Small-Scale Plasma Membrane Preparation for the Analysis of Candida albicans Cdr1-mGFPHis
Summary
This article presents a small-scale plasma membrane isolation protocol for the characterization of Candida albicans ABC (ATP-binding cassette) protein Cdr1, overexpressed in Saccharomyces cerevisiae. A protease-cleavable C-terminal mGFPHis double tag with a 16-residue linker between Cdr1 and the tag was designed to facilitate the purification and detergent-screening of Cdr1.
Abstract
The successful biochemical and biophysical characterization of ABC transporters depends heavily on the choice of the heterologous expression system. Over the past two decades, we have developed a yeast membrane protein expression platform that has been used to study many important fungal membrane proteins. The expression host Saccharomyces cerevisiae ADΔΔ is deleted in seven major endogenous ABC transporters and it contains the transcription factor Pdr1-3 with a gain-of-function mutation that enables the constitutive overexpression of heterologous membrane protein genes stably integrated as single copies at the genomic PDR5 locus. The creation of versatile plasmid vectors and the optimization of one-step cloning strategies enables the rapid and accurate cloning, mutagenesis, and expression of heterologous ABC transporters. Here, we describe the development and use of a novel protease-cleavable mGFPHis double tag (i.e., the monomeric yeast enhanced green fluorescent protein yEGFP3 fused to a six-histidine affinity purification tag) that was designed to avoid possible interference of the tag with the protein of interest and to increase the binding efficiency of the His tag to nickel-affinity resins. The fusion of mGFPHis to the membrane protein ORF (open reading frame) enables easy quantification of the protein by inspection of polyacrylamide gels and detection of degradation products retaining the mGFPHis tag. We demonstrate how this feature facilitates detergent screening for membrane protein solubilization. A protocol for the efficient, fast, and reliable isolation of the small-scale plasma membrane preparations of the C-terminally tagged Candida albicans multidrug efflux transporter Cdr1 overexpressed in S. cerevisiae ADΔΔ, is presented. This small-scale plasma membrane isolation protocol generates high-quality plasma membranes within a single working day. The plasma membrane preparations can be used to determine the enzyme activities of Cdr1 and Cdr1 mutant variants.
Introduction
The extraction of integral membrane proteins from their native lipid environment can dramatically affect their structure and function1,2,3,4. The complex lipid composition of biological membranes5 ensures that critically important protein-lipid interactions can occur6. Lipids maintain the structural integrity of membrane proteins, thus enabling them to function correctly in their membrane compartment destination(s)7,8. Therefore, a critical first step in the membrane protein purification is the extraction of the protein from its native environment without affecting its structure and/or function.
There are many obstacles to characterizing the structure of membrane proteins, most of which are related to their hydrophobic nature, and the difficulties of expressing properly folded and functional membrane proteins in the quantities required for X-ray crystallography or cryo-electron microscopy (cryo-EM)9,10,11,12. There are three types of membrane protein expression systems: homologous9, heterologous13,14,15, and in vitro expression systems16,17. The often-low expression levels, or the prohibitive costs, of many expression systems leave only a few hosts as the preferred option to produce membrane proteins. They include the bacterial host, Escherichia coli, the yeasts S. cerevisiae and Pichia pastoris, and higher eukaryotes such as Sf9 insect cells or mammalian cell lines18. All membrane protein expression technologies have advantages and disadvantages; however, S. cerevisiae is perhaps the best studied eukaryotic model organism suitable for membrane protein production. It is highly versatile with applications in genetic engineering, drug discovery, synthetic biology, and the expression of eukaryotic membrane proteins14,19,20,21.
In this study, a patented S. cerevisiae membrane protein expression technology21 was used, with S. cerevisiae ADΔ14 and ADΔΔ22 as the preferred hosts (Figure 1A), to overexpress and study the major C. albicans multidrug efflux pump Cdr1. Both the S. cerevisiae strains are derivatives of AD1-8u– 23 that have either the ura3 (ADΔ) or both the ura3 and his1 (ADΔΔ) genes deleted to eliminate any false positive uracil or histidine prototroph transformants arising through the unwanted integration at the URA3 or the HIS1 genomic loci. The deletion of the 7 major multidrug efflux pumps23, indicated in Figure 1A, makes ADΔΔ exquisitely sensitive to most xenobiotics. The gain-of-function mutant transcription factor Pdr1-3 causes the constitutive overexpression of heterologous membrane proteins such as Cdr1 (red octagons in Figure 1A) after integration of the heterologous-ORF-containing transformation cassette (Figure 1A) at the genomic PDR5 locus (blue rectangle in Figure 1A) via two homologous recombination events. Proper plasma membrane localization of C-terminally mGFPHis tagged proteins can be confirmed by confocal microscopy (Figure 1A), and the His tag can be used for nickel-affinity purification of the tagged protein. Cloning some fungal ABC transporters (e.g., Candida krusei ABC1) into pABC3-derived plasmids was, however, not possible because they could not be propagated in Escherichia coli due to cell toxicity. This prompted the development of the one-step cloning of membrane proteins14,24 tagged at either their N- or C-terminus with various affinity, epitope, or reporter tags directly into S. cerevisiae ADΔΔ (Figure 1C). S. cerevisiae ADΔΔ strains overexpressing various CDR1 mutants can also be created efficiently this way by using up to five individual PCR fragments that overlap by 25 bp (Figure 1C). Employing this protocol, many ORFs of interest can be cloned, expressed, and characterized at low cost and at high efficiency within a very short time span. The transformation efficiency reduces only ~2-fold with each additional PCR fragment.
If desired, expression levels can also be readily manipulated by primer design to predictably tune expression levels down to anywhere between 0.1%-50% of the usually high, constitutive expression levels25. The optimized, multifunctional, pABC314 derivative cloning vector, pABC3-XLmGFPHis26 (Figure 1B) contains a HRV-3C protease cleavage site (X; LEVLFQ|GP), a protease that performs better at 4 °C than the frequently used tobacco etch virus (TEV) protease27. L is a five amino acid (GSGGS) linker, mGFP is a monomeric mutant (A206K)28,29 version of the yeast enhanced green-fluorescence protein variant yEGFP330, and His is a three amino acid linker (GGS) followed by the six-histidine (HHHHHH) nickel-affinity protein purification tag.
This expression technology has been successfully used in drug discovery and the study of membrane proteins. The first structure for a fungal azole drug target, S. cerevisiae Erg1131, was solved using this technology. It also enabled the detailed characterization of C. albicans Cdr132,33,34 and the creation of a cysteine-deficient Cdr1 molecule35 suitable for cysteine-crosslinking studies to verify any future high-resolution structure. Many other ABC transporters from major human fungal pathogens (i.e., C. albicans, Candida glabrata, Candida auris, Candida krusei, Candida utilis, Cryptococcus neoformans, Aspergillus fumigatus, Penicillium marneffei, and the Fusarium solani species complex) have also been studied in detail using this expression platform24,36,37,38,39. This has enabled the generation of a panel of S. cerevisiae strains overexpressing efflux pumps that has been used in high-throughput screens to discover the novel fluorescent efflux pump substrate Nile red40 and specific41 and broad-spectrum14,33,42,43,44 efflux pump inhibitors. The use of this system also enabled the discovery of clorgyline as the first of its kind broad-spectrum fungal multidrug efflux pump inhibitor42.
Complete solubilization of membrane proteins and the creation of a homogeneous membrane protein-micelle preparation devoid of endogenous lipids, requires high detergent concentrations45. But unfortunately, this also often inactivates the membrane protein5,8,45,46. The properties of detergent monomers and their aggregation in solution are affected by the physical properties of the hydrophobic tail, the length and branching of the alkyl chain, the presence of an aromatic nucleus or fluoroalkyl side chain, or the number of polyoxyethylene units. Thus, detergent screening is an important first step to determine the most suitable detergent for membrane protein solubilization and purification.
C. albicans is a major human fungal pathogen of immunocompromised individuals that can cause serious, life threatening invasive infections47, and it can become resistant to azole antifungal drugs48,49. One of the main mechanisms of C. albicans multidrug resistance is the overexpression of Cdr150, which is a type II ATP-binding cassette (ABC) transporter51 of the ABCG subfamily located in the plasma membrane. Full-size fungal ABCG transporters (consisting of two nucleotide binding domains [NBDs] and two transmembrane domains [TMDs]) are more commonly known as pleiotropic drug resistance (PDR) transporters and are characterized by their unique inverted domain topology [NBD-TMD]2. PDR transporters are only found in plants52,53 and fungi54. Despite their importance, there are no structures for PDR transporters, although structures for human half-size ABCG transporters have recently been solved which helped create the first tentative model for Cdr133. Our recent experimental evidence suggests, however, that this model is flawed possibly because fungal PDR transporters have characteristic asymmetric NBDs resulting quite possibly in a unique transport mechanism. A high-resolution structure of Cdr1 is, therefore, required for both the rational design of novel efflux pump inhibitors that may help overcome efflux-mediated drug resistance, and to provide insights into the mechanism of action of this important ABC transporter family.
The objective of this study was to develop reliable protocols for the expression, solubilization, and purification of Cdr1 in the genetically modified S. cerevisiae expression host, with the ultimate aim of obtaining a high-resolution structure for Cdr1. As part of this process, a protease-cleavable mGFPHis double tag (Figure 1B) was designed with a 16-residue linker separating the tag from the C-terminus of Cdr1, which improved binding of the attached 6x His affinity tag to the nickel-affinity resin and enabled the monitoring of Cdr1 expression levels in living cells and during the entire purification process. A reproducible protocol for small-scale yeast plasma membrane protein preparations containing about 10% C. albicans Cdr1 (as estimated by Coomassie staining after SDS-PAGE) was also developed, which could be used for the biochemical characterization of Cdr1.
Protocol
1. Preparation of fresh or frozen stocks of transformation competent ADΔ and ADΔΔ cells
- Inoculate 25 mL of 2x YPCD [i.e., 2x YPD; 2% (w/v) yeast extract, 2% (w/v) peptone, 4% (w/v) dextrose), 0.079 % (w/v) CSM (complete supplement mixture)]35 medium with a single yeast colony and incubate overnight (o/n) for 16 h at 30 °C with shaking at 200 revolutions per minute (rpm).
- Inoculate 225 mL of 2x YPCD medium with the 25 mL o/n culture and check the cell optical density at 600 nm (OD600); the OD600 is usually ~0.5-1.0.
NOTE: At this stage make sure that all materials that are required for the following transformation experiment are readily available. - Grow the culture at 30 °C for a further ~6-8 h with shaking at 200 rpm until the cell density reaches an OD600 of ~6-8.
NOTE: The following steps are performed at room temperature (RT) unless otherwise stated. - Harvest these logarithmic-phase cells by centrifugation at 3,000 x g for 3 min.
- Resuspend the cells and wash them twice with sterile double-distilled water (ddH2O; i.e., 200 mL and then 20 mL).
- Harvest the cells at 3,000 x g for 3 min.
- Slowly (i.e., add 30 equal aliquots of frozen competent cell (FCC) solution every minute for 30 min) resuspend the cell pellet in X mL of FCC [5% (w/v) glycerol, 10% (v/v) dimethyl sulfoxide (DMSO)] on ice (where X = OD600; e.g., if OD600 = 6 resuspend in 6 mL, or if OD600 = 3 resuspend in 3 mL).
NOTE: The correct FCC composition is critical for the transformation success. - Keep cells on ice for 2 h before transformation or store aliquots at -80 °C until required.
NOTE: ADΔ and ADΔΔ cells are very sensitive to freezing. Thus, cells must be cooled down slowly to -80 °C: place ice-cold microcentrifuge tubes containing 50-600 µL cell aliquots into a plastic storage box (RT). Place the box in a larger polystyrene container (RT) and close the container with a fitting polystyrene lid. Then, put the container into the -80 °C freezer.
CAUTION: Slow freezing is critical for cell survival.
2. Transformation of ADΔ and ADΔΔ with CaCDR1-XLmGFPHis and confirmation of correct transformants by colony PCR and DNA sequencing
NOTE: Plasmid pABC3-CDR1-mGFPHis was created using conventional cloning strategies described in detail in Lamping et al., 201055 and is illustrated in Figure 1B. Wild-type C. albicans CDR1 was isolated as a PacI/NotI fragment from plasmid pABC3-CaCDR1A-GFP14 and cloned into pABC3-XLmGFPHis26.
- PCR amplify the entire CDR1 transformation cassette with a high-fidelity DNA polymerase and primer pair PDR5-pro/PDR5-ter35 using 1-10 ng of pABC3-CaCDR1-XLmGFPHis as a DNA template or, alternatively, digest 2 µg of pABC3-CaCDR1-XLmGFPHis to completion with 10 U restriction enzyme AscI at 37 °C (Figure 1A,B).
NOTE: The CDR1 transformation cassette (Figure 1A) comprises the PDR5 promoter – CaCDR1–mGFPHis – PGK1 terminator – URA3 selection marker – PDR5 downstream region. - Perform agarose gel electrophoresis and gel extract the ~8 kb transformation cassette.
NOTE: Gel purification of the ~8 kb CaCDR1 transformation cassette removes any possible undigested plasmid DNA, which could lead to incorrect transformants that have the entire plasmid, rather than the linear transformation cassette, integrated at the genomic PDR5 locus. - Denature the required amount of salmon sperm carrier DNA (2 mg/mL; 10 mM Tris, 1 mM EDTA; pH 7.5) for 10 min in a boiling water bath and keep on ice. Use screw-capped tubes for boiling salmon sperm DNA to avoid opening of the lid.
- Mix 50 µL of denatured salmon sperm DNA with 14 µL of the transformation cassette (500-2,000 ng) and keep the 64 µL of DNA mixture on ice until further use.
NOTE: Use 10 ng of an E. coli-yeast shuttle plasmid (e.g., pYES2) as a positive transformation control (Figure 2B). - Resuspend fresh or frozen competent cells quickly defrosted for 5 min in a 30 °C water bath and divide them into 50 µL aliquots in 1.5 mL microcentrifuge tubes.
- Harvest cells by centrifugation for 1 min in a microfuge at maximum speed (18,000 x g).
- Remove the supernatant and keep the cell pellet at RT.
- For each transformation, mix 296 µL combinations (RT) of 260 µL of 50% (w/v) polyethylene glycol (PEG 3350) and 36 µL of 1 M lithium acetate (LiAc), by repeat pipetting, with the appropriate 64 µL of ice-cold DNA mixtures.
- Add the appropriate mixture immediately to a 50 µL competent cell pellet aliquot.
- Resuspend the cell pellet in the 360 µL of PEG-LiAc-DNA mixture by thoroughly vortexing for about 30 s.
- Incubate the cell mixture in a 30 °C water bath for 1 h.
NOTE: ADΔ and ADΔΔ cells transform better at 30 °C than at 42 °C35. - Harvest the cells at 18,000 x g for 10 s. Discard the supernatant and resuspend the cell pellet in 80 µL of ddH2O.
- Spread the cells onto a CSM-URA agar plate35 [i.e., 0.67% (w/v) yeast nitrogen base without amino acids, 0.077% (w/v) CSM minus uracil, 2% (w/v) glucose, and 2% (w/v) agar].
- Incubate the plates for 2-3 days at 30 °C until uracil prototroph transformants are clearly visible.
NOTE: Expect ~100 transformants per µg of the linear ~8 kb CaCDR1 transformation cassette and ~4 x 104 transformants per µg pYES2. - Pick five independent transformants and spread them on a fresh CSM-URA plate to separate the uracil prototroph transformants from remnants of non-transformed host cells.
- Remove any possible petite mutants by growing the transformants on YPG-agar plates [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (v/v) glycerol, and 2% (w/v) agar] (Figure 2D).
NOTE: Petite mutants have defective mitochondria and, thus, cannot grow on non-fermentable carbon sources. They are quite common in S. cerevisiae56. S. cerevisiae ADΔ and ADΔΔ are particularly prone to acquire the petite phenotype. - Perform yeast colony PCR and confirm at least three independent transformants to be correctly integrated into the genomic PDR5 locus (Figure 1C) by amplifying the entire ~8 kb CaCDR1 transformation cassette with a specific DNA polymerase that is optimized for amplifying PCR products from impure DNA template sources. Use a set of primers that bind just outside the integration site and 1 µL aliquots of cell suspensions (in ddH2O) derived from single colonies as DNA templates.
NOTE: This particular DNA polymerase reliably amplifies ~8 kb PCR fragments from intact yeast cells. However, for reliable amplification, 45 PCR cycles are required, and the yeast cells must be resuspended at OD600 1-10 in ddH2O. - Confirm the correct ~8 kb PCR amplification product by DNA agarose gel electrophoresis of a 1 µL portion of the PCR reaction.
- Remove excess amplification primers from a 10 µL portion of the PCR reaction with an enzyme mixture of a single-strand DNA exonuclease and a phosphatase following the manufacturer's instructions before sequencing the entire ORF using portions of the treated DNA sample with appropriate primers.
3. Small-scale yeast plasma membrane isolation protocol
- Growing yeast cells
- Pre-culture a single yeast colony in 10 mL of YPD at 30 °C for ~7-8 h with shaking at 200 rpm.
- Inoculate 40 mL of YPD medium with the 10 mL pre-culture and incubate the cells at 30 °C o/n (~16 h) with shaking at 200 rpm until the cell density reaches an OD600 of 1-3.
- Harvesting yeast cells
- Harvest 40 OD units (ODU; e.g., 1 mL at an OD600 of 1 = 1 ODU) of logarithmic-phase cells at 4,200 x g for 5 min at 4 °C.
- Resuspend and wash cells twice with ice cold sterile ddH2O (i.e., 40 mL and then 1 mL; harvest cells in between steps by centrifugation at 4,200 x g for 5 min at 4 °C).
- Resuspend the pellet in 1 mL of ice cold sterile ddH2O and transfer the cell suspension into a pre-cooled (on ice) 1.5 mL microcentrifuge tube.
- Harvest cells at 3,300 x g for 3 min at 4 °C.
- Aspirate the supernatant.
- Resuspend the cell pellet in 0.5 mL homogenizing buffer [HB; 50 mM Tris, 0.5 mM EDTA, 20% (v/v) glycerol; pH 7.5] freshly supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF).
NOTE: Always add PMSF fresh because PMSF is inactivated quickly upon exposure to water.
CAUTION: PMSF is a serine protease inhibitor, which is extremely corrosive and destructive to tissues. It may cause irreversible eye damage. - Store the cell suspension at -80 °C or use immediately.
- Isolation of plasma membranes
- If frozen, defrost the cells on ice for ~1 h.
- Add ice-cold 0.5 mm diameter silica beads to the 0.5 mL cell suspension to reach a total volume of 1 mL.
- Break cells with 6 cycles of vortexing at maximum shaking intensity for 1 min interspersed with 3 min cooling periods on ice.
- Make a thin hole at the bottom of the tube with a heated scalpel blade.
- Collect the broken cell homogenate through the bottom of the tube fitted into another ice-cold 1.5 mL microcentrifuge tube with a 10 s low-speed (~200 rpm) spin.
NOTE: This ensures that the silica beads remain in the original tube. - Centrifuge the cell homogenate at 5,156 x g for 5 min at 4 °C to remove cell debris, unbroken cells, and nuclei.
- Transfer 450 µL of supernatant into an ice cold 1.5 mL microcentrifuge tube and add an additional 1 mL of ice-cold HB supplemented with fresh PMSF (1 mM).
NOTE: This dilution step is critical for the high-quality plasma membrane protein recovery. - Harvest plasma membranes at 17,968 x g for 1 h at 4 °C and resuspend the plasma membrane pellet, by repeat pipetting, in 100 µL of HB freshly supplemented with 1 mM PMSF. Loosen the cell pellet for proper plasma membrane homogenization by stirring the cell pellet with the 100 µL pipette tip before releasing the 100 µL of HB and up and down pipetting.
- Measure the protein concentration of the plasma membrane preparation with a protein assay kit that is compatible with buffers containing reducing agent and detergent.
- Store the plasma membranes at -80 °C or keep on ice for immediate use.
4. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
- Assemble the apparatus for preparing polyacrylamide gels.
- For two separating gels (7% polyacrylamide), mix 2.1 mL of 40% acrylamide/bis-acrylamide, 3 mL of 4x separating buffer (1.5 M Tris, 0.4% sodium dodecyl sulfate [SDS] (w/v); pH 8.8), and 6.9 mL of ddH2O. Add 8 µL of tetramethylethylenediamine (TEMED) and 60 µL of 10% ammonium persulphate (APS) to initiate polymerization of acrylamide.
CAUTION: Acrylamide/bis-acrylamide is very toxic. It causes skin irritation, peripheral neuropathy and is a carcinogen. TEMED is harmful if swallowed or inhaled. APS is harmful if swallowed. It causes serious eye and skin irritations. - Pour ~4-5 mL of this mixture into the assembled gel apparatus, up to ~2 cm from the top.
- Carefully layer ~1-2 mL of 0.1% SDS on top to create a planar meniscus.
- Allow the polyacrylamide to set for ~60 min at RT.
- Prepare a stacking gel mixture for two gels by mixing 0.5 mL of 40% acrylamide/bis-acrylamide, 1 mL of 4x stacking buffer (0.5 M Tris, 0.4% SDS (w/v); pH 6.8), and 6.9 mL of ddH2O. Add 2 µL of TEMED and 30 µL of 10% APS to initiate polymerization of acrylamide.
- Remove the 0.1% SDS layer from the polymerized separating gel and rinse with ddH2O to remove traces of SDS.
- Pour the stacking gel mix onto the separating gel.
- Place a comb into the stacking gel and remove any air bubbles from around the comb.
- Allow the stacking gel to set for ~60 min at RT.
- Remove the comb and rinse the gel slots with water. Put the gel into the gel tank and fill the gel tank to the top with 1x running buffer (24.8 mM Tris, 190 mM glycine, 0.1% SDS).
NOTE: Prepare 1x running buffer from a 10x buffer stock (248 mM Tris, 1.9 M glycine, 1% SDS (w/v) in ddH2O) stored at RT. - Mix 5-10 µL plasma membrane samples (i.e., 10-20 µg protein) with equal volumes of 2x protein loading dye [120 mM Tris-HCl (pH 6.8), 20% glycerol, 0.02% bromophenol blue, 4% SDS, 200 mM dithiothreitol (DTT)] and immediately load into individual gel slots submerged in running buffer.
CAUTION: DTT is harmful if swallowed. It causes serious eye and skin irritations.
NOTE: Make a 10 mL stock of 2x protein loading dye, aliquot and store at -20 °C. Do not heat the mixed samples but immediately load them into individual gel slots so that the GFP tag is not denatured, and the in-gel fluorescence signals can be detected. - Load protein molecular weight markers (10-245 kDa range) into a separate slot to enable the size estimation of individual protein fragments.
- Perform gel electrophoresis at 200 V until the blue loading dye reaches the bottom of the gel (usually 45-55 min).
- Examine the gel for in-gel GFP-fluorescence with a gel imaging system (excitation and emission wavelengths are 475-485 nm and 520 nm, respectively).
- Following in-gel fluorescence imaging, fix proteins by gentle agitation of the gel in ~10-20 mL Protein Gel Fixing Solution (40% ethanol, 10% acetic acid) for 15 min at RT.
- Rinse the gel twice for 10 min with 10 mL of ddH2O and visualize protein bands by placing the gel in 10 mL colloidal Coomassie stain solution with gentle shaking for ~1 h at RT.
- For improved visualization of protein bands, de-stain the gel once or twice in ~20 mL of ddH2O for ~1 h before recording images with the gel imaging system.
5. Determination of Cdr1 ATPase Activities 57
- Dilute plasma membrane samples >2.2 mg/mL to 1-2 mg/mL in HB.
- Equilibrate the ATPase assay cocktail (75 mM MES-Tris, 75 mM potassium nitrate, 0.3 mM ammonium molybdate, 7.5 mM sodium azide; pH 7.5) and Mg-ATP (28.8 mM ATP disodium salt, 28.8 mM MgCl2; pH 7.0) to 30 °C in a 30 °C incubator.
CAUTION: Sodium azide is highly toxic.
NOTE: Ensure all buffer stocks and bottles are phosphate free; i.e., wash glassware with 1% (vol/vol) HCl and rinse it several times with ddH2O. Also, keep it separate from other glassware that likely contains traces of phosphate commonly present in detergents used to wash glassware. - Add 90 µL of assay cocktail with or without Cdr1-ATPase inhibitor (20 µM of oligomycin) into individual wells of a 96-well microtiter plate.
NOTE: The assay is performed in triplicate. - Add 5 µL of the isolated plasma membranes (~5-10 µg protein) or phosphate standards (0-100 nmoles Pi) into the appropriate wells of the microtiter plate.
NOTE: Keep the first and last columns for two separate sets of phosphate standards. - Start the assay by adding 25 µL of prewarmed 28.8 mM Mg-ATP (6 mM final concentration) with a multi-channel pipette into individual wells and incubate at 30 °C for 30 min.
- Stop the reaction by adding 130 µL of development reagent (1.6% sodium L-ascorbate, 1% SDS, 12% ammonium molybdate in 6 M sulfuric acid).
CAUTION: Concentrated sulfuric acid reacts violently with water. It is corrosive and may cause skin and lung damage. - Incubate at RT for 10 min.
- Read the absorbance of the microtiter plate wells at 750 nm with a microtiter plate reader.
NOTE: Sticking to the 10 min incubation time for blue dye development (i.e., a reduced phosphor-molybdenum complex) is critical for assay accuracy because the blue dye development continues with time. - Obtain the Cdr1-specific ATPase activity (i.e., the oligomycin-sensitive ATPase activity) by subtracting the ATPase activity in the presence of oligomycin from the total ATPase activity in the absence of oligomycin.
6. Small scale detergent screen
- Combine the plasma membrane preparations (i.e., 2.5 mg of plasma membrane protein) of cells overexpressing Cdr1-mGFPHis with GTED-20 buffer [10 mM Tris, 0.5 mM EDTA, 20 % (w/v) glycerol; pH 7.5; freshly supplemented with 1 mM PMSF] containing 5 mg of the test detergent, to reach a total volume of 0.5 mL supplemented with 1% (w/v) detergent.
- Rotate the mixture at 4-8 °C for 2 h, with a rotation device.
- Centrifuge the mixture at 141,000 x g at 4 °C for 1 h.
- Transfer the supernatant containing solubilized material to a fresh microcentrifuge tube.
- Add 0.5 mL of GTED-20 buffer supplemented with 2% (w/v) SDS to the insoluble pellet fraction and incubate at 30 °C o/n in a shaking incubator to extract all detergent-insoluble membrane protein.
- Analyze and compare the supernatant and solubilized pellet fractions by SDS-PAGE.
- Photograph gels containing GFP-tagged proteins for in-gel GFP-fluorescence before Coomassie staining and quantify expression levels with the imaging system.
- Use the soluble membrane protein fraction for downstream applications such as fluorescence size exclusion chromatography (FSEC)58 to identify suitable detergent(s).
Representative Results
A high frequency of transformation of S. cerevisiae ADΔΔ (~4 x 104 transformants/µg) was achieved with pYES2 (Figure 2B). As expected, the no DNA (i.e., ddH2O only) control gave no transformants, and 1 µg of the linear CDR1–mGFPHis transformation cassette (Figure 1A) gave ~50 transformants (Figure 2C) with the optimized ADΔΔ transformation protocol. The CDR1–mGFPHis transformants were also tested for their ability to grow on a non-fermentable carbon source to eliminate any possible petite mutants with defective mitochondria. Three (yellow circles; Figure 2D) of the 25 uracil-prototroph transformants tested could not grow on YPG agar plates and were discarded from further investigations. The Cdr1-mGFPHis expressed by the newly created strain ADΔΔ-CDR1-mGFPHis is properly localized in the plasma membrane (Figure 1A) and was expressed at sufficient levels for the structural and functional characterization of Cdr1 (Figure 3). The design of the optimized double tag (Figure 1B) also enables the removal of the mGFPHis double tag following Nickel-affinity purification of Cdr1-mGFPHis to prevent possible interference from the tag in downstream applications. Preliminary results, however, have shown that the 3 amino acid linker between Cdr1 and the HRV-3C protease cleavage site may have to be extended to achieve faster, more effective, cleavage. Similar observations were recently reported for the N-terminally tagged nucleoside transporter Escherichia coli NupC, which required a 15 rather than the originally designed 3 amino acid linker for efficient cleavage of the detergent (DDM) solubilized NupC bound to the Nickel-affinity resin59. The 5 amino acid linker C-terminal of the HRV-3C cleavage site prevents steric interference of the mGFPHis double tag with the attached protein of interest, which we have previously observed for C. utilis ABC transporter Cdr139. The yEGFP3-A206K mutation was created to prevent artificial GFP-dimerization at high protein concentrations28, and the additional 3 amino acid linker between mGFP and the His Nickel-affinity tag ensures proper surface exposure of the His tag to maximize the binding efficiency of the tagged protein to the nickel affinity resin (data not shown).
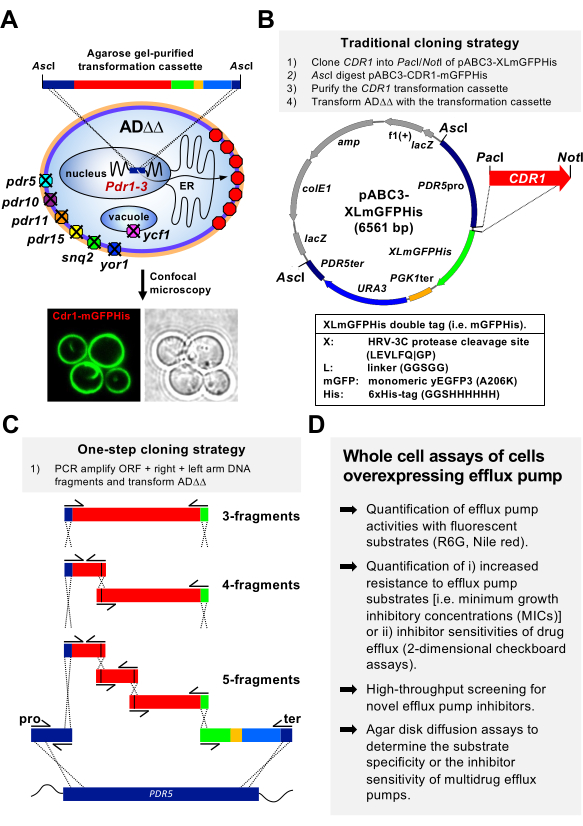
Figure 1: A yeast membrane protein expression platform for the efficient cloning and expression of fungal plasma membrane transporters. (A) A transformation cassette comprising the PDR5 promoter (blue), CDR1 (red) C-terminally tagged with XLmGFPHis (green), the PGK1 terminator (orange), the URA3 selection marker (light blue), and a piece of the PDR5 downstream region (blue) is used to transform the expression host S. cerevisiae ADΔΔ by integration at the genomic PDR5 locus. The gain-of-function mutant transcription factor Pdr1-3 causes the constitutive overexpression of Cdr1 (red octagons) in the plasma membrane (see confocal microscopy image underneath). (B) Plasmid pABC3-XLmGFPHis, which can be used in a traditional cloning strategy or as a PCR template to generate a transformation cassette. Improvements of plasmid pABC3-XLmGFPHis over pABC3-GFP14 are listed underneath the plasmid map. (C) An alternative more efficient one-step cloning strategy to integrate the transformation cassette at the genomic PDR5 locus of ADΔΔ. An ORF (red) can be PCR amplified using primers (arrows) that create overlaps with the left arm (blue; PDR5 promoter) and right arm (green; XLmGFPHis–PGK1–URA3–PDR5 downstream) fragments. Mutations (black lines) can be introduced into the ORF by primer design if needed. Homologous recombination events that direct the correct integration at the PDR5 locus are indicated by the crossed dashed lines. (D) Whole cell assays that can be used for the functional characterization of fungal multidrug efflux pumps. Please click here to view a larger version of this figure.
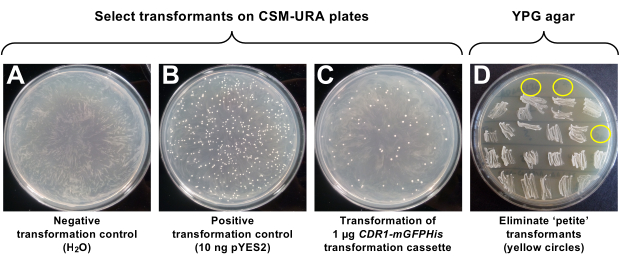
Figure 2: Transformation of ADΔΔ with the CDR1–mGFPHis transformation cassette and elimination of petite ADΔΔ-CDR1-mGFPHis transformants. Uracil prototroph transformants were selected by incubating transformed ADΔΔ cells on CSM-URA plates at 30 °C for 3 days. (A) Negative control (no DNA). (B) Positive control (10 ng of pYES2). (C) CDR1–mGFPHis transformants (1 µg of DNA). (D) Testing ADΔΔ-CDR1-mGFPHis transformants for a petite phenotype on YPG agar plates. Petite transformants that could not grow on YPG agar plates due to defective mitochondria are encircled in yellow. Please click here to view a larger version of this figure.
The GFP fluorescence is a reliable measure for Cdr1 expression levels as there was a linear relationship between the amount of Cdr1-mGFPHis (step 3.3.9) and in-gel fluorescence signals (Figure 3A). In this project, a reproducible optimized workflow was developed for the rapid generation of high-quality small-scale plasma membrane preparations for the biochemical characterization of plasma membrane proteins. It was possible to generate almost 0.5 mg plasma membrane protein (Figure 3C; left graph) showing the highest Cdr1 ATPase activity (˜400 nmol/min/mg; Figure 3C; right graph) when breaking 40 ODU of logarithmic phase (OD600nm of 1-3) cells (resuspended in 0.5 mL of HB) with the same volume (i.e., 0.5 mL) of silica beads and 6 cycles of vortexing for 1 min at maximum shaking intensity followed by 3 min cooling periods on ice. Further increase in the number of breakage cycles (step 3.3.3) reduced the quality of the isolated plasma membrane preparation (the Cdr1 ATPase activity dropped from 170 nmol/min/mg to ~60 nmol/min/mg; data not shown). Although there were only minor differences in the SDS-PAGE protein patterns of the plasma membrane samples isolated from cells broken with higher numbers of breakage cycles (data not shown), it is likely that the almost 3-fold reduced Cdr1 ATPase activities after 10 or more breakage cycles was caused by either: i) partial denaturation of Cdr1 due to exposure to elevated temperature; ii) post-translational modifications such as phosphorylation or dephosphorylation; or by iii) increased cross-contamination of the isolated plasma membrane vesicles with membrane fractions of other organelles. Breaking 40 ODU of cells in 0.5 mL of HB yielded the highest quality of plasma membranes (i.e., the most Cdr1 per 10 µg of plasma membrane protein; Figure 3B) with highest Cdr1 ATPase activity (Figure 3C; right graph). Higher (> 40 ODU/0.5 mL of HB) or lower (20 ODU/0.5 mL of HB) cell densities reduced the ATPase activity of the isolated plasma membranes (Figure 3C; right graph), although, their yield increased in proportion to increasing cell densities (Figure 3C; left graph). Thus, 40 ODU/0.5 mL of HB was the optimum cell density for the isolation of the highest quality of plasma membranes. So, using the optimized protocol for the small-scale isolation of plasma membrane preparations led to 2-3 times higher Cdr1 specific ATPase activities (~300 nmol/min/mg; Figure 3C; right graph) compared to the Cdr1 specific ATPase activities that were initially obtained (~100 nmol/min/mg; data not shown). These ATPase activities were also significantly higher35 than any previously reported Cdr1 ATPase activities (100-200 nmol/min/mg) that had been obtained with a more labor-intensive and time-consuming large-scale plasma membrane preparation protocol14,41,60.
The most common method to extract membrane proteins from biological membranes is solubilization with detergent. The challenge, however, is to find a suitable detergent that has the least detrimental effect on protein stability and/or folding characteristics. Labeling the protein with mGFPHis facilitates screening to select the best detergent(s) for protein extraction. In total, 31 detergents, listed in the Table of Materials, with various properties were tested for their ability to solubilize Cdr1 from crude plasma membranes of S. cerevisiae ADΔΔ-CDR1-mGFPHis cells. The results for a representative set of 16 test detergents (A-Q) are shown in Figure 3D. Lanes T, P, and S contain 10 µL aliquots of total (T) plasma membrane protein immediately after detergent solubilization (step 6.2) and the detergent insoluble pellet (P; solubilized o/n in 0.5 mL of GTED-20, 2% SDS; step 6.5) and the detergent soluble supernatant (S; step 6.4) fractions after separation by ultracentrifugation at 141,000 x g. The desired detergent should solubilize as much Cdr1-mGFPHis as possible without altering its structure and/or function. The crude plasma membrane proteins (5 mg/mL) were solubilized for 2 h with 1% (w/v) detergent (T) and aliquots of the soluble (S) and insoluble (P) fractions were analyzed by SDS-PAGE (Figure 3D) and later also by fluorescence-detection size-exclusion chromatography (FSEC)58 (Figure 4). We determined that a minimum detergent to protein ratio of 2:1 (w/w) was required for efficient solubilization of Cdr1. In fact, to determine the optimum detergent (DDM) concentration required for the solubilization of Cdr1-mGFPHis, the critical micelle concentration of the detergent (x CMC) and the detergent/membrane protein ratio (w/w) were investigated. Large differences in the solubilization efficiencies of Cdr1-mGFPHis were observed when using fixed concentrations of 10x or 80x CMC of DDM. The solubilization efficiencies varied between 40% and 80% or 60% and 90% depending on the amounts of crude plasma membranes used in the various solubilization experiments. However, choosing detergent to protein ratios of ≥2 (w/w) gave reproducibly good results with >85% of Cdr1-mGFPHis being solubilized, no matter the amount of plasma membrane protein used. So, if using the more common approach of simply choosing 1% or 2% (w/v) detergent for the solubilization of membrane proteins such as Cdr1-mGFPHis, keeping the detergent to protein ratio above 2 (w/w) is important [i.e., keep the plasma membrane protein concentration below 5 mg/mL when using 1% (i.e., 10 mg/mL) detergent].
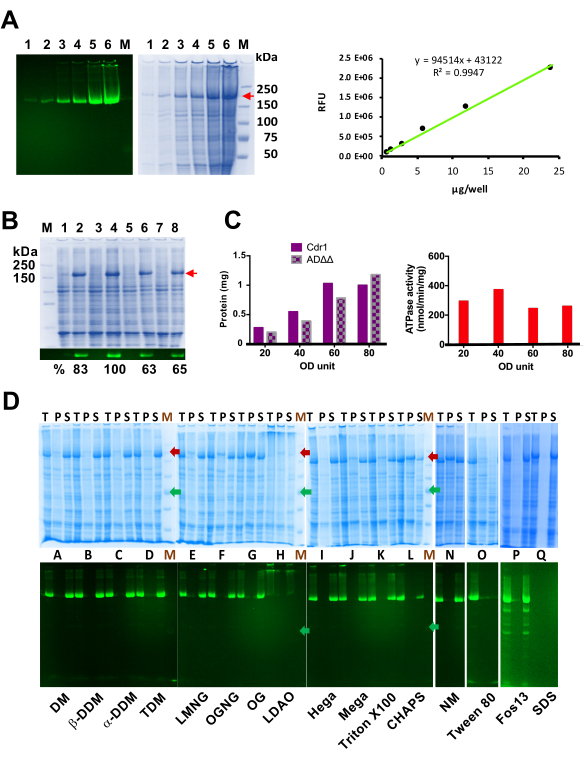
Figure 3: Quantification of Cdr1-mGFPHis expression levels, optimization of a small-scale plasma membrane isolation protocol and detergent screen for Cdr1-mGFPHis. (A) Quantification of Cdr1-mGFPHis with in-gel fluorescence; the Coomassie stained and in-gel fluorescence SDS-PAGE images of the same 0.7% polyacrylamide gel are shown on the left. Lanes 1 to 6 were loaded with 0.75, 1.5, 3, 6, 12, and 24 µg of ADΔΔ-CDR1-mGFPHis plasma membrane protein. Cdr1-mGFPHis is indicated with a red arrow. M = Precision Plus Protein Marker. The mGFP fluorescence intensities (shown on the right) were linear over the entire concentration range tested and there was minimal background fluorescence. (B) Effect of cell density at breakage on the quality of isolated plasma membranes. Coomassie stained polyacrylamide gel (7%) of plasma membrane protein samples (10 µg) and green fluorescence signals of Cdr1-mGFPHis of the same gel before Coomassie staining. M = Precision Plus Protein Marker. Lanes 1, 3, 5, and 7 are plasma membrane proteins of ADΔΔ and lanes 2, 4, 6, and 8 are plasma membrane proteins of ADΔΔ-CDR1-mGFPHis isolated from 20, 40, 60, and 80 ODU of cells, respectively. Cdr1-mGFPHis is indicated with a red arrow. The relative percentages of the green fluorescent signals are listed underneath. (C) Effect of cell density at breakage on the yield of isolated plasma membranes and the ATPase activity of Cdr1-mGFPHis. On the left, effect of the cell density (ODU/0.5 mL HB) on the amount of plasma membrane protein isolated from ADΔΔ and ADΔΔ-CDR1-mGFPHis (Cdr1) cells. On the right, effect of the cell density on the Cdr1 ATPase activity of the plasma membranes isolated from ADΔΔ-CDR1-mGFPHis cells. (D) Exemplary SDS-PAGE of 10 µL aliquots of the solubilization mixture (T; 0.5 mL), the pellet after solubilization (P; 0.5 mL), and the solubilized (supernatant) fractions (S; 0.5 mL) of detergent solubilized crude plasma membrane proteins of ADΔΔ-CDR1-mGFPHis (top) and in-gel fluorescence of Cdr1-mGFPHis before Coomassie staining of the same gel (bottom). M = broad MW pre-stained protein ladder (245, 180, 135, 100, 75, 63, and 48 kDa bands; the 180 kDa and 75 kDa bands are indicated with red and green arrows, respectively). Lanes A to L and N to Q are the T, S, and P fractions for DM (A), β-DDM (B), α-DDM (C), TDM (D), LMNG (E), OGNG (F), OG (G), LDAO (H), Hega (I), Mega (J), Triton-X100 (K), CHAPS (L), NM (N), Tween80 (O), Fos-choline-13 (P) and SDS (Q), respectively. Abbreviations are defined in the Table of Materials. Please click here to view a larger version of this figure.
Based on detergent screening results (Figure 3D), DDM (B and C) and Fos-choline-13 (P) appeared to be the best detergents for the solubilization of Cdr1-mGFPHis. However, the use of Fos-choline-13 seemed to cause partial proteolysis of Cdr1-mGFPHis (P in Figure 3D). LMNG (E), PCC-α-M (not shown) and possibly also DM (A) were the next best detergents. The detergent screen also showed that glucosides OGNG (F) and OG (G), CHAPS (L), NM (N), and Anzergents (not shown) were bad choices for the solubilization of Cdr1-mGFPHis as significant amounts of the protein were found in the insoluble pellet fractions (Figure 3D). SDS denatures Cdr1-XLmGFPHis, which is why no green fluorescence signals are visible in the Q lanes (Figure 3D).
The suitability of a detergent for the solubilization of properly folded native Cdr1-mGFPHis particles was also assessed by FSEC of the detergent solubilized plasma membrane protein (step 6.4 supernatant). Examples of chromatograms obtained for 100 µL crude plasma membrane protein from ADΔΔ-CDR1-mGFPHis (2 mg/mL) solubilized with 1% detergent are shown in Figure 4. The peak at 9 mL elution volume represents mostly aggregated Cdr1-mGFPHis that elutes in the void volume, whereas properly solubilized and correctly folded Cdr1-mGFPHis particles are represented by the Gaussian-shaped peak at 15.5 mL elution volume. The broad shoulder between 12 and 14 mL elution volume possibly contains less well-defined Cdr1-mGFPHis micelle particles and/or indicate partial misfolding or protein aggregates. Chromatograms of maltosides and LMNG extracted proteins showed most of Cdr1-mGFPHis eluting as a nicely shaped Gaussian peak at 15.5 mL with a small shoulder eluting slightly earlier at 14 mL (Figure 4A). These samples had no Cdr1-mGFPHis eluting in the void volume. The blank sample is the buffer plus DDM control that gave no mGFP signal. The chromatograms in Figure 4B highlight the importance of the type of sugar headgroup for the quality of the detergent solubilized Cdr1-mGFPHis particles. Glucose containing detergents (OG, NG, OGNG) performed worse than sucrose containing detergents (DDS), which in turn performed worse than maltose containing detergents (NM, DMNG, DDM). Cdr1-mGFPHis solubilized with glucose containing detergents eluted as an aggregated (OG) or broad aggregated peak (NG, OGNG), which was to be expected from the large proportion of Cdr1-mGFPHis precipitate in the pellet fractions for OGNG (F) and OG (G) observed in Figure 3D. Although the DMNG chromatogram (green; Figure 4B) was qualitatively similar to the DDM chromatogram (blue; Figure 4B), the higher peaks for DDM indicate that a large portion of DMNG solubilized Cdr1-mGFPHis may actually be denatured. Figure 4C shows the FSEC chromatograms for the zwitterionic Fos-cholines (Fos-choline-8, Fos-choline-10, Fos-choline-13) and two non-ionic detergents (digitonin, Triton-X100), and Figure 4D shows how loading twice as much (200 µL) detergent solubilized protein had no noticeable effect on the quality of the chromatogram for Fos-choline-8, -10, and -13 and DDM. Only digitonin (orange; Figure 4C) gave a similar chromatogram to DDM (blue; Figure 4C) and, although Fos-choline-13 gave a symmetrical sharp shaped peak, it again eluted with a significantly lower elution volume (14 mL) than that for DDM (15.5 mL). Overall, there was a clear trend of better solubilization by detergents with longer aliphatic side-chains of 12 or 13 carbon residues; e.g., DDM > DM > NM (12, 10, or 9 carbons), Fos-choline-13 > 10 > 8 (13, 10, or 8 carbons), and LMNG > DMNG > OGNG (12, 10, or 8 carbons), respectively. Thus, detergents with hydrophobic tails of less than 12 carbons were far less suitable detergents for solubilization of Cdr1-mGFPHis than detergents with longer hydrophobic tails of 12 or 13 carbons, and non-ionic detergents (DDM, LMNG) seemed generally more suitable for solubilization of Cdr1-mGFPHis than zwitterionic detergents (Figure 3D, Figure 4).
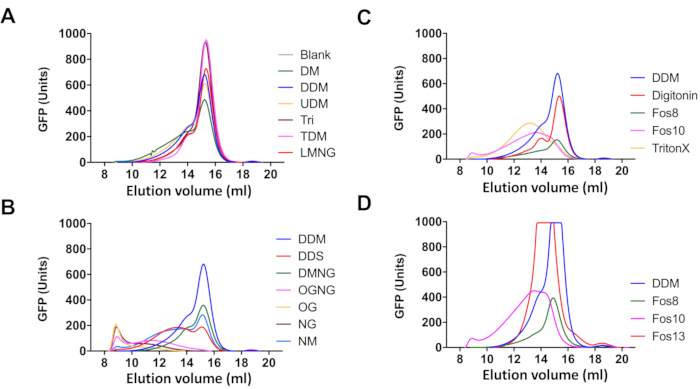
Figure 4: Detergent screening for Cdr1-mGFPHis solubilization using FSEC. Chromatograms of 100 µL of solubilized ADΔΔ-CDR1-mGFPHis crude plasma membrane (2 mg/mL) with 1% of the detergents indicated and separated using a Superose 6-increase 10/300 GL size exclusion column. The chromatograms depict the relative mGFP fluorescence units (FU) of one column volume (CV; 25 mL) of collected elution buffer. Please click here to view a larger version of this figure.
Discussion
Despite recent progress in the structural analysis of membrane proteins, no 3D structure for Cdr1, or any other PDR transporter, is currently available. So, gaining knowledge of the Cdr1 structure and its biochemical features is important, as this will not only provide insight into rational design of novel drugs to overcome efflux-mediated drug resistance, but also into the mechanism of function of an important subfamily of ABC proteins.
One of the main requirements for the structural characterization of membrane proteins is the expression of correctly folded and intact membrane protein in quantities required for X-ray crystallography or cryo-EM. Important criteria when selecting an expression system are ease of use, growth rates and costs, and also the ability to express proteins with post-translational modifications that can be critical for the function and/or stability of a protein61,62.
Over the past two decades a S. cerevisiae membrane protein expression technology60 has been optimized14 to facilitate the one-step cloning of membrane proteins of interest24 tagged at either their N- or C-terminus with various affinity, epitope, or reporter tags and, if desired, their expression levels predictably repressed to anywhere between 50% to as low as 0.1% of the maximum expression level25. This versatile plasma membrane protein expression platform enables researchers to characterize fungal efflux pumps in great detail. The development and optimization of whole cell assays of pump function enabled the successful characterization of the substrate specificity24 and the inhibitor sensitivity of a number of major fungal multidrug efflux pumps, and they were employed in high-throughput drug screens to identify novel42,43,44, or confirm existing14,33,36, broad-spectrum efflux pump inhibitors or to develop novel efflux pump inhibitors specific for Cdr141.
Although the existing expression platform has been used successfully to express fungal ABC transporters from an array of fungi, including Basidomycota (C. neoformans Mdr1),14 filamentous fungi such as P. marneffei (Abc1)37 and many Saccharomycotina species (e.g., S. cerevisiae Pdr5, C. glabrata Cdr1 and Cdr2, C. utilis Cdr1, C. krusei Abc1, Abc11 and Abc12 and C. albicans Cdr1 and Cdr2),14,24,32,36,39,60,63 there has been less success expressing high levels of properly folded human ABC transporters64. Preliminary results indicate that this is due to the specific lipid environment that these transporters require for proper expression and function in yeast. Indications are that this may also be true for plant ABC transporters, although this has yet to be confirmed experimentally.
The major reasons for the significantly higher Cdr1 ATPase specific activities (i.e., preparations with less contaminating membranes) obtained with the optimized small-scale plasma membrane isolation protocol are two-fold: i) the gentler manual breakage of cells compared to using a bead-beater for larger scale plasma membrane preparations; and ii) the harvesting of cells at mid-log phase, which reduces possible mitochondrial contamination due to glucose repression of mitochondrial enzymes. The 3 min cooling periods between each of the six breakage cycles could be extended without any noticeable effect on the quality of the plasma membranes. Repeat freeze-thaw cycles are, however, to be avoided because the Cdr1 ATPase activities of the isolated plasma membranes reduced by ~10% with every additional freeze-thaw cycle. This is why it is recommended to split the plasma membrane samples into smaller aliquots to reduce the need for multiple freeze-thaw cycles. The mGFPHis double tag improves purification yield and allows efficient one-step detergent screening. By using this construct, correct localization, proper folding/trafficking and thermostability can be readily detected; and the double tag can be removed, if necessary, by cleavage with a commercially available protease, although the 3 amino acid-linker between the protein of interest and the tag may have to be extended for efficient removal of the tag.
The combination of several features in these protocols, namely, the use of a DNA polymerase specifically designed for the colony PCR amplification of the 8 kb transformation cassette; the optimized high efficiency yeast transformation method; the ability to create frozen competent yeast stocks; and the optimized small-scale plasma membrane preparation protocol demonstrate the creation of an optimized membrane protein expression platform amenable for the high-throughput cloning, expression, and characterization of fungal ABC efflux pumps.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors gratefully acknowledge funding from the New Zealand Marsden Fund (Grant UOO1305), and a block grant from Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (M. Niimi). They wish to thank the University of Otago for providing G. Madani with a PhD Scholarship. The authors also wish to express their gratitude to Professor Stefan Raunser and his colleagues, Dr Amir Apelbaum, and Dr Deivanayagabarathy Vinayagam, for their support and supervision during a 6-month visit of G. Madani at the Max Planck Institute of Molecular Physiology (MPIMP), Dortmund, Germany. The authors also thank the German Academic Exchange Service (DAAD) for providing G. Madani with a research grant (57381332) to visit the MPIMP.
Materials
| 2-(N-Morpholino)ethane-sulphonic acid (MES) | Sigma-Aldrich | M3671 | |
| 2-Amino-2-(hydroxymethyl)-1,3-propanediol (Tris base; ultra-pure) | Merck | 77-86-1 | |
| 2,2-Didecylpropane-1,3-bis-β-D-maltopyranoside | Anatrace | NG310S | LMNG |
| 2,2-Dihexylpropane-1,3-bis-β-D-glucopyranoside | Anatrace | NG311S | OGNG (MNG-OG) |
| 2,2-Dioctylpropane-1,3-bis-β-D-maltopyranoside | Anatrace | NG322S | DMNG |
| 4-Trans-(4-trans-propylcyclohexyl)-cyclohexyl α-D-maltopyranoside | Glycon Biochemicals GmbH | D99019-C | PCC-α-M |
| 40% Acrylamide/Bis-acrylamide (37.5:1) | Bio-Rad | 1610148 | |
| Acetic acid (glacial) | Merck | 64-19-7 | |
| Agar | Formedium | 009002-18-0 | |
| Ammonium molybdate | Sigma-Aldrich | 13106-76-8 | |
| Ammonium persulphate (APS) | Bio-Rad | 1610700 | |
| ATP disodium salt | sigma-Aldrich | A-6419 | |
| Bromophenol blue | SERVA Electrophoresis GmbH | 34725-61-6 | |
| CHAPS | Anatrace | C316S | |
| CHAPSO | Anatrace | C317S | |
| CSM | Formedium | DCS0019 | |
| CSM minus uracil | Formedium | DCS0161 | |
| Cyclohexyl-1-butyl-β-D-maltopyranoside | Anatrace | C324S | CYMAL-4 |
| Cyclohexyl-1-heptyl-β-D-maltopyranoside | Anatrace | C327S | CYMAL-7 |
| Cyclohexyl-methyl-β-D-maltopyranoside | Anatrace | C321S | CYMAL-1 |
| Digitonin | Sigma-Aldrich | 11024-24-1 | |
| Dithiothreitol (DTT) | Roche Diagnostics | 10197785103 | |
| DMSO | Merck | 67-68-5 | |
| Ethanol | Merck | 459836 | |
| Ethylenediaminetetraacetic acid disodium salt (EDTA; Titriplex III) | Merck | 6381-92-6 | |
| ExoSAP-IT PCR Product Cleanup Reagent | Applied Biosystems | 78205 | A blend of exonuclease and phosphatase |
| Glucose | Formedium | 50-99-7 | |
| Glycerol | Merck | 56-81-5 | |
| Glycine | Merck | G8898 | |
| HEPES | Formedium | 7365-45-9 | |
| Hydrochloric acid | Merck | 1003172510 | |
| KOD Fx Neo | TOYOBO Co | KFX-201 | Use for reliable colony PCR |
| lithium acetate (LiAc) | Sigma-Aldrich | 546-89-4 | |
| Magnesium chloride hexa-hydrate | sigma-Aldrich | M2393 | |
| MES | Formedium | 145224-94-8 | |
| n-Decanoyl-N-hydroxyethyl-glucamide | Anatrace | H110S | HEGA-10 |
| n-Decanoyl-N-methyl-glucamide | Anatrace | M320S | MEGA-10 |
| n-Decyl-phosphocholine | Anatrace | F304S | Fos-choline-10 |
| n-Decyl-β-D-maltopyranoside | Anatrace | D322S | DM |
| n-Dodecyl-N,N-dimethyl-3-ammonio-1-propanesulphonate | Anatrace | AZ312S | Anzergent 3-12 |
| n-Dodecyl-N,N-dimethylamine-N-oxide | Anatrace | D360S | LDAO |
| n-Dodecyl-α-D-maltopyranoside | Anatrace | D310HA | α-DDM |
| n-Dodecyl-β-D-maltopyranoside | Anatrace | D310S | β-DDM |
| n-Nonyl-β-D-glucopyranoside | Anatrace | N324S | NG |
| n-Nonyl-β-D-maltopyranoside | Anatrace | N330S | NM |
| n-Octadecyl-N,N-dimethyl-3-ammonio-1-propanesulphonate | Anatrace | AZ318S | Anzergent 3-18 |
| n-Octyl-N,N-dimethyl-3-ammonio-1-propanesulphonate | Anatrace | AZ308S | Anzergent 3-8 |
| n-Octyl-phosphocholine | Anatrace | F300S | Fos-choline-8 |
| n-Octyl-β-D-glucopyranoside | Anatrace | O311S | OG |
| n-Tetradecyl-phosphocholine | Anatrace | F312S | Fos-choline-14 |
| n-Tetradecyl-β-D-maltopyranoside | Anatrace | T315S | TDM |
| n-Tridecyl-phosphocholine | Anatrace | F310S | Fos-choline-13 |
| n-Tridecyl-β-D-maltopyranoside | Anatrace | T323S | – |
| n-Undecyl-β-D-maltopyranoside | Anatrace | U300S | UM (UDM) |
| N,N,N’,N’-tetramethyl-ethylenediamine (TEMED) | Sigma-Aldrich | T9281 | |
| Octylphenoxypolyethoxyethanol | Sigma-Aldrich | 9002-93-1 | TRITON X-100 |
| Oligomycin | Sigma-Aldrich | 75351 | |
| Peptone | Formedium | 3049-73-7 | |
| phenylmethylsulfonyl fluoride (PMSF) | Roche Diagnostics | 329-98-6 | |
| Phusion Hot Start Flex DNA Polymerase | New England Biolabs | M0535S | High-fidelity DNA polymerase |
| polyethylene glycol (PEG 3350) | Sigma-Aldrich | 25322-68-3 | |
| polyoxyethylenesorbitan monooleate | Sigma-Aldrich | 9005-65-6 | TWEEN 80 |
| Potassium nitrate | Sigma-Aldrich | P8394 | |
| Protein Assay Kit | Bio-Rad | 5000122 | RC DC Protein Assay Kit II |
| QC Colloidal Coomassie Stain | Bio-Rad | 1610803 | |
| Prism Ultra Protein Ladder (10-245 kDa) | Abcam | AB116028 | |
| Sodium azide | Sigma-Aldrich | 71289 | |
| Sodium dodecyl sulphate | Sigma-Aldrich | 151-21-3 | SDS |
| Sodium L-ascorbate BioXtra | Sigma-Aldrich | 11140 | |
| Sucrose Monododecanoate | Anatrace | S350S | DDS |
| Sulphuric acid | Sigma-Aldrich | 339741 | |
| Yeast extract | Formedium | 008013-01-2 | |
| Yeast nitrogen base without amino acids | Formedium | CYN0402 | |
| Equipment (type) | |||
| Centrifuge (Eppendorf 5804) | Eppendorf | ||
| Centrifuge (Beckman Ultra) | Beckman | ||
| Centrifuge (Sorvall RC6) | Sorvall | ||
| FSEC apparatus (NGC Chromatography Medium Pressure system equipped with a fluorescence detector, an autosampler, a fractionator) | Bio-Rad | ||
| Gel imaging (GelDoc EZ Imager) | Bio-Rad | ||
| Microplate reader (Synergy 2 Multi-Detection) | BioTek Instruments | ||
| PCR thermal cycler (C1000 Touch) | Bio-Rad | ||
| Power supply (PowerPac) | Bio-Rad | ||
| SDS PAGE (Mini-PROTEAN Tetra) | Bio-Rad | ||
| Shaking incubator (Multitron) | Infors HT, Bottmingen | ||
| Superose 6 Increase 10/300 GL | GE Healthcare Life Sciences | GE17-5172-01 | |
| UV/Visible spectrophotometer (Ultraspec 6300 pro) | Amersham BioSciences UK Ltd | ||
References
- Arachea, B. T., et al. Detergent selection for enhanced extraction of membrane proteins. Protein Expression and Purification. 86 (1), 12-20 (2012).
- Guo, Y. Be cautious with crystal structures of membrane proteins or complexes prepared in detergents. Crystals (Basel). 10 (2), 86 (2020).
- Lewinson, O., Orelle, C., Seeger, M. A. Structures of ABC transporters: handle with care. FEBS Letters. 594 (23), 3799-3814 (2020).
- Luckey, M. . Membrane Structural Biology: With Biochemical and Biophysical Foundations. , 69-105 (2014).
- Opekarova, M., Tanner, W. Specific lipid requirements of membrane proteins–a putative bottleneck in heterologous expression. Biochimica et Biophysica Acta. 1610 (1), 11-22 (2003).
- Qiu, W., et al. Structure and activity of lipid bilayer within a membrane-protein transporter. Proceedings of the National Academy of Sciences of the United States of America. 115 (51), 12985-12990 (2018).
- Dowhan, W. Molecular basis for membrane phospholipid diversity: why are there so many lipids. Annual Review of Biochemistry. 66, 199-232 (1997).
- Lee, A. G. Lipid-protein interactions in biological membranes: a structural perspective. Biochimica et Biophysica Acta. 1612 (1), 1-40 (2003).
- Ahn, J. H., Pan, J. G., Rhee, J. S. Homologous expression of the lipase and ABC transporter gene cluster, tliDEFA, enhances lipase secretion in Pseudomonas spp. Applied and Environmental Microbiology. 67 (12), 5506-5511 (2001).
- Newby, Z. E., et al. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nature Protocols. 4 (5), 619-637 (2009).
- Parker, J. L., Newstead, S. Membrane protein crystallisation: current trends and future perspectives. Advances in Experimental Medicine and Biology. 922, 61-72 (2016).
- Wiener, M. C. A pedestrian guide to membrane protein crystallization. Methods. 34 (3), 364-372 (2004).
- Grisshammer, R. Understanding recombinant expression of membrane proteins. Current Opinion in Biotechnology. 17 (4), 337-340 (2006).
- Lamping, E., et al. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot Cell. 6 (7), 1150-1165 (2007).
- Macauley-Patrick, S., Fazenda, M. L., McNeil, B., Harvey, L. M. Heterologous protein production using the Pichia pastoris expression system. Yeast. 22 (4), 249-270 (2005).
- Focke, P. J., et al. Combining in vitro folding with cell free protein synthesis for membrane protein expression. Biochimie. 55 (30), 4212-4219 (2016).
- Reckel, S., et al. Strategies for the cell-free expression of membrane proteins. Methods in Molecular Biology. 607, 187-212 (2010).
- Pandey, A., Shin, K., Patterson, R. E., Liu, X. Q., Rainey, J. K. Current strategies for protein production and purification enabling membrane protein structural biology. Biochemistry and Cell Biology. 94 (6), 507-527 (2016).
- Harvey, C. J. B., et al. HEx: A heterologous expression platform for the discovery of fungal natural products. Science Advances. 4 (4), (2018).
- Kingsman, S. M., Kingsman, A. J., Dobson, M. J., Mellor, J., Roberts, N. A. Heterologous gene expression in Saccharomyces cerevisiae. Biotechnology & Genetic Engineering Reviews. 3, 377-416 (1985).
- Monk, B. C., et al. Yeast membrane protein expression system and its application in drug screening. US patent. , (2002).
- Sagatova, A. A., Keniya, M. V., Wilson, R. K., Monk, B. C., Tyndall, J. D. Structural insights into binding of the antifungal drug fluconazole to Saccharomyces cerevisiae lanosterol 14alpha-demethylase. Antimicrobial Agents and Chemotherapy. 59 (8), 4982-4989 (2015).
- Decottignies, A., et al. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. The Journal of Biological Chemistry. 273 (20), 12612-12622 (1998).
- Lamping, E., Zhu, J. Y., Niimi, M., Cannon, R. D. Role of ectopic gene conversion in the evolution of a Candida krusei pleiotropic drug resistance transporter family. Génétique. 205 (4), 1619-1639 (2017).
- Lamping, E., Niimi, M., Cannon, R. D. Small, synthetic, GC-rich mRNA stem-loop modules 5′ proximal to the AUG start-codon predictably tune gene expression in yeast. Microbial Cell Factories. 12, 74 (2013).
- James, J. E., Lamping, E., Santhanam, J., Cannon, R. D. PDR transporter ABC1 is involved in the innate azole resistance of the human fungal pathogen Fusarium keratoplasticum. Frontiers in Microbiology. , (2021).
- Ullah, R., et al. Activity of the human rhinovirus 3C protease studied in various buffers, additives and detergent solutions for recombinant protein production. PLoS One. 11 (4), 0153436 (2016).
- von Stetten, D., Noirclerc-Savoye, M., Goedhart, J., Gadella, T. W., Royant, A. Structure of a fluorescent protein from Aequorea victoria bearing the obligate-monomer mutation A206K. Acta Crystallographica Section F. Structural Biology and Crystalization Communications. 68, 878-882 (2012).
- Zacharias, D. A., Violin, J. D., Newton, A. C., Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296 (5569), 913-916 (2002).
- Cormack, B. P., et al. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology (Reading). 143, 303-311 (1997).
- Monk, B. C., et al. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proceedings of the National Academy of Sciences of the United States of America. 111 (10), 3865-3870 (2014).
- Holmes, A. R., et al. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrobial Agents and Chemotherapy. 52 (11), 3851-3862 (2008).
- Tanabe, K., et al. FK506 resistance of Saccharomyces cerevisiae Pdr5 and Candida albicans Cdr1 involves mutations in the transmembrane domains and extracellular loops. Antimicrobial Agents and Chemotherapy. 63 (1), 01146 (2019).
- Tanabe, K., et al. Chimeras of Candida albicans Cdr1p and Cdr2p reveal features of pleiotropic drug resistance transporter structure and function. Molecular Microbiology. 82 (2), 416-433 (2011).
- Madani, G., Lamping, E., Cannon, R. D. Engineering a cysteine-deficient functional Candida albicans Cdr1 molecule reveals a conserved region at the cytosolic apex of ABCG transporters important for correct folding and frafficking of Cdr1. mSphere. 6 (1), (2021).
- Lamping, E., et al. Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrobial Agents and Chemotherapy. 53 (2), 354-369 (2009).
- Panapruksachat, S., et al. Identification and functional characterization of Penicillium marneffei pleiotropic drug resistance transporters ABC1 and ABC2. Medical Mycology. 54 (5), 478-491 (2016).
- Wada, S., et al. Phosphorylation of Candida glabrata ATP-binding cassette transporter Cdr1p regulates drug efflux activity and ATPase stability. The Journal of Biological Chemistry. 280 (1), 94-103 (2005).
- Watanasrisin, W., et al. Identification and characterization of Candida utilis multidrug efflux transporter CuCdr1p. FEMS Yeast Research. 16 (4), (2016).
- Ivnitski-Steele, I., et al. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Analytical Biochemistry. 394 (1), 87-91 (2009).
- Niimi, K., et al. Specific interactions between the Candida albicans ABC transporter Cdr1p ectodomain and a D-octapeptide derivative inhibitor. Molecular Microbiology. 85 (4), 747-767 (2012).
- Holmes, A. R., et al. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrobial Agents and Chemotherapy. 56 (3), 1508-1515 (2012).
- Reis de Sa, L. F., et al. Synthetic organotellurium compounds sensitize drug-resistant Candida albicans clinical isolates to fluconazole. Antimicrobial Agents and Chemotherapy. 61 (1), 01231 (2017).
- Tanabe, K., et al. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochemical and Biophysical Research Communications. 364 (4), 990-995 (2007).
- le Maire, M., Champeil, P., Moller, J. V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochimica et Biophysica Acta. 1508 (1-2), 86-111 (2000).
- Seddon, A. M., Curnow, P., Booth, P. J. Membrane proteins, lipids and detergents: not just a soap opera. Biochimica et Biophysica Acta. 1666 (1-2), 105-117 (2004).
- Pfaller, M. A. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clinical Infectious Diseases. 22, 89-94 (1996).
- Cannon, R. D., et al. Efflux-mediated antifungal drug resistance. Clinical Microbiology Reviews. 22 (2), 291-321 (2009).
- Sanglard, D., et al. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrobial Agents and Chemotherapy. 39 (11), 2378-2386 (1995).
- Prasad, R., De Wergifosse, P., Goffeau, A., Balzi, E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Current Genetics. 27 (4), 320-329 (1995).
- Lamping, E., Madani, G., Lee, H. J., Niimi, M., Cannon, R. D., Prasad, R. . Candida albicans: Cellular and Molecular Biology. , 379-406 (2017).
- Crouzet, J., Trombik, T., Fraysse, A. S., Boutry, M. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Letters. 580 (4), 1123-1130 (2006).
- Kang, J., et al. Plant ABC Transporters. Arabidopsis Book. 9, 0153 (2011).
- Lamping, E., et al. Fungal PDR transporters: phylogeny, topology, motifs and function. Fungal Genetics and Biology. 47 (2), 127-142 (2010).
- Lamping, E., Cannon, R. D. Use of a yeast-based membrane protein expression technology to overexpress drug resistance efflux pumps. Methods in Molecular Biology. 666, 219-250 (2010).
- Day, M. Yeast petites and small colony variants: for everything there is a season. Advances in Applied Microbiology. 85, 1-41 (2013).
- Niimi, K., et al. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a D-octapeptide derivative. Antimicrobial Agents and Chemotherapy. 48 (4), 1256-1271 (2004).
- Kawate, T., Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 14 (4), 673-681 (2006).
- Hao, Z., et al. A novel and fast purification method for nucleoside transporters. Frontiers in Molecular Biosciences. 3, 23 (2016).
- Nakamura, K., et al. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrobial Agents and Chemotherapy. 45 (12), 3366-3374 (2001).
- Bernaudat, F., et al. Heterologous expression of membrane proteins: choosing the appropriate host. PLoS One. 6 (12), 29191 (2011).
- Byrne, B. Pichia pastoris as an expression host for membrane protein structural biology. Current Opinion in Structural Biology. 32, 9-17 (2015).
- Holmes, A. R., et al. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Molecular Microbiology. 62 (1), 170-186 (2006).
- Keniya, M. V., et al. Drug resistance is conferred on the model yeast Saccharomyces cerevisiae by expression of full-length melanoma-associated human ATP-binding cassette transporter ABCB5. Molecular Pharmaceutics. 11 (10), 3452-3462 (2014).

