Measurement of Protein Import Capacity of Skeletal Muscle Mitochondria
Summary
Mitochondria are key metabolic organelles that exhibit a high level of phenotypic plasticity in skeletal muscle. The import of proteins from the cytosol is a critical pathway for organelle biogenesis, essential for the expansion of the reticulum and the maintenance of mitochondrial function. Therefore, protein import serves as a barometer of cellular health.
Abstract
Mitochondria are key metabolic and regulatory organelles that determine the energy supply as well as the overall health of the cell. In skeletal muscle, mitochondria exist in a series of complex morphologies, ranging from small oval organelles to a broad, reticulum-like network. Understanding how the mitochondrial reticulum expands and develops in response to diverse stimuli such as alterations in energy demand has long been a topic of research. A key aspect of this growth, or biogenesis, is the import of precursor proteins, originally encoded by the nuclear genome, synthesized in the cytosol, and translocated into various mitochondrial sub-compartments. Mitochondria have developed a sophisticated mechanism for this import process, involving many selective inner and outer membrane channels, known as the protein import machinery (PIM). Import into the mitochondrion is dependent on viable membrane potential and the availability of organelle-derived ATP through oxidative phosphorylation. Therefore its measurement can serve as a measure of organelle health. The PIM also exhibits a high level of adaptive plasticity in skeletal muscle that is tightly coupled to the energy status of the cell. For example, exercise training has been shown to increase import capacity, while muscle disuse reduces it, coincident with changes in markers of mitochondrial content. Although protein import is a critical step in the biogenesis and expansion of mitochondria, the process is not widely studied in skeletal muscle. Thus, this paper outlines how to use isolated and fully functional mitochondria from skeletal muscle to measure protein import capacity in order to promote a greater understanding of the methods involved and an appreciation of the importance of the pathway for organelle turnover in exercise, health, and disease.
Introduction
Mitochondria are organelles that exist in complex morphologies in different cell types and are recognized to possess an increasing array of functions that are critical for cellular health. As such, they can no longer be whittled down merely to energy-producing organelles. Mitochondria are key metabolic regulators, determinants of cell fate, and signaling hubs, the functions of which can serve as useful indicators of overall cellular health. In skeletal muscle cells, electron microscopy studies reveal the presence of geographically distinct subsarcolemmal (SS) and intermyofibrillar (IMF) mitochondria, which exhibit a degree of connectivity1,2,3,4 that is now recognized to be highly dynamic and adaptable to changes in skeletal muscle activity levels, as well as with age and disease. Mitochondrial content and function in muscle can be assessed in numerous ways5,6, and traditional methods of organelle isolation have been applied to better understand the respiratory and enzymatic capacities (Vmax) of mitochondria distinct from the influence of the cellular milieu7,8. In particular, these traditional methods have revealed subtle biochemical distinctions between mitochondria isolated from subsarcolemmal and intermyofibrillar regions, belying possible functional implications for metabolism in these subcellular regions8,9,10,11.
The biogenesis of mitochondria is unique in requiring the contribution of gene products from both nuclear and mitochondrial DNA. However, the vast majority of these are derived from the nucleus since mtDNA transcription only leads to the synthesis of 13 proteins. Since mitochondria normally comprise >1000 proteins involved in diverse metabolic pathways, biogenesis of the organelle requires a tightly regulated means of import and assembly of precursor proteins from the cytosol into the various mitochondrial sub-compartments to maintain proper stoichiometry and function12,13. Nuclear-encoded proteins destined for mitochondria normally carry a mitochondrial targeting sequence (MTS) that targets them to the organelle and facilitates their sub-compartmental localization. Most matrix-bound proteins contain a cleavable N-terminal MTS, while those destined for the outer or inner mitochondrial membrane usually have internal targeting domains14. The import process is carried out by a set of diverse channels that provide multiple avenues for entry into the organelle13. The translocase of the outer membrane (TOM) complex shuttles precursors from the cytosol into the intermembrane space, where they are recognized by the translocase of the inner membrane (TIM) complex. This complex is responsible for importing nuclear-encoded precursors into the matrix, where proteases cleave the N-terminal targeting presequence. Proteins destined for the outer membrane can be directly inserted into this membrane through the TOM complex, while those destined for the inner membrane are inserted by a TIM protein, specifically TIM22. Following their import, proteins are further processed by resident proteases and chaperones and often combine to form larger complexes, such as those found in the electron transport chain.
Mitochondrial protein import itself also serves as a measurement of mitochondrial health, as this process relies on the presence of membrane potential and a source of energy in the form of ATP15. For example, when the membrane potential is dissipated, protein kinase PINK1 cannot be taken up by the organelle, and this leads to phosphorylation signals that trigger the onset of the degradation of the organelle through a pathway called mitophagy16,17. Under similar circumstances, when the import is impeded, the protein ATF5 cannot enter the organelle, and it subsequently translocates to the nucleus, where it serves as a transcription factor for the up-regulation of UPR gene expression18,19. Thus, measuring protein import efficiency can provide comprehensive insight into the health of the organelle, while the gene expression response can be used to indicate the degree of retrograde signaling to the nucleus.
Despite its obvious importance for the biogenesis of mitochondria and for cellular health in general, the import pathway in mammalian mitochondria is remarkably understudied. In this report, we describe the specific steps involved in measuring the import of precursor proteins into skeletal muscle mitochondria and provide data to illustrate the adaptive response of the import system to changes in muscle and disuse, illustrating the contribution of the protein import to the adaptive plasticity of skeletal muscle.
Protocol
All animals used in these experiments are maintained in the animal care facility at York University. The experiments are conducted in accordance with the Canadian Council on Animal Care guidelines with approval from the York University Animal Care Committee (Permit: 2017-08).
1. Functional isolation of subsarcolemmal and intermyofibrillar mitochondria from skeletal muscle
- Reagent preparation:
- Prepare all the buffers and media as mentioned in Table 1.
- Set the buffers to pH 7.4 and store at 4 °C (up to 2 weeks), except nagrase protease.
- Prepare fresh nagarse protease (Table 1) each time.
- Tissue removal and mincing
NOTE: A flowchart of the steps below for the isolation of mitochondria is provided in Figure 1.- Fill a glass scintillation vial with ~20 mL of Buffer 1 (Table 1) and place it on ice.
- While the mouse is under anesthetic, harvest skeletal muscles (tibialis anterior, quadriceps, gastrocnemius etc.), approximately 500-1000 mg, and place the muscles in the scintillation vial containing chilled Buffer 1.
NOTE: Gaseous isoflurane is used as an anesthetic at 2.5% at a flow rate of 0.4 L of O2/min. A pinch test is performed to ensure that the animal is non-responsive. - Following tissue collection, euthanize the animal via cervical dislocation.
- Pre-chill a watch glass on ice. Remove any fat or connective tissue from the muscles and mince the tissue on the watch glass until it is a homogenous slurry.
- Place the minced tissue in a pre-chilled 50 mL plastic centrifugation tube (see Table of Materials) and record the exact weight.
- Dilute the minced tissue 10-fold with Buffer 1 + ATP (Table 1).
- Homogenize the muscle sample using an 8 mm twin-blade homogenizer (see Table of Materials) at a power output of 9.8 Hz for 10 s, ensuring no visible chunks of muscle are remaining.
- Centrifuge the samples at 800 x g for 10 min using a high-speed centrifuge (see Table of Materials).
NOTE: The purpose of this step is to separate SS and IMF mitochondrial fractions. The supernate contains SS mitochondria, while the pellet contains IMF mitochondria.
- SS mitochondrial isolation
- Filter the supernate through a single layer of cheesecloth into another set of 50 mL plastic centrifugation tubes to remove any large debris or contaminants.
- Centrifuge the supernate at 9,000 x g for 10 min at 4 °C using a high-speed centrifuge.
- Discard the supernate and resuspend the pellet in 3.5 mL of Buffer 1 + ATP.
NOTE: Resuspend with a P1000 and do this carefully. Mitochondria are fragile organelles. Perform gently and avoid touching the pellet with the pipette tip. - Centrifuge the sample at 9,000 x g for 10 min at 4 °C using a high-speed centrifuge.
- Discard the supernate unless further processing to isolate a cytosolic fraction devoid of nuclei, mitochondria, and other organelles is desired. Resuspend the pellet carefully in roughly 95 µL of the resuspension medium (Table 1) using a P200 pipette.
NOTE: Similar to step 1.3.3, avoid touching the pellet with the pipette tip. The volume of the resuspension medium can be altered depending on the size of the pellet. This is the SS mitochondrial fraction, which can be stored on ice until the IMF fraction is isolated. If desired, the cytosolic fraction, devoid of organelles, can be isolated by centrifuging the supernate at 100,000 x g in an ultracentrifuge (see Table of Materials) and thereafter discarding the pellet.
- IMF mitochondrial isolation
- Dilute the pellet from step 1.2.8 by 10-fold in Buffer 1 + ATP. Resuspend using a Teflon pestle by gently mixing the pellet and buffer together until the sample is consistent.
- Homogenize the sample using an 8 mm twin-blade homogenizer (see Table of Materials) at a power output of 9.8 Hz for 10 s.
- Centrifuge the sample at 800 x g for 10 min at 4 °C using a high-speed centrifuge.
- Discard the supernate and dilute the IMF mitochondrial pellet 10-fold using Buffer 2 and resuspend using the Teflon pestle by gently mixing until it is homogeneous.
- Add 25 µL/g of tissue to 10 mg/mL nagarse protease (Table 1) and place the centrifugation tube on its side on ice, gently mixing every minute by rocking the tube side to side.
NOTE: The addition of nagarse helps liberate IMF mitochondria by performing limited digestion of the myofibrils. - After 5 min, add 20 mL of Buffer 2 (Table 1) to dilute the nagarse protease to the point of inactivity and stop the digestion.
- Immediately centrifuge the sample at 5,000 x g for 5 min at 4 °C using a high-speed centrifuge.
- Discard the supernate and dilute the pellet 10-fold using Buffer 2 and resuspend using a Teflon pestle by gently mixing.
- Centrifuge the sample at 800 x g for 15 min at 4 °C to pellet the fibrous material.
- Pour the supernate in a new set of 50 mL plastic centrifuge tubes, careful not to disrupt the pellet, which can be discarded.
- Centrifuge the sample at 9,000 x g for 10 min at 4 °C as in step 1.4.7.
- Discard the supernate, add 3.5 mL of Buffer 2 and gently resuspend the pellet using a P1000 pipette.
- Centrifuge the sample at 9,000 x g for 10 min at 4 °C as in step 1.4.7.
- Discard the supernate and resuspend the pellet gently with roughly 180 µL of resuspension medium using a P200 pipette.
NOTE: The volume of the resuspension medium can be altered depending on the size of the pellet. This is the IMF mitochondrial fraction, which can be stored on ice until it is to be used in the import assay.
2. Mitochondrial protein import
- In vitro transcription
NOTE: These subsequent steps outline how to prepare a radiolabelled protein for import. The import pathway under investigation may dictate the choice of target protein, for example, to evaluate import into the mitochondrial matrix, ornithine carbamyl transferase, OCT, and malate dehydrogenase, MDH, are commonly used, while Tom40 is commonly used as an indication of import into the outer mitochondrial membrane. For the following steps, plasmid DNA encoding the protein of choice is required. The transcription of plasmid DNA can be done prior to the mitochondrial isolation on a separate day, and the mRNA collected from this experiment can be stored and used in future translation and import experiments.- Linearize the DNA: Combine 40 µL of plasmid DNA (5 µg/µL), 5 µL of 10x enzyme buffer and 5 µL of restriction enzyme and incubate at 37 °C for 30 min.
NOTE: The restriction enzymes utilized may vary based on the plasmid, which may require a different enzyme buffer, and experimental conditions DNA can be used in any amount/volume, as the template can be scaled up or down. - Purify and precipitate the linearized DNA using phenol and ethanol. Carry out all the subsequent steps (2.1.3-2.1.15) in sterile 1.5 mL tubes, and use a tabletop centrifuge (see Table of Materials) for the centrifugation steps.
- Add the appropriate volume of sterile H2O so that the final volume equals 400 µL. Then, add 400 µL of phenol.
- Mix vigorously by inversion for ~10 s, and centrifuge at 17,000 x g for 1 min at 4 °C. Withdraw and save the upper phase.
- Add 400 µL of phenol:chloroform:isoamylalcohol (25:24:1, v:v:v).
- Mix vigorously by inversion for ~10 s, and centrifuge at 17,000 g for 1 min at 4 °C. Withdraw and save the upper phase.
- Add 400 µL of chloroform:isoamylalcohol (24:1, v:v).
- Mix vigorously by inversion for ~10 s, and centrifuge at 17,000 g for 1 min at 4 °C. Withdraw and save the upper phase.
- Add 40 µL of 3M sodium acetate (pH 7.0), and add 1 mL of 95% ethanol.
- Place sterile 1.5 mL tubes in -80 °C freezer on their side for 15 min.
- Centrifuge the samples at 17,000 x g for 10 min at 4 °C.
- Discard the supernate and wash the pellet with 300 µL of 80% ethanol.
- Centrifuge the samples at 17,000 x g for 2 min at 4 °C.
- Discard the supernate and air-dry the pellet. Once the pellet is dry, it should be clear.
- Resuspend the pellet in 20 µL of sterile H2O
- Measure the DNA concentration using a spectrophotometer (see Table of Materials) and ensure that the A260:280 ratio is above 1.8.
- Dilute the DNA to a concentration of 0.8 µg/µL in sterile H2O.
- Transcribe the DNA: Combine plasmid (0.8 µg/µL, 60.8 µL), sterile H2O (8.4 µL), 10 mM NTP (5.2 µL), 10 mM ATP ( 10.0 µL), 1 mM M-7-G (11.6 µL), the mix as mentioned step 2.1.19 (15.6 µL), RNAsin (5.2 µL) and an appropriate RNA polymerase (4.8 µL) to form "one reaction mix" (total volume = 121.6 µL) and incubate for 90 min at the optimum temperature for the polymerase (37 °C for T7 polymerase; 40 °C for SP6 polymerase)
- To prepare the mix add 200 µL of 1M HEPES (pH 7.9), 100 µL of 1 M MgAc2, 500 µL of 4 M KAc, 100 µL of 20 mM spermidine, 100 µL of 0.5 M DTT, and 200 µL of sterile H20.
NOTE: The reaction can be scaled up or down in volume as long as the provided proportions of reagents are maintained. The RNA polymerase used is dictated by the bacterial promoter sequence within the plasmid being used. - Then, purify and precipitate the mRNA using phenol and ethanol as in steps 2.1.2-2.1.15. Bring the volume up to 400 µL with sterile H2O (add 280 µL) and proceed as described in steps 2.1.2-2.1.15. Resuspend the final pellet in 20 µL of sterile H2O.
- Measure the concentration of mRNA using a spectrophotometer, ensure that the A260:280 ratio is ~2.0.
- Dilute the mRNA to 2.8 µg/µL in sterile H2O, and store at -20 °C in 50 µL aliquots.
- Linearize the DNA: Combine 40 µL of plasmid DNA (5 µg/µL), 5 µL of 10x enzyme buffer and 5 µL of restriction enzyme and incubate at 37 °C for 30 min.
- In vitro translation
NOTE: The translation of the desired DNA must be carried out on the same day of the import experiment. This experiment requires the use of radiolabeled amino acids, and therefore the user must have a permit for the use of radioisotopes, and all handling and disposal must be in accordance with the institution's policies/requirements. Steps involving radioisotope safety measures have been omitted or described briefly, as these will likely differ based on the institution.- Prepare enough translation reaction mix (Table 2) to supply ~20 µL per sample of mitochondria to be used in the import experiment and include the same volume for a translation lane.
- Incubate the reaction for 30 min at 30 °C (time may vary with mRNA, 25-60 min).
- Record the 35S-methionine use as per the institution's radioactive safety requirements and dispose of anything that has come into contact with the radioisotope per the institution's guidelines.
- Protein import
NOTE: Freshly isolated mitochondria are required for the next steps. Refer to the mitochondrial isolation protocol. Other isolation methods can be used as long as the mitochondria are viable and functional. For the subsequent steps, the mitochondrial samples utilized are resuspended in a resuspension buffer as described above (Table 1). The protein concentration of the mitochondrial sample must also be measured prior to starting the import assay.- Roughly 15 min following the start of the translation reaction, aliquot 90 µg of mitochondria into sterile 1.5 mL tubes and pre-incubate at 30 °C for ~10 min.
NOTE: Aliquot more mitochondria than what will actually be used in the experiment; only 75 µg will be actually used. However, aliquoting in excess is not a requirement. - In a fresh set of sterile 1.5 mL tubes, combine 75 µg of mitochondria with 18 µL of the translation reaction and incubate at 30 °C for desired time.
- Keep the remaining volume of the translation reaction on ice; this will be utilized later.
NOTE: Import is a time-dependent process, so it may be prudent to start with a time-course experiment ranging in incubation times from 1-60 min. A typical reaction time is 30 min. - During this time, prepare the sucrose cushion by combining 1 g of sucrose, 200 µL of 2.5 M KCl, 10 µL 1 M MgCl2, 100 µL of 1 M HEPES (pH 7.4), and bring the volume to 5 mL using sterile H2O.
- Prepare a new set of sterile 1.5 mL tubes corresponding to each tube in the import reaction. Aliquot 600 µL of the sucrose cushion into each tube and keep on ice until the import reaction is over.
- To terminate the import reaction after the appropriate incubation time, remove the tube from 30 °C, place it on ice and then carefully transfer the import reaction on top of the tube with the sucrose cushion.
NOTE: This step needs to be done very slowly to ensure that the mitochondria are not damaged/ruptured. The sucrose cushion helps slow down the migration of the mitochondria and "cushions" its descent to the bottom of the tube during the subsequent centrifugation step. - Centrifuge the samples + sucrose cushion at 17,000 x g for 15 min at 4 °C.
- Prepare the breaking buffer by combining 25 mL of 2.4 M sorbitol, 2 mL of 1 M HEPES (pH 7.4), and 72 mL of double-distilled H2O.
- Prepare lysis buffer for SDS-PAGE gel electrophoresis: Combine 50 mL of heated glycerol, 11.5 g of SDS, 31.25 mL of 1 M Tris (pH 6.8) and bring the volume to 500 mL with double-distilled H2O.
- For sample preparation, mix 475 µL of lysis buffer with 25 µL of β-mercaptoethanol to be used in a later step.
NOTE: Lysis buffer can be made in advance and stored at room temperature; however, the addition of β-mercaptoethanol is to be done fresh. - Using a P1000 pipette, carefully remove the supernate and do not disturb the pellet. Dispose of the supernate in an appropriate radioactive waste container.
- For import into the outer membrane, perform this using Tom40, resuspend the pellet in 50 µL of freshly prepared 0.1 M sodium carbonate (pH 11.5) and incubate on ice for 30 min.
- Following the incubation, centrifuge the sample 14,000 x g for 5 min at 4 °C. This step is only done for outer membrane import reactions.
- To prepare the samples for SDS-PAGE electrophoresis, add 20 µL of breaking buffer, 20 µL of lysis buffer, and β-mercaptoethanol and resuspend well. Add 5 µL of sample dye.
- Prepare the control/translation lane by combining 3 µL of the remaining translation reaction from step 2.3.3 with 37 µL of lysis buffer and 5 µL of sample dye. Mix well.
- Boil the samples for 5 min at 95 °C and spin down gently at a low speed to avoid pelleting mitochondria.
- Apply the samples to a SDS polyacrylamide gel.
NOTE: The gel percentage and subsequent run time will depend on which protein is being imported. For example, OCT is 37 kDa, and its mature processed band is slightly smaller; therefore, a 12% gel allows good separation of these bands. - Once electrophoresis is complete, remove the gel and place it in double-distilled H2O.
- Boil the gel in 5% TCA in a fume hood for 5 min, continuously stirring/moving the gel to avoid cracking.
NOTE: TCA helps solidify the gel for visualization; this can be done in a metal tray over a Bunsen burner. Use enough TCA to generously cover the gel, ~½ full. - Remove the gel and place it back in double-distilled H2O. Agitate it on a rotating plate for 1 min at ~50 rpm.
- Wash the gel in 10 mM Tris on a rotating plate for 5 min at ~50 rpm, again using enough to generously cover the gel, ~½ of the container.
- Precipitate the protein by washing the gel in 1 M Salicyclic acid on a rotating plate for 30 min, again using enough Salicyclic acid to generously cover the gel, ~½ of the container being used.
- Dehydrate the gel as described in steps 2.3.24-2.3.31.
NOTE: This can be done in a number of ways. A gel dryer (see Table of Materials) provides a time-efficient option (described below); however other commercially available methods/kits can be used that may be more cost-effective but more time-consuming. Gel drying kits can be utilized as an alternative that does not require the application of heat or suction but instead allows the gel to air-dry between cellophane sheets to prevent distortion. - Place a large sheet of blotting paper down on the porous bed of the gel dryer. Place one sheet of paper towel in the area where the gel will be applied.
- Cut a 15 cm x 20 cm piece of blotting paper and place it on the paper towel.
- Scoop out the gel from the container using a second piece of 15 cm x 20 cm blotting paper and lay it down flat on top of the first piece.
- Cut a slightly larger piece of plastic wrap, ~20 cm x 25 cm, and place it down on gel, ensuring no creases or bubbles under the wrap.
NOTE: This may take a few attempts, but it is imperative that there are no trapped air pockets. - Gently lay down the plastic cover of the gel dryer over top of the wrap, again ensuring that there are no bubbles or creases.
- Turn on the pump/vacuum and ensure that the plastic cover has formed a seal. Test the seal by lifting a corner of the plastic cover and wait for it to re-seal.
- Close the gel dryer and run for 90 min. Set the temperature to start at 30 °C, gradually reach 80 °C, and then return to 30 °C at the end of the run.
- Once dehydrated, the gel will solidify and feel paper-thin. Wrap the gel in the plastic wrap used in the drying process.
- Place the dehydrated gel in a film cassette with a phosphorus film (see Table of Materials), with the white side facing the gel. Exposure times vary but can be 24-48 h for visualization of bands.
- Visualize using autoradiography using any suitable imager that is capable of phosphorus imaging.
- Roughly 15 min following the start of the translation reaction, aliquot 90 µg of mitochondria into sterile 1.5 mL tubes and pre-incubate at 30 °C for ~10 min.
Representative Results
We have extensively illustrated that this protocol is a valid assay for determining the rate of import into functional and intact isolated skeletal muscle mitochondria. In comparison to untreated conditions, the import of typical precursor proteins such as malate dehydrogenase (MDH) into the matrix is sensitive to membrane potential because it can be inhibited by valinomycin, a respiratory chain uncoupler (Figure 2A). Import is also impeded when mitochondrial inner and outer membranes are solubilized in the presence of the detergent Triton X-100. The import process is sensitive to the presence of external ATP, which serves to unfold precursor proteins for translocation across the membranes, and is tightly controlled by the rate of respiration and ATP provision (data not shown20). Distinct differences in the import are also observed between intermyofibrillar and subsarcolemmal mitochondrial fractions, in part due to variations in protein import machinery expression, as well as the respiratory rate between these mitochondria (Figure 2B).
Mitochondria in skeletal muscle are highly dynamic organelles that respond readily to changes in energy demand. Mitochondrial content in the muscle increases following periods of chronic exercise or in response to electrical stimulation-induced contractile activity (see for review21). For example, 7 days of chronic contractile activity of rat skeletal muscle enhances import into the OM and matrix by 1.6-fold and 1.4-fold, respectively22 (Figure 3A, 3B). These changes in mitochondrial content are brought about, in part, by alterations in the capacity of the mitochondrial protein import system. Indeed, a close relationship between the import rate of precursor proteins and a good estimate of mitochondrial content, as measured by the complex IV marker cytochrome oxidase under control or denervated conditions, can be illustrated23 (Figure 3C).
The adaptability of the import system in muscle to alterations in contractile activity suggests that exercise could be used as a treatment to resolve defects in the import pathway if identified. During the investigation of mitochondrially-mediated apoptosis using Bax-Bak double knockout animals, we noticed that the reduced mitochondrial content in the muscles of these experimental animals was accompanied by a decrease in the import of precursor proteins into the matrix. We then investigated the possibility that exercise could restore this import capacity. Indeed, following 6 weeks of voluntary wheel run training, protein import was restored to control levels in the knockout animals24, illustrating the adaptive plasticity of the import pathway to rescue mitochondrial content and function (Figure 4).
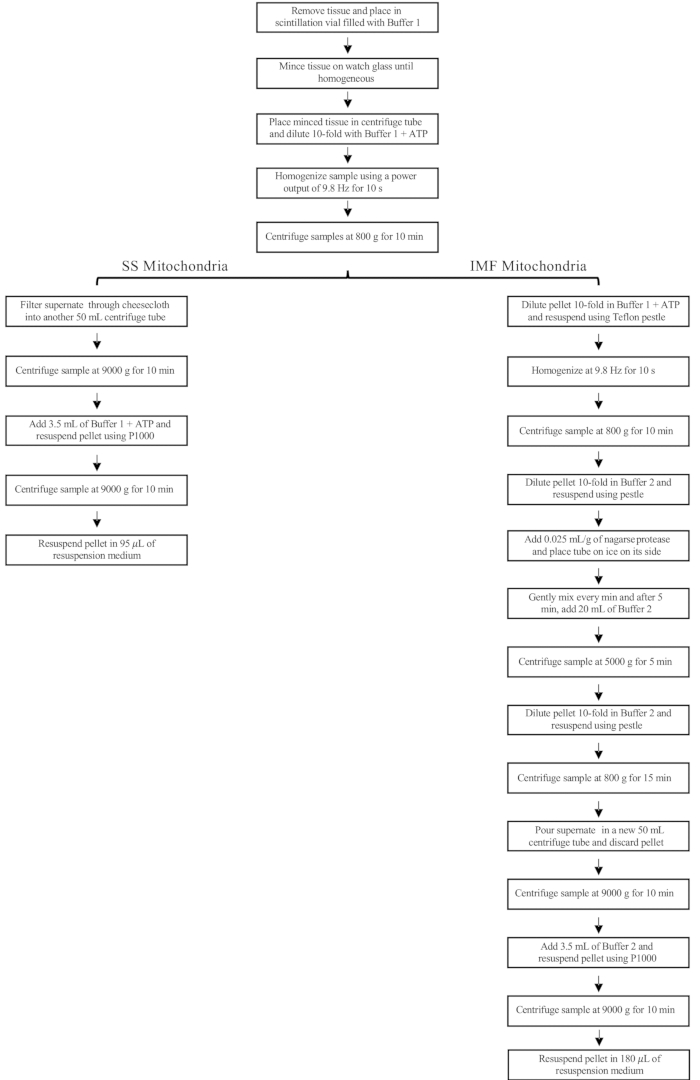
Figure 1: Schematic of the workflow for isolating SS and IMF mitochondria from skeletal muscle based on the previous work8. This protocol allows the isolation of functional mitochondria based on their geographic location within skeletal muscle. SS mitochondria are more rapidly and easily liberated, while the IMF mitochondria require a further digestion step with protease to untangle them from the myofibrils. Note that the isolation of these subfractions can be done in tandem. An updated, similar procedure has recently been published25. Please click here to view a larger version of this figure.
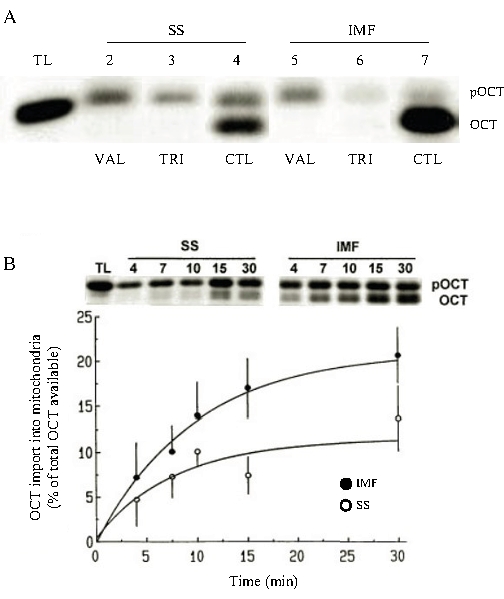
Figure 2: Protein import into the mitochondrial matrix. (A) Normal rates of import for MDH are shown in SS and IMF mitochondria (lanes 4 and 7) and the translation product of precursor MDH (lane 1). The lower band represents the imported mature MDH. Addition of valinomycin inhibits MDH protein import into the matrix of SS and IMF mitochondria, as this uncoupler dissipates the membrane potential (lanes 2 and 5). Triton-X is a detergent that solubilizes the inner membrane, thereby inhibiting MDH import into these subfractions (lanes 3 and 6). Protein import was carried out for increasing time durations, 4 min, 7 min, 10 min, 15 min, and 30 min. These data illustrate that import is a time-dependent process and also that SS and IMF mitochondria have different rates or capacities for import (B). TL, translation lane; VAL, valinomycin; TRI, Triton-X; CTL, control; SS, subsarcolemmal; IMF, intermyofibrillar. This figure was modified from Takahashi M & Hood DA20. Please click here to view a larger version of this figure.
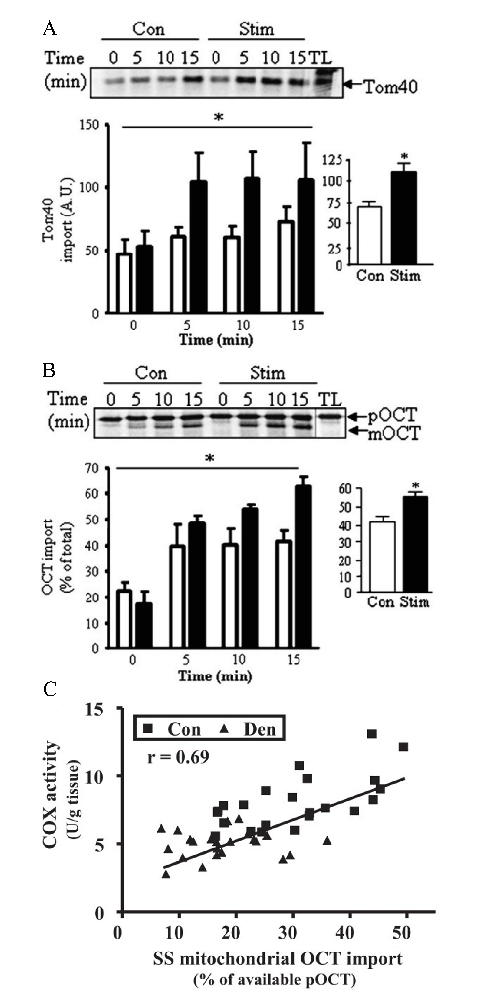
Figure 3: Protein import is adaptable and tightly linked to estimates of mitochondrial content. Sprague Dawley rats were subjected to electrical stimulation to induce contractile activity, a model of exercise training. (A) Tom40 import into the OM was 1.6-fold higher in muscle from chronically stimulated animals compared to controls at any given time point. (B) OCT import into the mitochondrial matrix was increased at every time point of incubation, and overall, this resulted in a 1.4-fold increase in mitochondria from chronically stimulated muscle. (C) Import is positively correlated with an index of mitochondrial content, as assessed by COX activity, r = 0.69. These measurements were taken from animals that were subjected to denervation, which has been shown to decrease mitochondrial content and the rate of import. Con, control; Den, denervated; Stim, stimulated; TL, translation lane; * p < 0.05. Figures 3A and 3B were adapted from Joseph A-M & Hood DA22 and 3C from Singh B & Hood DA23. Please click here to view a larger version of this figure.
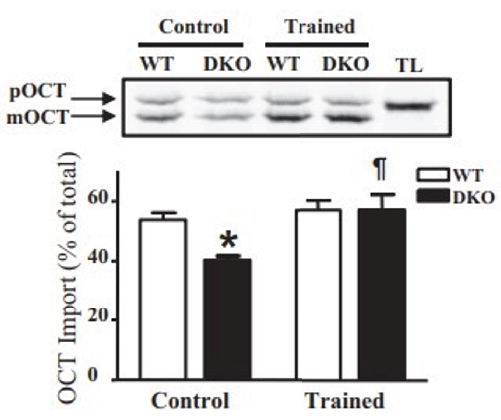
Figure 4: Training rescues import defects in vivo. Bax/Bak double knockout animals exhibit reduced protein import into the mitochondrial matrix by 37%. Six weeks of voluntary wheel running rescued the import defect to control levels. WT, wildtype; DKO, double knockout; TL, translation lane; * p < 0.05 main effect of genotype; ¶ p < 0.05 main effect of training. This figure was adapted from Zhang Y et al.24. Please click here to view a larger version of this figure.
| Buffer 1: | Buffer 1 + ATP: | Buffer 2: | Resuspension medium: | Nagarse protease |
| 100 mM KCl | 100 mM KCl | 100 mM KCl | 100 mM KCl | 10 mg/mL in Buffer 2 |
| 5 mM MgSO4 | 5 mM MgSO4 | 5 mM MgSO4 | 10 mM MOPS | NOTE: Make fresh and keep on ice |
| 5 mM EDTA | 5 mM EDTA | 5 mM EGTA | 0.2% BSA | |
| 50 mM Tris | 50 mM Tris | 50 mM Tris | ||
| 1 mM ATP | 1mM ATP |
Table 1: Buffers and resuspension media.
| % of the reaction volume | 1 Reaction mix | |
| Reticulocyte Lysate | 64.10% | 11.8 µL |
| Amino acids (-methionine) | 2.20% | 0.4 µL |
| Sterile H2O | 21.60% | 3.97 µL |
| 35S-methionine | 7.20% | 1.33 µL |
| mRNA | 5.40% | 1.0 µL |
| Total Volume | 18.5 µL | |
| NOTE: | ||
| 1) Add 35S-methionine last; | ||
| 2) Thaw the lysate slowly on ice, and limit freeze/thaw cycles to two. It is recommended that the lysate be aliquoted upon arrival | ||
| 3) The volume of mRNA can be adjusted to optimize translational efficiency by altering the sterile H2O volume accordingly. | ||
Table 2: Translation reaction mix
Discussion
Mitochondria are uniquely dependent on the expression and coordination of both the nuclear and mitochondrial genomes for their synthesis and expansion within cells. However, the nuclear genome encodes the vast majority (99%) of the mitochondrial proteome, and this underscores the importance of the protein import machinery in supporting mitochondrial biogenesis. Import also serves as an important signaling event, as failure to import can promote the initiation of the unfolded protein response and/or mitophagy15,16,26. Since import relies on the expression of multiple functional channels and chaperones, an intact membrane potential, as well as the availability of ATP, the evaluation of mitochondrial protein import can provide valuable insight into the health and energy status of the organelle.
The evaluation of protein import capacity into mitochondria requires the isolation of the organelle from the surrounding tissue using standard differential centrifugation techniques. Import is evaluated in the same manner as one would evaluate the Vmax of mitochondrial enzyme activities, in a highly controlled manner dependent on time and substrate concentration and independent of cellular cytosolic influences. The isolation method presented here, or similar renditions of it, have been used for many years to evaluate mitochondrial respiration and enzyme activities8,9,10,11. This technique is not without its limitations, as it disrupts the normal morphology of mitochondria as they exist in situ27, which in skeletal muscle is highly complex, ranging from small spheroid structures, to a highly branched reticular network3, depending on their subsarcolemmal or intermyofibrillar location within the muscle cell. In addition, the use of the protease nagarse has come under scrutiny3,28. An update to this method, using trypsin instead of nagarse, has recently been published25, and trypsin has been used by others to isolate muscle mitochondria29. Indeed, any alternative isolation method that yields functionally intact mitochondria can be used, including techniques that do not employ proteases30 or those designed for small biopsy-sized samples from human muscle31. The method presented here has the advantage of isolating SS mitochondria quickly and easily, but a greater yield and purity of mitochondria can be obtained through the isolation of the IMF subfraction. If done carefully, this isolation protocol can result in functionally intact organelles with a high respiratory control ratio, indicative of appropriate rates of state 3 and 4 respiration8.
It is also imperative for the measurement of mitochondrial protein import that these organelles maintain their membrane potential, as import across the inner membrane is dependent on this electrophoretic proton motive force, which serves to attract positively charged precursor sequences and help mediate the transport of the precursor into the negatively charged matrix space. Under such conditions, comparisons of mitochondrial function and import can be evaluated between physiologically relevant experimental situations, such as exercise, aging, and muscle disuse, for example. In this respect, previous work has shown that import is a highly adaptable pathway that responds to altered states of muscle use and disuse and is sensitive to inhibition via excessive ROS emission10,22,23,32. This plasticity is due, in part, to adaptive changes in the expression of the protein import machinery components. Since this technique relies on the isolation of mitochondria, any regulation on the import pathway that may stem from cytosolic factors or inter-organelle crosstalk is removed from the interpretation of the experiment. This is both a limitation and a strength of the technique, as conclusions can be made of the capacity of the import pathway itself (similar to the Vmax of enzyme activity), but transient or external signals that may occur with the experimental model may be lost. To circumvent this, it is possible to incubate the import reaction with a sample of the cytosolic fraction, isolated as described above, in order to evaluate changes in the cytosolic environment that may influence import rates, as done previously22. In addition, import machinery components are subject to acute, post-translational modifications that can alter their functions. Recent work has shown that phosphorylation of specific TOM import receptors can be linked to mitophagy33. Indeed, an area of research that warrants more attention is the acute modulation of the import process via intrinsic signaling pathways mediated by ROS or by covalent modification of import receptors and chaperones, as documented in yeast and other lower organisms34,35.
Protein import into mitochondria represents a gateway to adaptive organelle growth and is a sensitive indicator of mitochondrial health. Understanding how this process is regulated can shed light on the regulation of mitochondrial biogenesis, UPR signaling, and the initiation of mitophagy. Mitochondrial protein import is not a widely studied process in mammalian experimental models, and the development of the vast potential of research in this area could help us achieve a greater understanding of diseases in which mitochondrial dysfunction is apparent or represent an attractive therapeutic target to promote mitochondrial health.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Dr. G.C. Shore of McGill University, Dr. A. Strauss of the Washington School of Medicine, and Dr. M.T. Ryan of La Trobe University for the original donations of expression plasmids that were used for this research. This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC) to D. A. Hood. D. A. Hood is also the holder of a Canada Research Chair in Cell Physiology.
Materials
| 0.2% BSA | Sigma | A2153 | |
| 35S-methionine | Perkin Elmer | NEG709A500UC | Purchase requires a valid radioisotope permit |
| ATP | Sigma | A7699 | |
| Blotting paper; Whatman 3MM CHR Paper | Thermo Fisher | 05-714-5 | |
| Cassette for film | Kodak | Kodak Xomatic | |
| Centrifugation Tube | Thermo Fisher | 3138-0050 | |
| Chloroform | Thermo Fisher | C298-4 | |
| DTT | Sigma | D9779-5G | |
| EDTA | BioShop | EDT002 | |
| EGTA | Sigma | E4378 | |
| Gel Dryer | BioRad | Model 583 | |
| Gel Drying Kit | Sigma or BioRad | Z377570-1PAK or OW-GDF-10 | Various options are commercially available through many companies, these are just as few examples. |
| Glycerol | Caledon Laboratory Chemicals | 5350-1-40 | |
| HEPES | Sigma | H3375 | |
| High Speed Centrifuge | Beckman Coulter | Avanti J-25 Centrifuge | |
| Homogenizer | IKA | T25 Digital Ultra Turrex | |
| Isoamylalcohol, or 3-methylbutanol | Sigma | I9392 | |
| KAc | BioShop | POA301.500 | |
| KCl | Sigma | P3911 | |
| M7G | New England Biolab | S1404S | Dilute with 1000ul 20mM HEPES to make 1mM stock |
| MgCl | BioShop | MAG510 | |
| MgSO4 | Thermo Fisher | M65-500 | |
| MOPS | BioShop | MOP001 | |
| NaCl | BioShop | SOD001 | |
| NTP | Thermo Fisher | R0191 | |
| OCT Plasmid | – | – | Donated from Dr. G. C. Shore, McGill University, Montreal, Canada; alternative available through Addgene, plasmid #71877 |
| pGEM4Z/hTom40 Plasmid | – | – | Donated from Dr. M. T. Ryan, La Trobe University, Melbourne, Australia |
| pGMDH Plasmid | – | – | Donated from Dr. A. Strauss, Washington University School of Medicine |
| Phenol | Sigma | P4557 | |
| Phenol:Chloroform:Isoamyalcohol | Sigma | P3803 | Can also be made with the ratio provided |
| Phosphorus Film | Fujifilm | BAS-IP MS 2025 | |
| Rabbit reticulocyte lysate | Promega | L4960 | Avoid freeze-thaw; aliquot lysate upon arrival; amino acids are provided in the kit as well |
| RNAsin | Promega | N2311 | |
| Rotor for High Speed Centrifuge | Beckman Coulter | JA-25.50 | |
| SDS | BioShop | SDS001.500 | Caution: harmful if ingested or inhaled, wear a mask. |
| Sodium acetate | Bioshop | SAA 304 | |
| Sodium Carbonate | VWR | BDH9284 | |
| Sodium salicylate | Millipore Sigma | 106601 | |
| Sorbitol | Sigma | S6021 | |
| SP6 RNA Polymerase | Promega | P1085 | |
| Spectrophotometer | Thermo Fisher | Nanodrop 2000 | |
| Spermidine | Sigma | S-2626 | |
| Sucrose | BioShop | SUC507 | |
| T7 RNA Polymerase | Promega | P2075 | |
| Tabletop Centrifuge | Thermo Fisher | AccuSpin Micro 17 | |
| Trichloroacetic acid | Thermo Fisher | A322-500 | |
| Tris | BioShop | TRS001 | |
| β-mercaptoethanol | Sigma | M6250-100ML |
References
- Kirkwood, S. P., Munn, E. A., Brooks, G. A. Mitochondrial reticulum in limb skeletal muscle. The American Journal of Physiology. 251 (3), 395-402 (1986).
- Glancy, B., et al. Power grid protection of the muscle mitochondrial reticulum. Cell Reports. 19 (3), 487-496 (2017).
- Vincent, A. E., et al. Quantitative 3D mapping of the human skeletal muscle mitochondrial network. Cell Reports. 26 (4), 996-1009 (2019).
- Ogata, T., Yamasaki, Y. Ultra-high-resolution scanning electron microscopy of mitochondria and sarcoplasmic reticulum arrangement in human red, white, and intermediate muscle fibers. Anatomical Record. 248 (2), 214-223 (1997).
- Hood, D. A., Tryon, L. D., Carter, H. N., Kim, Y., Chen, C. C. W. Unravelling the mechanisms regulating muscle mitochondrial biogenesis. Biochemical Journal. 473, 2295-2314 (2016).
- Perry, C. G. R., Kane, D. A., Lanza, I. R., Neufer, P. D. Methods for assessing mitochondrial function in diabetes. Diabetes. 62, 1032-1036 (2013).
- Holloszy, J. O. Biochemical adaptations in muscle. The Journal of Biological Chemistry. 242 (9), 2278-2282 (1967).
- Cogswell, A. M., Stevens, R. J., Hood, D. A. Properties of skeletal muscle mitochondria from subsarcolemmal and intermyofibrillar isolated regions. The American Journal of Physiology. 264, 383-389 (1993).
- Koves, T. R., Noland, R. C., Bates, A. L., Henes, S. T., Muoio, D. M., Cortright, R. N. Subsarcolemmal and intermyofibrillar mitochondria play distinct roles in regulating skeletal muscle fatty acid metabolism. American Journal of Physiology – Cell Physiology. 288, 1074-1082 (2005).
- Bizeau, M. E., Willis, W. T., Hazel, J. R. Differential responses to endurance training in subsarcolemmal and intermyofibrillar mitochondria. Journal of Applied Physiology. 85 (4), 1279-1284 (1998).
- Krieger, D. A., Tate, C. A., McMillin-Wood, J., Booth, F. W. Populations of rat skeletal muscle mitochondria after exercise and immobilization. Journal of Applied Physiology: Respiratory, Environmental and Exercise Physiology. 48 (1), 23-28 (1980).
- Calvo, S. E., Clauser, K. R., Mootha, V. K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Research. 44 (1), 1251-1257 (2016).
- Wiedemann, N., Pfanner, N. Mitochondrial machineries for protein import and assembly. Annual Review of Biochemistry. 86 (1), 685-714 (2017).
- Backes, S., Herrmann, J. M. Protein translocation into the intermembrane space and matrix of mitochondria: mechanisms and driving forces. Frontiers in Molecular Biosciences. 4, 83 (2017).
- Harbauer, A. B., Zahedi, R. P., Sickmann, A., Pfanner, N., Meisinger, C. The protein import machinery of mitochondria – A regulatory hub in metabolism, stress, and disease. Cell Metabolism. 19 (3), 357-372 (2014).
- Jin, S. M., Lazarou, M., Wang, C., Kane, L. A., Narendra, D. P., Youle, R. J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. The Journal of Cell Biology. 191 (5), 933-942 (2010).
- Matsuda, N., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of Cell Biology. 189 (2), 211-221 (2010).
- Fiorese, C. J., Schulz, A. M., Lin, Y. -. F., Rosin, N., Pellegrino, M. W., Haynes, C. M. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Current biology. 26 (15), 2037-2043 (2016).
- Quiros, P. M., et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of Cell Biology. 216 (7), 2027-2045 (2017).
- Takahashi, M., Hood, D. A. Protein import into subsarcolemmal and intermyofibrillar skeletal muscle mitochondria. Differential import regulation in distinct subcellular regions. The Journal of Biological Chemistry. 271 (44), 27285-27291 (1996).
- Hood, D. A., Memme, J. M., Oliveira, A. N., Triolo, M. Maintenance of skeletal muscle mitochondria in health, exercise, and aging. Annual Review of Physiology. 81, (2019).
- Joseph, A., Hood, D. A. Mitochondrion plasticity of TOM complex assembly in skeletal muscle mitochondria in response to chronic contractile activity. Mitochondrion. 12 (2), 305-312 (2012).
- Singh, K., Hood, D. A. Effect of denervation-induced muscle disuse on mitochondrial protein import. American Journal of Physiology-Cell Physiology. 300 (1), 138-145 (2011).
- Zhang, Y., et al. Altered mitochondrial morphology and defective protein import reveal novel roles for Bax and/or Bak in skeletal muscle. American Journal of Physiology. Cell Physiology. 305 (5), 502-511 (2013).
- Lai, N., Kummitha, C., Rosca, M., Fujioka, H., Tandler, B., Hoppel, C. Isolation of mitochondrial subpopulations from skeletal muscle: optimizing recovery and preserving integrity. Acta Physiologica. 25 (2), 13182 (2019).
- Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M., Haynes, C. M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 337 (6094), 587-590 (2012).
- Picard, M., Taivassalo, T., Gouspillou, G., Hepple, R. T. Mitochondria: Isolation, structure and function. Journal of Physiology. 589 (18), 4413-4421 (2011).
- Kras, K. A., Willis, W. T., Barker, N., Czyzyk, T., Langlais, P. R., Katsanos, C. S. Subsarcolemmal mitochondria isolated with the proteolytic enzyme nagarse exhibit greater protein specific activities and functional coupling. Biochemistry and Biophysics Reports. 6, 101-107 (2016).
- Sánchez-Duarte, E., et al. Nicorandil affects mitochondrial respiratory chain function by increasing complex III activity and ROS production in skeletal muscle mitochondria. Journal of Membrane Biology. 253 (4), 309-318 (2020).
- Iñigo, M. R., et al. Estrogen receptor-α in female skeletal muscle is not required for regulation of muscle insulin sensitivity and mitochondrial regulation. Molecular Metabolism. 34 (2020), 1-15 (2020).
- Newsom, S. A., Stierwalt, H. D., Ehrlicher, S. E., Robinson, M. M. Substrate-specific respiration of isolated skeletal muscle mitochondria after 1 h of moderate cycling in sedentary adults. Medicine and Science in Sports and Exercise. 53 (7), 1375-1384 (2021).
- Takahashi, M., Chesley, A., Freyssenet, D., Hood, D. A. Contractile activity-induced adaptations in the mitochondrial protein import system. The American Journal of Physiology. 274 (5), 1380-1387 (1998).
- Kravic, B., et al. In mammalian skeletal muscle, phosphorylation of TOMM22 by protein kinase CSNK2/CK2 controls mitophagy. Autophagy. 8627, 01-65 (2017).
- Opalińska, M., Meisinger, C. Metabolic control via the mitochondrial protein import machinery. Current Opinion in Cell Biology. 33, 42-48 (2015).
- Gerbeth, C., et al. Glucose-induced regulation of protein import receptor tom22 by cytosolic and mitochondria-bound kinases. Cell Metabolism. 18 (4), 578-587 (2013).

