP mix (10 mM each of dATP, dCTP, dGTP, and dTTP) | 1 μL | ||
| EmGFP forward primer* (5’-GGCATGGACGAGCTGTACAA-3’; 10 μM) | 1 μL | ||
| miRNA reverse primer* (5’- CTCTAGATCAACCACTTTGT-3’; 10 μM) | 1 μL | ||
| Pfu DNA polymerase (5 units/μL) | 0.5 μL | ||
| H2O | 40.5 μL | ||
| Total volume | 50 μL | ||
| * Provided in the miRNA expression kit. | |||
| Thermocycling conditions | |||
| 94 °C | 5 min | ||
| 30 cycles of the following: | |||
| 94 °C | 45 s | ||
| 58 °C | 1 min | ||
| 72 °C | 2 min | ||
| 72 °C | 10 min | ||
Table 3: Reaction setup and the thermocycling conditions.
Discussion
This protocol provides a detailed guide to gene silencing in the developing chick retina by transgenic expression of artificial miRNAs using in ovo electroporation and the Tol2 transposon system.
The following factors are of critical importance in performing this technique successfully. First, it is critical to use miRNA sequences that are confirmed to exert robust knockdown effects. Before applying them for in ovo electroporation, test individual pre-miRNA sequences for gene suppression efficiency in in vitro assays (step 1.4) and chain two pre-miRNA sequences that show the highest knockdown activities (step 1.5). Second, it is important not to damage the optic vesicle and surrounding nervous tissues when inserting the micropipette needle for injection of the DNA solution. Excessive damage on embryos could lead to embryonic malformation and death. To avoid tissue damages, introduce the micropipette into the optic vesicle in the correct orientation. Smaller and sharper tips of the micropipette needle would make penetration of the optic vesicle smoother and result in less tissue damage. However, needles with very small tips have difficulty with loading and releasing the DNA solution. In the study described here, needles with a 10-20 µm diameter tip opening were used. Third, make sure to fill the entire optic vesicle with the DNA solution (as visualized by the spread of fast green). Fourth, place the electrodes in the optimal positions to target DNAs into the desired area of the optic vesicle (e.g., temporal side, nasal side). Negatively charged DNA molecules will move towards the anode side; therefore the precise placement and orientation of the electrodes critically affect the final results. Finally, use the optimal DNA concentrations and electroporation parameters.
If no fluorescent signal is observed after 24 h of electroporation, the following should be considered: (1) Confirm that the electroporated plasmids have correct sequences. (2) Recheck the concentration and purity of individual plasmids. (3) Make sure that the DNA solution fills the lumen of the optic vesicle and it does not diffuse away into the neural tube. (4) Check the electroporation parameters used are correct. (5) Check that the electrodes are in contact with the vitelline membrane while the electric pulses are applied.
The combination of in ovo electroporation and a transposon-mediated gene integration method in this protocol offers several distinct advantages. First, it supports the stable production of miRNAs and thus persistent suppression of target genes. Development of the chick retina starts at E1.5 and continues until hatching (Retinal ganglion cells are produced over the period from E2 to E9). For loss-of-function analysis of gene function, expression of artificial miRNAs should be stably maintained during this period. Expression of RNAi sequences using conventional expression vectors is generally transient because the expression construct fails to be efficiently integrated into chromosomes. In the method described here, however, chromosomal integration of the expression cassette is mediated by the activity of co-electroporated transposase, which results in stable expression of the transgenes.
Second, in this method, the same CAGGS promoter induces co-cistronic expression of multiple miRNA sequences and the EmGFP marker in transfected cells, thus circumventing the risk of promoter interference, which is one of the common complications with retroviral vectors containing two promoters to attain dual gene expression25.
Finally, unlike replication-competent retroviral vectors, the transgene that is electroporated by this method is not transferred to neighboring cells. Therefore, the area of transfection can be more precisely controlled by using the appropriate types of electrodes and by placing them in the correct positions.
Despite the advantages described above, the current method also has limitations that are inherent to in ovo electroporation. For example, the rates of transfection and gene suppression may vary between embryos, and a relatively large number of embryos may need to be transfected for analysis. In addition, electroporation into the embryonic retina in ovo at later developmental stages (e.g., E4-E5) has experimental difficulties because the eyes are located in close proximity to the heart at those stages, which increases the risk of cardiac arrest after electroporation.
The system described here allows rapid and robust gene suppression in the developing chick retina. This approach can also be applied to other parts of the chick embryo, including the neural and non-neural tissues. We, therefore, expect that this system can contribute to the elucidation of molecular mechanisms of chick development.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The pT2K-CAGGS and pCAGGS-T2TP vectors were kindly provided by Yoshiko Takahashi (Kyoto University, Kyoto, Japan) and Koichi Kawakami (National Institute of Genetics, Mishima, Japan), respectively. We thank Michael Berberoglu for his crucial reading of the manuscript. This work was supported by grants from the Royal Society and Biotechnology and Biological Sciences Research Council (BBSRC) (UK) to M.N.
Materials
| 18 G needle, 2" | VWR | 89219-320 | |
| AP-TAG kit A and AP-TAG kit B | GenHunter Corp | Q201 and Q202 | Plasmid vectors for making AP fusion proteins (https://www.genhunter.com/products/ap-tag-kit-a.html, https://www.genhunter.com/products/ap-tag-kit-b.html) |
| Block-iT RNAi Designer | Invitrogen | An online tool to choose target sequences and design pre-miRNA sequences (https://rnaidesigner.thermofisher.com/rnaiexpress/) | |
| BSA 10 mg | Sigma-Aldrich | A2153 | |
| C115CB cables | Sonidel | C115CB | https://www.sonidel.com/product_info.php?products_id¼254 |
| C117 cables | Sonidel | C117 | https://www.sonidel.com/product_info.php?products_id¼252 |
| Capillary tubes with omega dot fiber (Micropipette needles) | FHC | 30-30-1 | 1 mm O.D. 0.75 mm I.D |
| CUY21 square wave electroporator | Nepa Gene | CUY21 | |
| Diethanolamine (pH 9.8) | Sigma-Aldrich | D8885 | |
| Dissecting microscope | |||
| Egg incubator | Kurl | B-Lab-600-110 | https://www.flemingoutdoors.com/kuhl%2D%2D-600-egglaboratory-incubator%2D%2D-b-lab-600-110.html |
| Electrode holder | Sonidel | CUY580 | https://www.sonidel.com/product_info.php?products_id¼85 |
| Electrodes | Nepa Gene | CUY611P3-1 | https://www.sonidel.com/product_info.php?products_id¼94 |
| Electromax DH10B | Invitrogen | 18290-015 | Electrocompetent E. coli cells |
| Fast green FCF | Sigma-Aldrich | F7258 | |
| Fertilized chicken eggs (Gallus gallus) | Obtained from commercial vendors (e.g. Charles River) or local farmers | ||
| Gooseneck fiber light source | |||
| FuGene 6 transfection reagent | Promega | E2691 | |
| Hamilton syringe (50 μL) | Sigma-Aldrich | 20715 | Hamilton Cat No 80901 |
| Hanks' balanced salt solution | Sigma-Aldrich | H6648 | |
| Heavy mineral oil | Sigma-Aldrich | 330760 | |
| HEPES | GIBCO | 15630080 | |
| L-Homoarginine | Sigma-Aldrich | H10007 | |
| MgCl2 | Sigma-Aldrich | 13112 | |
| Micromanipulator | Narishige (Japan) | MM3 | http://products.narishige-group.com/group1/MM-3/electro/english.html |
| Micropipette puller | Shutter Instrument | P97 | |
| p-Nitrophenylphosphate | Sigma-Aldrich | 20-106 | |
| PBS | Sigma-Aldrich | D8662 | |
| pCAGGS-T2TP vector | Tol2 transposase expression plasmid. A generous kind gift of Koichi Kawakami (National Institute of Genetics, Japan). Also available from Addgene. | ||
| Pfu | ThermoFisher | F566S | |
| Picospritzer (Optional) | Parker | Pressure microinjection system | |
| Plasmid maxi kit | Qiagen | 12163 | Plasmid maxiprep kit |
| pT2K-CAGGS vector | Tol2 transposon vector. Kindly provided by Yoshiko Takahashi (Kyoto University, Japan) | ||
| PVC tubing | VWR (UK) | 228-3830 | |
| Spectinomycin | Sigma-Aldrich | S9007-5 | |
| T4 DNA ligase | Promega | M1801 | |
| The BLOCK-iT Pol II miR RNA expression kit with EmGFP | Invitrogen | K493600 | Contains the miRNA expression vector (pcDNA6.2-GW/EmGFP-miRNA), a control vector (pcDNA6.2-GW/EmGFP-miRNA-negative control plasmid), accessory reagents, and instructions (https://www.thermofisher.com/order/catalog/product/K493600?SID.srch-hj-K4936-00) |
| Thermal cycler |
References
- Muramatsu, T., Mizutani, Y., Ohmori, Y., Okumura, J. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochemical and Biophysical Research Communications. 230, 376-380 (1997).
- Funahashi, J., et al. Role of Pax-5 in the regulation of a mid-hindbrain organizer’s activity. Development, Growth & Differentiation. 41 (1), 59-72 (1999).
- Harada, H., Omi, M., Nakamura, H. In ovo electroporation methods in chick embryos. Methods in Molecular Biology. 1650, 167-176 (2017).
- Hu, W. Y., Myers, C. P., Kilzer, J. M., Pfaff, S. L., Bushman, F. D. Inhibition of retroviral pathogenesis by RNA interference. Current Biology. 12 (15), 1301-1311 (2002).
- Katahira, T., Nakamura, H. Gene silencing in chick embryos with a vector-based small interfering RNA system. Development, Growth & Differentiation. 45 (4), 361-367 (2003).
- Harpavat, S., Cepko, C. L. RCAS-RNAi: a loss-of-function method for the developing chick retina. BMC Developmental Biology. 6, 2 (2006).
- Nakamura, H., Funahashi, J. Introduction of DNA into chick embryos by in ovo electroporation. Methods. 24, 43-48 (2001).
- Koga, A., Iida, A., Hori, H., Shimada, A., Shima, A. Vertebrate DNA transposon as a natural mutator: the medaka fish Tol2 element contributes to genetic variation without recognizable traces. Molecular Biology and Evolution. 23 (7), 1414-1419 (2006).
- Kawakami, K., Shima, A., Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proceedings of the National Academy of Sciences of the United States of America. 97 (21), 11403-11408 (2000).
- Kawakami, K., et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Developmental Cell. 7 (1), 133-144 (2004).
- Kawakami, K., Imanaka, K., Itoh, M., Taira, M. Excision of the Tol2 transposable element of the medaka fish Oryzias latipes in Xenopus laevis and Xenopus tropicalis. Gene. 338 (1), 93-98 (2004).
- Sato, Y., et al. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Biologie du développement. 2 (2), 616-624 (2007).
- Kawakami, K., Noda, T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Génétique. 166 (2), 895-899 (2004).
- Hou, X., et al. Conditional knockdown of target gene expression by tetracycline regulated transcription of double strand RNA. Development, Growth & Differentiation. 53, 69-75 (2011).
- Nakamoto, C., et al. Nel positively regulates the genesis of retinal ganglion cells by promoting their differentiation and survival during development. Molecular Biology of the Cell. 25 (2), 234-244 (2014).
- Nakamoto, M., Nakamoto, C., Mao, C. -. A. . iRetinal Development: Methods and Protocols. Vol. 2092 Methods in Molecular Biology. 8, 91-108 (2020).
- BLOCK-iT PolII miR RNAi Expression Vector Kits, User Manual Pol II miR RNAi Expression Vector Kits. Invitrogen Available from: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets/LSG/manuals/blockit_miRNAexpressionvector_man.pdf&title=BLOCK-iT&trade (2021)
- Flanagan, J. G., et al. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues, and embryos. Methods in Enzymology. 327, 19-35 (2000).
- Hamburger, V., Hamilton, H. I. A series of normal stages in the development of the chick embryo. Journal of Morphology. 88, 49-92 (1951).
- Matsuhashi, S., et al. New gene, nel, encoding a M(r) 93 K protein with EGF-like repeats is strongly expressed in neural tissues of early stage chick embryos. Developmental Dynamics. 203 (2), 212-222 (1995).
- Matsuhashi, S., et al. New gene, nel, encoding a Mr 91 K protein with EGF-like repeats is strongly expressed in neural tissues of early stage chick embryos. Developmental Dynamics. 207 (2), 233-234 (1996).
- Jiang, Y., et al. In vitro guidance of retinal axons by a tectal lamina-specific glycoprotein Nel. Molecular and Cellular Neuroscience. 41 (2), 113-119 (2009).
- Nakamura, R., Nakamoto, C., Obama, H., Durward, E., Nakamoto, M. Structure-function analysis of Nel, a Thrombospondin-1-like glycoprotein involved in neural development and functions. Journal of Biological Chemistry. 287 (5), 3282-3291 (2012).
- Nakamoto, C., Durward, E., Horie, M., Nakamoto, M. Nell2 regulates the contralateral-versus-ipsilateral visual projection as a domain-specific positional cue. Development. 146 (4), (2019).
- Yee, J. K., et al. Gene expression from transcriptionally disabled retroviral vectors. Proceedings of the National Academy of Sciences of the United States of America. 84 (15), 5197-5201 (1987).

