Inter-Brain Synchrony in Open-Ended Collaborative Learning: An fNIRS-Hyperscanning Study
Summary
The protocol for conducting fNIRS hyperscanning experiments on collaborative learning dyads in a naturalistic learning environment is outlined. Further, a pipeline to analyze the Inter-Brain Synchrony (IBS) of oxygenated hemoglobin (Oxy-Hb) signals is presented.
Abstract
fNIRS hyperscanning is widely used to detect the neurobiological underpinnings of social interaction. With this technique, researchers qualify the concurrent brain activity of two or more interactive individuals with a novel index called inter-brain synchrony (IBS) (i.e., phase and/or amplitude alignment of the neuronal or hemodynamic signals across time). A protocol for conducting fNIRS hyperscanning experiments on collaborative learning dyads in a naturalistic learning environment is presented here. Further, a pipeline of analyzing IBS of oxygenated hemoglobin (Oxy-Hb) signal is explained. Specifically, the experimental design, the process of NIRS data recording, data analysis methods, and future directions are all discussed. Overall, implementing a standardized fNIRS hyperscanning pipeline is a fundamental part of second-person neuroscience. Also, this is in line with the call for open-science to aid the reproducibility of research.
Introduction
Recently, to reveal the concurrent brain activity across the interactive dyads or members of a group, researchers employ the hyperscanning approach1,2. Specifically, electroencephalogram (EEG), functional magnetic resonance imaging (fMRI), and functional near-infrared spectroscopy (fNIRS) are used to record the neural and brain activities from two or more subjects simultaneously3,4,5. Researchers extract a neural index entailing concurrent brain coupling based on this technique, which refers to inter-brain synchrony (IBS) (i.e., phase and/or amplitude alignment of the neuronal or hemodynamic signals across time). A large variety of hyperscanning research found IBS during social interaction between multiple individuals (e.g., player-audience, instructor-learner, and leader-follower)6,7,8. Furthermore, IBS holds specific implications of effective learning and instruction9,10,11,12,13,14. With the surging of hyperscanning research in naturalistic learning scenarios, establishing a standard protocol of hyperscanning experiments and the pipeline of data analysis in this field is necessary.
Thus, this paper provides a protocol for conducting fNIRS-based hyperscanning of collaborative learning dyads and a pipeline for analyzing IBS. fNIRS is an optical imaging tool, which radiates near-infrared light to assess the spectral absorption of hemoglobin indirectly, and then hemodynamic/oxygenation activity is measured15,16,17. Compared with fMRI, fNIRS is less prone to motion artifacts, allowing measurements from subjects who are doing real-life experiments (e.g., imitation, talking, and non-verbal communication)18,7,19. In comparison with EEG, fNIRS holds higher spatial resolution, allowing researchers to detect the location of brain activity20. Thus, these advantages in spatial resolution, logistics, and feasibility qualify fNIRS to conduct hyperscanning measurement1. Using this technology, an emerging research body detects an index term as IBS-the neural alignment of two (or more) people's brain activity-in different forms of naturalistic social settings9,10,11,12,13,14. In those studies, various methods (i.e., Correlation analysis and Wavelet Transform Coherence (WTC) analysis) are applied to calculate this index; meanwhile, a standard pipeline on such analysis is essential but lacking. As a result, a protocol for conducting fNIRS-based hyperscanning and a pipeline using WTC analysis to identify IBS is presented in this work
This study aims to evaluate IBS in collaborative learning dyads using the fNIRS hyperscanning technique. First, a hemodynamic response is recorded simultaneously in each dyads' prefrontal and left temporoparietal regions during a collaborative learning task. These regions have been identified as associated with interactive teaching and learning9,10,11,12,13,14. Second, the IBS is calculated on each corresponding channel. The fNIRS data recording process consists of two parts: resting-state session and collaborative session. The resting-state session lasts for 5 min, during which both the participants (sitting face-to-face, apart from one another by a table (0.8 m)) are required to remain still and relax. This resting-state session is served as the baseline. Then, in the collaborative session, the participants are told to study the entire learning materials together, eliciting understanding, summarizing the rules, and making sure all learning materials are mastered. Here, the specific steps of conducting the experiment and fNIRS data analysis are presented.
Protocol
All recruited participants (40 dyads, mean age 22.1 ± 1.2 years; 100% right-handed; normal or corrected-to-normal vision) were healthy. Before the experiment, participants gave informed consent. Participants were financially compensated for their participation. The study was approved by the University Committee of Human Research Protection (HR-0053-2021), East China Normal University.
1. Preparation steps before adopting data
- Homemade NIRS caps
- Adopt elastic swimming cap to place optode holder grid.
NOTE: Considering that the head sizes of the participants are different, two sizes of caps are used. Small caps are prepared for participants with a head circumference of 55.4 ± 1.1 cm, and large caps are for the participants with a head circumference of 57.9 ± 1.2 cm. - Anchor the location of the EEG electrodes (inion, Cz, T3, T4, Fpz, and P5) as reference optodes according to the standard international 10-10 system on elastic swimming caps (see Table of Materials).
- First, place the standard 10-10 EEG cap (see Table of Materials) on the head mold, and put the elastic swimming cap on the EEG cap. Second, mark reference optodes (inion, Cz, T3, T4, Fpz, and P5) with chalk on each cap. Finally, cut two holes about 15 mm in diameter to place the two reference optodes (i.e., Fpz and P5, Figure 1).
NOTE: Specifically, a 3 x 5 optode probe set and a 4 x 4 optode probe set are separately placed over the prefrontal area (reference optode is placed at Fpz, Figure 1B) and left temporoparietal regions (reference optode is placed at P5, Figure 1B).
- First, place the standard 10-10 EEG cap (see Table of Materials) on the head mold, and put the elastic swimming cap on the EEG cap. Second, mark reference optodes (inion, Cz, T3, T4, Fpz, and P5) with chalk on each cap. Finally, cut two holes about 15 mm in diameter to place the two reference optodes (i.e., Fpz and P5, Figure 1).
- Cut holes to place the other optodes. Arrange a swimming cap with two grid holders directly on the head mold. Then, mark the location of other optodes with chalk. After that, cut the rest holes to make sure the grid holder fits in.
- Mount two probe sets (i.e., 3 x 5 and 4 x 4) to the swimming caps (see Table of Materials).
NOTE: The NIRS measurement system (see Table of Materials) provides these standard probe sets (i.e., 3 x 5 and 4 x 4) with standard holder sockets ensuring the 30 mm optode separation. - Open the probe set monitor window at the NIRS measurement system and select four probe sets arranged in 3 x 5 and 4 x 4 for each person, separately.
NOTE: The probe arrangements of the two caps should correspond to the structures in the probe set window (i.e., the exact location of the receiver probe numbers and the respective emitter).
- Adopt elastic swimming cap to place optode holder grid.
- Preparation of the experiment
- Before recording data, ensure the NIRS system is keeping a stable operating temperature by starting the system for at least 30 min.
NOTE: The stable operating temperature ranged from 5 °C to 35 °C. - Set the measurement mode to event-related measurement. Ensure the triggers receiver is active (i.e., the RS232 serial input).
NOTE: The experiment is programmed in commercially available psychology software (see Table of Materials). The absorption of near-infrared light (two wavelengths: 695 and 830 nm) is measured with a sampling rate of 10 Hz. - Prepare the lighted fiber optic probe, which can be used to move hair aside.
- Set the experiment environment with one table with two chairs to keep participants' seats face-to-face.
- Before recording data, ensure the NIRS system is keeping a stable operating temperature by starting the system for at least 30 min.
2. Adopting data by instructing participants
- Prepare the participants
- Instruct the participants, including the details of NIRS measurement methods.
NOTE: All the participants were healthy and were financially compensated for participation. No participants withdrew from the experiment halfway through. The laser beam of the NIRS may be harmful to the participants' eyes, and they were instructed not to look directly into those laser beams. - Make the participants sit face-to-face (apart from a table (0.8 m)) to make sure they can see each other directly. Adjust the chair-to-table distance (i.e., nearly 0.3 m) to make the participants sit comfortably.
- Turn on the laser button, and place the caps with the probe sets on the participants' heads.
NOTE: The 3 x 5 probe sets cover the forehead of the participants (middle probe of the bottom row is placed on Fpz); the 4 x 4 probe sets cover the left temporoparietal cortex (the third probe of the third row is placed on P5). - Put the four optical fiber bundles loosely on the holder's arms without contact with the participants or chairs.
NOTE: Here, the NIRS measurement system has four bundles of optical fibers. Additionally, ensure that the participants do not feel too heavy to pull off the caps. - Let the probe tips touch participants' scalp by carefully pushing each spring load probe further into its socket.
- Perform signal calibration.
- First, check the quality of the signal by clicking the Auto Gain in the probe set monitor window of the fNIRS machine. Then, a channel's poor signal and sufficient signal are marked in yellow and green in the probe set monitor window, respectively.
NOTE: For a channel with insufficient signals, lighted fiber optic probes are used to move the hair under the probe's tip to one side. - Then, push the probes further into their sockets to get sufficient signals. Repeat this process until all the channels are marked in green in the probe set monitor window of the NIRS measurement system, indicating that the quality of signals is accessible.
- First, check the quality of the signal by clicking the Auto Gain in the probe set monitor window of the fNIRS machine. Then, a channel's poor signal and sufficient signal are marked in yellow and green in the probe set monitor window, respectively.
- Instruct the participants, including the details of NIRS measurement methods.
- Run the experiment
- Start the experiment with a 5 min rest state, which serves as the baseline. Then, two participants are required to co-learn the learning materials.
- After the experiment, click on Text File Out to export the raw light intensity data and save the data as a text file.
NOTE: No filters are applied in the NIRS measurement system. - Use the three-dimensional (3D) digitizer (see Table of Materials) to determine the locations of emitters, receivers, and other references (i.e., inion, nasion, Cz, and left and right ears) for each participant.
- Obtain the MNI coordinates for the recording channels using the commercially available numeric computing platform21 (see Table of Materials). Supplementary Table S1 shows the corresponding anatomical locations of each channel.
- Clean probes and probe holders with ethanol. Wash caps with mild detergent and let the caps air dry.
3. Data analysis
- Data preprocessing
NOTE: Previous research has adopted variable non-commercial software packages (e.g., Homer222, AnalyzIR23, or nirs LAB24) with numeric computing platforms (see Table of Materials) on fNIRS data analysis, and they are all available on the website. Here Homer2 was used to do the preprocessing of the NIRS data. Additionally, both fNIRS recording data collected in the rest and collaborative learning phases share the same preprocessing and analysis pipeline.- Copy the dataset from the fNIRS machine. Convert the original data formation to the proper formation (i.e., convert cvs file to nirs file).
- Convert raw data to optical density (OD) data with "hmrIntensity2OD" function provided in the Numeric computing platform (see Table of Materials).
- Delete the bad channels. Then average the OD value for each participant on each channel and full sample points, respectively.
NOTE: Here, 46 averaged OD values are obtained.- Calculate the standard deviation (SD) for each participant.
- Mark as unusable and remove the channels with very low or high OD (which exceeded 5 SDs) from the analysis for each participant.
NOTE: This step can be performed before and/or after fNIRS data preprocessing. In this data analyses pipeline, the bad channels are detected before the fNIRS data preprocessing.
- Convert the OD time data into Oxy-Hb, DeOxy-Hb, and combined signal based on the modified Beer-Lambert Law25.
NOTE: Reference25 says, "All data analysis steps are conducted on Oxy-Hb data, which is an indicator of the change in regional cerebral blood flow having higher signal-to-noise ratio26. Additionally, previous research employed fNIRS hyperscanning in teaching and learning scenarios mainly focused on Oxy-Hb concentration11,12,13,14." - Calibrate Oxy-Hb time series from motion artifacts by the channel-by-channel wavelet-based method.
NOTE: Specifically, the Daubechies 5 (db5) wavelet with tuning parameter at 0.1 (see details in Homer2 manual)27,28 is adopted in removing motion artifacts. - Apply the band-pass filter (i.e., 0.01-1 Hz) on the calibrated Oxy-Hb data to reduce the high-frequency noise and slow drift.
- Conduct principal components analysis (PCA) on the OxyHb signal to remove non-neural global components (e.g., blood pressure, respiration, and blood flow variation)29.
NOTE: The PCA analysis proposed by Zhang and colleagues29 is adopted here.- First, decompose the signal.
NOTE: The specific formula of decomposition of fNIRS signal is: H = UΣVT. Here, temporal and spatial patterns of fNIRS data are presented in two matrices (i.e., U and V). U is a 2D (sample point x principal component) matrix. V is also a 2D (principal component x principal component) matrix. The column in V indicates one principal component (PC), and the strength of that PC for a particular channel is estimated in each entry of the column. The relative importance of each PC is represented by the value of the diagonal matrix Σ. - Second, conduct spatial smoothing.
NOTE: Gaussian kernel convolution is employed to remove localized signals and get the global component. - Third, reconstruct the signal.
NOTE: To calculate the global component of the fNIRS data, the smoothed spatial pattern matrix V* is plugged back into the decomposition formula: HGlobal = UΣ(V*)T. Then, localized derived neuronal signal can be obtained using original data H to subtract HGlobal : HNeuronal = H – HGlobal.
- First, decompose the signal.
- Inter-brain synchrony
NOTE: To reveal brain coupling in second-person neuroscience, wavelet transform coherence (WTC) is adopted here. Briefly, WTC measures the correlation between two-time series as a function of frequency and time. The specific formula of wavelet coherence of two-time series x and y is:
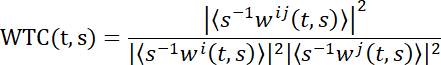
T and s denote the time and wavelet scale separately, ‹·› indicates a smoothing operation in scale and time. W represents the continuous wavelet transform. Then, a 2D (time x frequency) WTC matrix is generated30. Several toolboxes are used to calculate the WTC value. Here the toolbox created by Grinsted and colleagues was used30.- Adopt the WTC function of the numeric computing platform (see Table of Materials).
NOTE: Here, the default setting of the mother wavelet (i.e., Generalized Morse Wavelet with its parameters beta and gamma) is used. Mother wavelet converts each time series into the frequency and time domain. - Set the default setting on the other parameters (i.e., MonteCarloCount, representing the number of surrogate data sets in the significance calculation).
- Calculate the WTC value for two corresponding channels (the same channel in two participants) in a numeric computing platform (see Table of Materials). Following the same procedure, 46 WTC matrices are generated from 46 channels.
- Determine the frequency band of interest (FOI), which is sensitive to collaborative learning.
NOTE: Here, a cluster-based permutation approach is adopted to detect such FOI31, which offers a solution to multiple comparisons in multi-channel and multi-frequency data.- Perform Time-average of the WTC values in the resting and collaborative learning phases, respectively, for each channel combination. Then, conduct paired sample t-tests along with the entire frequency (frequency range: 0.01-1Hz32) on these time-averaged WTC values (collaborative learning vs. rest). Next, identify frequency bins at which the task effect is significant (collaborative learning > rest, p < 0.05).
- Obtain significant frequency neighboring points (≥2) as observed clusters and corresponding T values.
- Conduct a series of paired sample t-tests on permuted data to generate the T values for each cluster qualified in step 3.2.4.2 for 1000 times.
NOTE: For forming permuted data, participants are randomly assigned to form new two-member pairs. As the length of datasets varied across dyads for each random pair, the longer dataset is trimmed to the same length as the shorter one33. - Compare the averaged cluster-based T values from original pairs with the T values of 1000 permutations.
NOTE: The p values evaluated by this formula34:
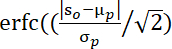 , where S0 denotes observed averaged-cluster t-value, μp and σp indicate the mean and standard deviation of permutation values.
, where S0 denotes observed averaged-cluster t-value, μp and σp indicate the mean and standard deviation of permutation values. - Average WTC values in the identified FOI in each channel in each dyad. Then, apply fisher z transformation to the WTC values to get a normal distribution of WTC values. Use this value to index the IBS for further statistical analysis.
- Adopt the WTC function of the numeric computing platform (see Table of Materials).
Representative Results
Figure 1 illustrates the experimental protocol and probe location. The fNIRS data recording process consists of two parts: resting-state session (5 min) and collaborative session (15-20 min). The collaborative learning dyads are required to relax and to keep still in the resting-state session. After that, participants are told to co-learning the learning material (Figure 1A). Their prefrontal and left temporoparietal regions are covered by the corresponding probe set (Figure 1B).
Figure 2 illustrates the fNIRS data analysis pipeline. The fNIRS data analysis is applied to all fNIRS data recorded from each participant and each channel. First, optode density in channel 33 for a certain dyad is visualized in Figure 2A. Optode density is recorded in 46 channels (CHs) of each collaborative learning dyad by the fNIRS measurement system. Second, With the operation clarified in steps 3.1.5 and 3.1.7, viable data are prepared for WTC analysis. Here, the red curve represents the data extracted by the wavelet-based motion artifacts removing method; the blue curve represents the data extracted by both the Wavelet-based motion artifacts removing method and PCA. Visualized difference between two curves suggests PCA is efficient in removing non-neural signals (Figure 2B). Third, the WTC matrix is visualized in Figure 2C. The color map varies from blue to yellow, representing the value of IBS raged from 0 to 1 (correlation coefficients as a function of time and frequency). Here, 1 denotes the largest coherence between two fNIRS signals, and 0 denotes no coherence is detected. A red rectangle in the plot marks significant coefficients. Additionally, results show a strong coherence around 1 Hz, representing the dyad's cardiac rhythm coherence. Finally, with the operation stated in steps 3.2.4, the comparison between the observed T value and the distribution of random T value (i.e.,1000 times) shows significant results (t (38) = 3.31, FDR corrected p < 0.05, Cohen's d = 1.05) in identified FOI (0.015 Hz-0.021 Hz) (Figure 2D)
Figure 3 presents the critical steps of the cluster-based permutation approach used to detect the collaborative learning relevant frequency band.
Taken together, following the data analysis pipeline, the frequency band (raged from 0.015 Hz to 0.021 Hz), which sensitive to collaborative learning, is identified by cluster-based permutation approach. Further, for each channel, the time-averaged IBS value is compared between the rest and the collaborative learning phases using a series of paired sample t-tests. For solving the multiple comparison problem, all the observed p-values in 46 channels are corrected by FDR methods35,36. The results show that the IBS at channel 33 reaches significance during collaborative learning (FDR corrected p < 0.05). No other corresponding channels indicated significant effects (p > 0.05).
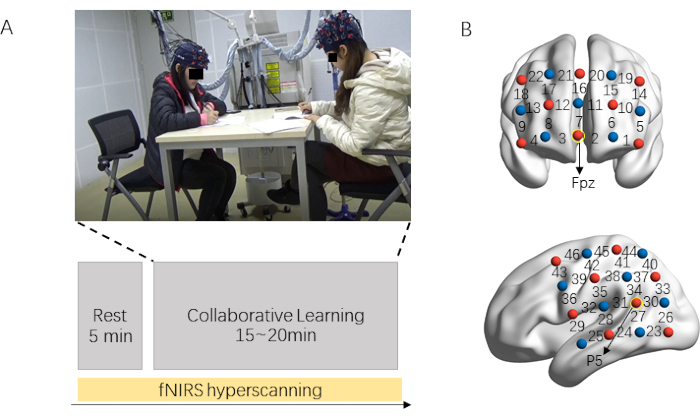
Figure 1: Experimental protocol and probe location. (A) Experimental procedure. Brain activity from dyads is acquired simultaneously using fNIRS. The resting-state session lasts for 5 min, in which dyads are required to relax and keep still. After that, participants are told to co-learn the learning material (15-20 min). (B) Optodes probe set. Two probe sets cover the prefrontal and left temporoparietal regions. Please click here to view a larger version of this figure.
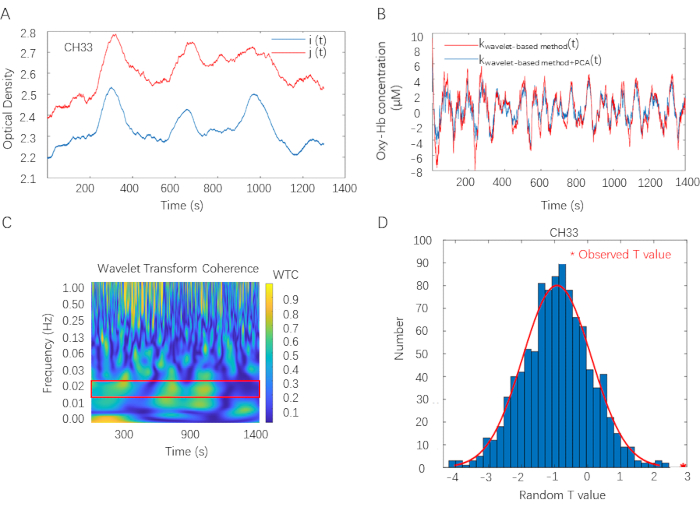
Figure 2: Overview of the fNIRS data analysis. (A) Optode density in channel 33 for one exemplary dyad. Optode density is recorded in 46 channels (CHs) of each collaborative learning dyad. i, j, Optode density of two participants of a collaborative learning dyad; t, time. (B) Data preprocess procedure. Wavelet-based motion artifacts removing method and PCA are applied on Oxy-Hb data in sequence. Here, the red curve represents the data extracted by the wavelet-based motion artifacts removing method; the blue curve represents the data extracted by both the Wavelet-based motion artifacts removing method and PCA. kwavelet-based method, data extracted by the Wavelet-based motion artifacts eliminating process. kwavelet-based method + PCA, data extracted by both Wavelet-based motion artifacts removing method and PCA. (C) WTC plot in channel 33 for one exemplary dyad. The color map varies from blue to yellow, representing the value of IBS ranged from 0 to 1 (correlations coefficients as a function of time and frequency). Here, 1 denotes the largest coherence between two fNIRS signals, and 0 indicates that no coherence is detected. A red rectangle in the plot marks significant coefficients. WTC estimates IBS on two clean Oxy-Hb time series. (D) Cluster-based permutation approach. Compare the observed T value with the distribution of random T values in identified FOI (0.015 Hz-0.021 Hz). Please click here to view a larger version of this figure.
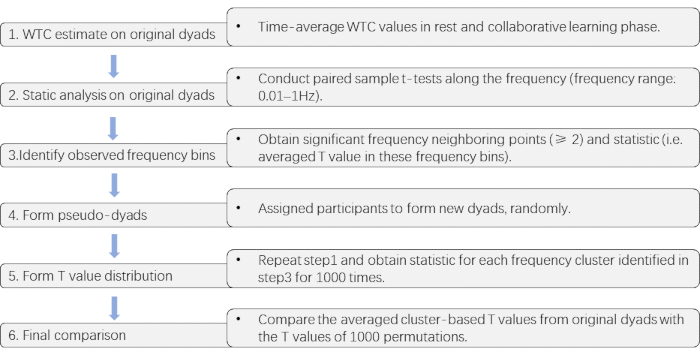
Figure 3: Flowchart of identifying the collaborative learning-related FOI. Please click here to view a larger version of this figure.
Supplementary Table S1. Please click here to download this Table.
Discussion
First, in the present protocol, the specific steps of conducting fNIRS hyperscanning experiments in a collaborative learning scenario are stated. Second, the data analysis pipeline that assesses the IBS of hemodynamic signals in collaborative learning dyads is also presented. The detailed operation on conducting fNIRS hyperscanning experiments would promote the development of open-science. Furthermore, the analysis pipeline is provided here to increase the reproducibility of hyperscanning research. In the following, the critical issues of experiment design, conducting an experiment, data analysis in (fNIRS) hyperscanning experiments are all highlighted. Additionally, possible solutions to present limitations are also discussed.
Experimental design
The experimental design for the fNIRS hyperscanning study is flexible. Here, the fNIRS hyperscanning technique is applied in the collaborative learning scenario. Two participants were asked to learn specific rules of the figure matrix together, and their brain activities were recorded by fNIRS simultaneously. This approach allows researchers to explore real-time concurrent neural dynamics (i.e., IBS) in collaborative learning dyads. According to previous research, IBS has been detected in teaching and learning scenarios and tracks the effective teaching mode11. Neural alignment detected in collaborative learning dyads may serve as a potential neural mechanism underpinning successful learning and provides implications for designing effective collaborative learning patterns. Meanwhile, critical issues on experimental design need to be addressed: the experiment time is limited to 30 min in this experiment. Two reasons account for this setting: First, wearing caps with fNIRS optodes on the head is not comfortable, participants cannot stand for a long time. Second, it's hard to ask participants to keep still during co-learning for a long time. The limited experiment time would allow good-quality signals to be obtained.
Conducting the experiment
The most challenging part of doing fNIRS hyperscanning in a collaborative learning scenario is getting high-quality brain signals. Based on the present protocol, three critical steps are highlighted: making appropriate caps, placing optodes, and conducting spatial registration of corresponding channels. First, since head circumference varies across participants, making caps that fit different individuals is essential. Second, when placing an appropriate cap on the participants' heads, ensure the tips of optodes can directly contact scalp skin. To achieve this goal, practicing this operation before the experiment is needed. Third, conducting spatial registration with a 3D digitizer can identify the corresponding anatomical locations of NIRS channels (CHs) on the cerebral cortex37,38,39. This protocol suggests completing spatial registration for all participants to get averaged and robust results. Along this line, previous research asked participants to conduct a pre-test to ensure accurate hemodynamic signals can be obtained. Specifically, participants performed a classical finger-thumb tapping task with their right hand, during which fNIRS recorded hemodynamic dynamics. Participants who detected a significant fNIRS signal (p < 0.05) in the left motor cortex are qualified to participate in the study. This technique ensures recorded signals are usable on all participants40.
Data analysis
The data analysis process in this protocol consists of two parts: preprocess and WTC analysis. Three critical data analysis steps should be highlighted here: First, conducting the principal component spatial filter algorithm (PCA) on the neural data. Zhang and couleage29 proposed this approach for the separation of the global and local effects. Although fNIRS allows relatively free movement and communication, PCA is necessary to extract accurate signals from systemic changes (e.g., breathing rate, blood pressure, heart rate, breathing rate, and autonomic nervous system activity). The protocol here suggests PCA is efficient in removing the global effects. This method is widely used in fNIRS hyperscanning studies13. Altogether, non-neural components can be removed successfully using spatial filtering. Second, WTC is adopted to identify the IBS of collaborative learning dyads. WTC is an approach of assessing the correlation coefficients between two-time series as a function of time and frequency41. This method can reveal locally phase-locked behavior that might not be detected with a traditional approach such as Fourier analysis30. And this method is widely used to estimate IBS in fNIRS hyperscanning with varied paradigms, such as cooperative and competitive behaviors4,42, studying action monitoring43, imitation44, verbal communication8, non-verbal communication19, teaching and learning activity11,12,13,14 and mother-child social interaction45.
Meanwhile, other techniques, such as Granger Causality Analyze (GCA), correlation analysis, and phase synchrony analysis, are used in hyperscanning research. GCA is a method for revealing directed (causal) information between two time-series data46. This method has once been used to test the direction of information flow between instructor and learner12. Correlation analysis is also adopted in the fNIRS-based hyperscanning field to estimate IBS in dyads who conduct cooperative or competitive tasks47,48. Compared to WTC analysis, this method only characterizes the covaried features of two fNIRS time series along time stream and missed potential information in frequency.
Additionally, other approaches that quantified phase synchrony with Phase locking value (PLV) were used in EEG hyperscanning studies. PLV estimates the consistency of the phase difference between two signals49. However, Burgess suggested PLV shows bias on detecting hyperconnectivity that doesn't exist, especially when small samples are employed50. Third, adopting a non-parametric statistical test to detect the collaborative learning-related frequency is essential. At first, task-related FOI is selected by either following suggestions in previous research or according to specific experiment design (i.e., how long for one task trial in an experiment). Recently, to obtain robust and reproductive results in the FOI selecting process, non-parametric statistical test approaches are adopted. Here, this technique operated efficiently. The collaborative learning-related FOI (0.015-0.021 Hz) is identified, and similar frequency bands have been identified in fNIRS hyperscanning research in teaching scenario13 and in verbal communication paradigms8. It is necessary to apply this technique in the multi-brain data analysis pipeline. All in all, establishing suitable algorithms and methods for the analysis of hyperscanning data will be a prominent field.
Limitation and future direction
Several limitations can be improved in the future to obtain reproductive and robust IBS within a realistic social interaction context from a multi-brain. First, the weight of the fiber is too heavy and uncomfortable to wear for a long time; thus, the time of the experiment is limited to 30 min. In the future, if recording the multi-brain activity in the classroom, it is hard to ask students to wear fNIRS caps during one school period (i.e., 50 min). Thus, wearable fNIRS settings are required in actual lecturing and learning scenario. Second, although the fNIRS shows higher tolerance to head motion than fMRI, this technique can only detect the brain activity of the surface cortex15. Thus, fNIRS hyperscanning cannot be used in the reward-related neural mechanism exploring paradigm, in which the amygdala plays a crucial role51. Meanwhile, the limited number of sources and detectors in the fNIRS setup suggests not the whole brain cortex would be measured. That means researchers have to select the region of interest (ROI) to measure. Third, PCA is adopted to eliminate the system contaminants. While this technique is efficient, in the future, adding short-channels that account for extra-cerebellar blood flow, which may contaminate fNIRS signals, is also an efficient approach29,39. Fourth, the data analysis procedure in this protocol can be applied in other naturalistic fNIRS hyperscanning studies. The next step is to develop fNIRS-specific data analysis packages with the standard guideline. Fifth, in this protocol, WTC is employed to identify the concurrent brain activity (i.e., IBS). With the development of a technique for calculating covaried neural activity, other methods such as graph theory and GCA also can be used. Sixth, it is necessary to recruit control conditions, such as talking conditions that require dyads to talk on specific topics to exclude confounding effects. Meanwhile, to reveal which learning activity in collaborative learning (i.e., knowledge co-construction52) would lead to the IBS. And whether these detected IBS can be used to track the learning performance of collaborative learning dyads are also important. Finally, it is also urgent to provide a framework to explain the mechanism of IBS. Researchers try to discern whether this is only the epiphenomenon or a neural mechanism of social interaction by Hamilton53. To achieve this goal, on the one hand, Hamilton proposed a xGLM approach that models brain activity, behavior data, and physiological data together to explore the reliable explanation of brain coupling53. On the other hand, Novembre and Lannetti suggested conducting multi-brain stimulation (MBS) to reveal the mechanism of concurrent brain activity54.
Conclusion
fNIRS hyperscanning leads to a paradigm shift from traditional experiment design to realistic social interaction scenarios in social neuroscience. The IBS extracted by this method provides a new view to explain the neurobiological mechanism of social interactions. Finally, the established standardized pipeline of collecting and analyzing data would be the milestone for generating valid results and advancing the recent hyperscanning experiment.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work is supported by the ECNU Academic Innovation Promotion Program for Excellent Doctoral Students (YBNLTS2019-025) and the National Natural Science Foundation of China (31872783 and 71942001).
Materials
| EEG caps | Compumedics Neuroscan,Charlotte,USA | 64-channel Quik-Cap | We choose two sizes of cap(i.e.medium and large). |
| NIRS measurement system with probe sets and probe holder grids | Hitachi Medical Corporation, Tokyo, Japan | ETG-7100 Optical Topography System | The current study protocol requires an optional second adult probe set for 92 channels of measurement in total. |
| Numeric computing platform | The MathWorks, Inc., Natick, MA | MATLAB R2020a | Serves as base for Psychophysics Toolbox extensions (stimulus presentation), Homer2 (fNIRS preprocess analysis), and "wtc" function(WTC computation). |
| Psychology software | psychology software tools,Sharpsburg, PA,USA | E-prime 2.0 | we apply E-prime to start the fNIRS measurement system and send triggers which marking the rest phase and collaborative learning phase for fNIRS recording data |
| Swimming caps | Zoke corporation,Shanghai,China | 611503314 | We first placed the standard 10-20 EEG cap on the head mold, and placed the swimming cap on the EEG cap. Second, we marked (inion, Cz, T3, T4, PFC and P5) with chalk. |
| Three-dimensional (3-D) digitizer | Polhemus, Colchester, VT, USA; | Three-dimensional (3-D) digitizer | Anatomical locations of optodes in relation to standard head landmarks were determined for each participant using a Patriot 3D Digitizer |
References
- Babiloni, F., Astolfi, L. Social neuroscience and hyperscanning techniques: past, present and future. Neuroscience & Biobehavioral Reviews. 44, 76-93 (2014).
- Schilbach, L., et al. Toward a second-person neuroscience. Behavior Brain Science. 36, 393-414 (2013).
- Montague, P. Hyperscanning: simultaneous fMRI during linked social interactions. NeuroImage. 16, 1159-1164 (2002).
- Cui, X., Bryant, D. M., Reiss, A. L. NIRS-based hyperscanning reveals increased interpersonal coherence in superior frontal cortex during cooperation. NeuroImage. 59 (3), 2430-2437 (2012).
- Dikker, S., et al. Brain-to-brain synchrony tracks real-world dynamic group interactions in the classroom. Current Biology. 27 (9), 1375-1380 (2017).
- Abrams, D. A., et al. Inter-subject synchronization of brain responses during natural music listening. European Journal of Neuroscience. 37 (9), 1458-1469 (2013).
- Pan, Y., et al. Instructor-learner brain coupling discriminates between instructional approaches and predicts learning. NeuroImage. 211, 116657 (2020).
- Jiang, J., et al. Leader emergence through interpersonal neural synchronization. Proceedings of the National Academy of Sciences of the United States of America. 112 (14), 4274-4279 (2015).
- Bevilacqua, D., et al. Brain-to-brain synchrony and learning outcomes vary by student-teacher dynamics: Evidence from a real-world classroom electroencephalography study. Journal of Cognitive Neuroscience. 31 (3), 401-411 (2019).
- Dikker, S., et al. Morning brain: real-world neural evidence that high school class times matter. Social Cognitive and Affective Neuroscience. 15 (11), 1193-1202 (2020).
- Pan, Y., Guyon, C., Borragán, G., Hu, Y., Peigneux, P. Interpersonal brain synchronization with instructor compensates for learner’s sleep deprivation in interactive learning. Biochemical Pharmacology. , 114111 (2020).
- Pan, Y., Novembre, G., Song, B., Li, X., Hu, Y. Interpersonal synchronization of inferior frontal cortices tracks social interactive learning of a song. NeuroImage. 183, 280-290 (2018).
- Zheng, L., et al. Enhancement of teaching outcome through neural prediction of the students’ knowledge state. Human Brain Mapping. 39 (7), 3046-3057 (2018).
- Zheng, L., et al. Affiliative bonding between teachers and students through interpersonal synchronisation in brain activity. Social Cognitive and Affective Neuroscience. 15 (1), 97-109 (2020).
- Kleinschmidt, A., et al. Simultaneous recording of cerebral blood oxygenation changes during human brain activation by magnetic resonance imaging and near-infrared spectroscopy. Journal of Cerebral Blood Flow & Metabolism. 16 (5), 817-826 (1996).
- Strangman, G., Culver, J. P., Thompson, J. H., Boas, D. A. A quantitative comparison of simultaneous BOLD fMRI and NIRS recordings during functional brain activation. NeuroImage. 17 (2), 719-731 (2002).
- Huppert, T. J., Hoge, R. D., Diamond, S. G., Franceschini, M. A., Boas, D. A. A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. NeuroImage. 29 (2), 368-382 (2006).
- Holper, L., Scholkmann, F., Wolf, M. Between-brain connectivity during imitation measured by fNIRS. NeuroImage. 63, 212-222 (2012).
- Hirsch, J., Zhang, X., Noah, J. A., Ono, Y. Frontal temporal and parietal systems synchronize within and across brains during live eye-to-eye contact. NeuroImage. 157, 314-330 (2017).
- Wilcox, T., Biondi, M. fNIRS in the developmental sciences. Wiley Interdisciplinary Reviews: Cognitive Science. 6 (3), 263-283 (2015).
- Ye, J. C., Tak, S., Jang, K. E., Jung, J., Jang, J. NIRS-SPM: statistical parametric mapping for nearinfrared spectroscopy. NeuroImage. 44 (2), 428-447 (2009).
- Huppert, T. J., Diamond, S. G., Franceschini, M. A., Boas, D. A. HomER: A review of time-series analysis methods for near-infrared spectroscopy of the brain. Applied Optics. 48 (10), 280-298 (2009).
- Santosa, H., Zhai, X., Fishburn, F., Huppert, T. The NIRS Brain AnalyzIR toolbox. Algorithms. 11 (5), 73 (2018).
- Xu, Y., Graber, H. L., Barbour, R. L. nirsLAB: a computing environment for fNIRS neuroimaging data analysis. Biomedical Optics. , (2014).
- Cope, M., Delpy, D. T. System for long-term measurement of cerebral blood and tissue oxygenation on newborn infants by near infra-red transillumination. Medical and Biological Engineering and Computing. 26 (3), 289-294 (1988).
- Hoshi, Y. Functional near-infrared spectroscopy: current status and future prospects. Journal of Biomedical Optics. 12 (6), 062106 (2007).
- Molavi, B., Dumont, G. A. Wavelet-based motion artifact removal for functional near-infrared spectroscopy. Physiological Measurement. 33 (2), 259 (2012).
- Cooper, R., et al. A systematic comparison of motion artifact correction techniques for functional near-infrared spectroscopy. Frontiers in Neuroscience. 6, 147 (2012).
- Zhang, X., Noah, J. A., Hirsch, J. Separation of the global and local components in functional near-infrared spectroscopy signals using principal component spatial filtering. Neurophotonics. 3 (1), 015004 (2016).
- Grinsted, A., Moore, J. C., Jevrejeva, S. Application of the cross wavelet transform and wavelet coherence to geophysical time series. Nonlinear Processes in Geophysics. 11, 561-566 (2004).
- Maris, E., Oostenveld, R. Nonparametric statistical testing of EEG-and MEG-data. Journal of Neuroscience Methods. 164 (1), 177-190 (2007).
- Nozawa, T., Sasaki, Y., Sakaki, K., Yokoyama, R., Kawashima, R. Interpersonal frontopolar neural synchronization in group communication: an exploration toward fNIRS hyperscanning of natural interactions. NeuroImage. 133, 484-497 (2016).
- Reindl, V., Gerloff, C., Scharke, W., Konrad, K. Brain-to-brain synchrony in parent-child dyads and the relationship with emotion regulation revealed by fNIRS-based hyperscanning. NeuroImage. 178, 493-502 (2018).
- Theiler, J., Eubank, S., Longtin, A., Galdrikian, B., Farmer, J. D. Testing for nonlinearity in time series: the method of surrogate data. Physica D: Nonlinear Phenomena. 58 (1-4), 77-94 (1992).
- Genovese, C. R., Lazar, N. A., Nichols, T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 15 (4), 870-878 (2002).
- Nichols, T., Hayasaka, S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Statistical Methods in Medical Research. 12 (5), 419-446 (2003).
- Tsuzuki, D., et al. Virtual spatial registration of stand-alone fNIRS data to MNI space. NeuroImage. 34 (4), 1506-1518 (2007).
- Singh, A. K., Okamoto, M., Dan, H., Jurcak, V., Dan, I. Spatial registration of multi-channel multi-subject fNIRS data to MNI space without MRI. NeuroImage. 27 (4), 842-851 (2005).
- Noah, J. A., et al. Comparison of short-channel separation and spatial domain filtering for removal of non-neural components in functional near-infrared spectroscopy signals. Neurophotonics. 8 (1), 015004 (2021).
- Noah, J. A., et al. Real-time eye-to-eye contact is associated with cross-brain neural coupling in angular gyrus. Frontiers in Human Neuroscience. 14 (19), (2020).
- Torrence, C., Compo, G. P. A practical guide to wavelet analysis. Bulletin of the American Meteorological Society. 79 (1), 61-78 (1998).
- Osaka, N., Minamoto, T., Yaoi, K., Azuma, M., Osaka, M. Neural synchronization during cooperated humming: a hyperscanning study using fNIRS. Procedia-Social and Behavioral Sciences. 126, 241-243 (2014).
- Dommer, L., Jäger, N., Scholkmann, F., Wolf, M., Holper, L. Between-brain coherence during joint n-back task performance: a two-person functional near-infrared spectroscopy study. Behavioural Brain Research. 234 (2), 212-222 (2012).
- Holper, L., Scholkmann, F., Wolf, M. Between-brain connectivity during imitation measured by fNIRS. Neuroimage. 63, 212-222 (2012).
- Nguyen, T., et al. The effects of interaction quality on neural synchrony during mother-child problem solving. Cortex. 124, 235-249 (2020).
- Seth, A. K., Barrett, A. B., Barnett, L. Granger causality analysis in neuroscience and neuroimaging. Journal of Neuroscience. 35 (8), 3293-3297 (2015).
- Funane, T., et al. Synchronous activity of two people’s prefrontal cortices during a cooperative task measured by simultaneous near-infrared spectroscopy. Journal of Biomedical Optics. 16 (7), 077011 (2011).
- Liu, T., Saito, H., Oi, M. Role of the right inferior frontal gyrus in turn-based cooperation and competition: a near-infrared spectroscopy study. Brain and Cognition. 99, 17-23 (2015).
- Lachaux, J. P., Rodriguez, E., Martinerie, J., Varela, F. J. Measuring phase synchrony in brain signals. Human Brain Mapping. 8 (4), 194-208 (1999).
- Burgess, A. P. On the interpretation of synchronization in EEG hyperscanning studies: a cautionary note. Frontiers in Human Neuroscience. 7, 881 (2013).
- Burgos-Robles, A., et al. Amygdala inputs to prefrontal cortex guide behavior amid conflicting cues of reward and punishment. Nature Neuroscience. 20 (6), 824-835 (2017).
- Mende, S., Proske, A., Narciss, S. Individual preparation for collaborative learning: Systematic review and synthesis. Educational Psychologist. , 1-25 (2020).
- Hamilton, A. F. D. C. Hyperscanning: Beyond the hype. Neuron. 109 (3), 404-407 (2021).
- Novembre, G., Iannetti, G. D. Hyperscanning alone cannot prove causality. Multibrain stimulation can. Trends in Cognitive Sciences. 25 (2), 96-99 (2021).

