Efficient and Cost Effective Electroporation Method to Study Primary Cilium-Dependent Signaling Pathways in the Granule Cell Precursor
Summary
Here, we present a reproducible in vitro electroporation protocol for genetic manipulation of primary cerebellar granule cell precursors (GCPs) that is cost-effective, efficient, and viable. Moreover, this protocol also demonstrates a straightforward method for the molecular study of primary cilium-dependent Hedgehog signaling pathways in primary GCP cells.
Abstract
The primary cilium is a critical signaling organelle found on nearly every cell that transduces Hedgehog (Hh) signaling stimuli from the cell surface. In the granule cell precursor (GCP), the primary cilium acts as a pivotal signaling center that orchestrates precursor cell proliferation by modulating the Hh signaling pathway. The investigation of primary cilium-dependent Hh signaling machinery is facilitated by in vitro genetic manipulation of the pathway components to visualize their dynamic localization to the primary cilium. However, transfection of transgenes in the primary cultures of GCPs using the currently known electroporation methods is generally costly and often results in low cell viability and undesirable transfection efficiency. This paper introduces an efficient, cost-effective, and simple electroporation protocol that demonstrates a high transfection efficiency of ~80-90% and optimal cell viability. This is a simple, reproducible, and efficient genetic modification method that is applicable to the study of the primary cilium-dependent Hedgehog signaling pathway in primary GCP cultures.
Introduction
Cerebellar GCPs are widely used to study the machinery of the Hh signaling pathway in neuronal progenitor cell-types owing to their high abundance and high sensitivity to the Hh signaling pathway in vivo1,2,3,4. In GCPs, the primary cilium acts as a pivotal Hh signal transduction hub5 that orchestrates the proliferation of the precursor cells6,7,8. In vitro visualization of Hh signaling components on the primary cilium is often challenging due to their low endogenous basal levels. Hence, transgene modification of protein expression levels and fluorophore tagging of the gene of interest are useful approaches to study the pathway at molecular resolution. However, genetic manipulation of GCP primary cultures using liposome-based transfection approaches often result in low transfection efficiency, hindering further molecular investigations9. Electroporation increases the efficiency but commonly requires exorbitant vendor-specific and cell type-restricted electroporation reagents10.
This paper introduces a high-efficiency and cost-effective electroporation method to manipulate the Hh signaling pathway components in GCP primary cultures. Using this modified electroporation protocol, a green fluorescent protein (GFP)-tagged Smoothened transgene (pEGFP-Smo) was efficiently delivered to GCPs and achieved high cell survival and transfection rates (80-90%). Furthermore, as evidenced by the immunocytochemical staining, the transfected GCPs showed high sensitivity to Smoothened agonist-induced activation of the Hh signaling pathway by trafficking EGFP-Smo to the primary cilia. This protocol shall be directly applicable and beneficial for experiments that involve in vitro genetic modification of cell types that are difficult to transfect, such as human and rodent primary cell cultures, as well as human induced pluripotent stem cells.
Protocol
All animal-related procedures were carried out in compliance with animal handling guidelines and the protocol approved by the Department of Health, Hong Kong. Animal experiment licenses following Animal (Control of Experiments) Ordinance (Cap. 340) were obtained from the Department of Health, Hong Kong Government. The animal work was carried out in compliance with the animal safety ethics approved by HKBU Research Office and Laboratory Safety Committee. Refer to the Table of Materials for details about all materials used in this protocol.
1. Preexperiment preparation
- Preparation of culture media and buffers
- Serum-free medium (SFM)
- To prepare 50 mL of SFM, add 500 µL of 100x L-glutamine substitute, 500 µL of Penicillin-Streptomycin, 1 mM of sodium pyruvate, and 12.5 µL of 1 M KCl (final 250 µM) to 49 mL of Neurobasal medium.
- Split the SFM into 2 aliquots of 10 mL and 40 mL in 50 mL conical tubes and store them at 4 °C for up to one month.
- Digestion blocking medium: 10% FBS in SFM
- Add 1.1 mL of heat-inactivated FBS to a 10 mL aliquot of SFM to prepare digestion blocking medium.
- GCP culture medium: SFM with B27
- To prepare GCP culture medium, add 800 µL of serum-free B-27 supplement to a 40 mL aliquot of SFM.
NOTE: B-27-supplemented SFM can be stored at 4 °C for up to one month. However, freshly prepared media produce optimal outcomes.
- To prepare GCP culture medium, add 800 µL of serum-free B-27 supplement to a 40 mL aliquot of SFM.
- Dissection buffer: EBSS with glucose + HEPES
- Add 6 g/L of glucose to calcium- and magnesium-free Earle's Balanced Salt Solution (EBSS) containing 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid).
- Sterilize the solution by passing through a 0.2 µm syringe filter and store it at 4 °C for long-term storage.
- Digestion Buffer
NOTE: Prepare freshly before use.- Prepare 2 mL of digestion buffer for the digestion of 2-4 cerebellar tissues and 4 mL for 4-10 tissues. To prepare 4 mL of digestion buffer, dissolve 1.5 mg of L-cysteine (final concentration 200-400 µg/mL) in 4 mL of EBSS. Invert the tube repeatedly until the powder is fully dissolved.
- Sterilize the solution using a 0.2 µm syringe filter and a 5 mL syringe, and transfer the solution to a sterile 35 mm cell culture dish.
- Add 4 µL of papain (1:1,000 dilution from a stock concentration of 20 units/mg) and 40 µL of DNase I (1:100 dilution from a 10 mg/mL stock for a final concentration of 0.1 mg/mL).
- Incubate the solution at 37 °C in a CO2 incubator for at least 30 min or until use.
NOTE: This step is critical to activate the papain. For optimal outcomes, do not exceed 45 min at 37 °C before use. Ensure the solution turns transparent and there are no white precipitates before use.
- Serum-free medium (SFM)
- Precoating coverslips
- To prepare the coverslips for cell attachment, incubate autoclaved 12 mm glass coverslips with 100 µg/mL of poly-D-lysine (PDL, 1 mg/mL in sterile dH2O) for at least 1 h at 37 °C. Keep the coverslips in the same PDL solution at 4 °C until use.
- On the day of primary cell culture, collect the PDL in a clean conical tube and rinse the coverslips three times with sterile dH2O thoroughly to remove residual PDL.
NOTE: The PDL can be stored at 4 °C for reuse. - Transfer the PDL-coated glass coverslips onto a 24-well plate by placing one coverslip in each well.
- Remove excess water and add 200-300 µL of Matrigel (reconstituted according to the instructions in the product datasheet in serum-free DMEM-F12) to fully cover the coverslips.
- Incubate the coverslips in the Matrigel for 1 h at 37 °C in the CO2 incubator.
NOTE: Remove the Matrigel before cell seeding. This Matrigel can be collected and stored at 4 °C for multiple reuses.
- Preparation of preplating culture dish
- Coat a 60 mm cell culture dish with 2 mL of 100 µg/mL PDL (reconstitute the 1 mg/mL PDL stock solution in sterile dH2O) by incubation at 37 °C for 1 h.
- Immediately before cell seeding, remove and collect the used PDL in a clean conical tube and store it at 4 °C for multiple reuses. Rinse and wash the PDL-coated dish three times with sterile dH2O. Air-dry the culture dish before cell seeding.
- Use one 60 mm cell culture dish to seed cells harvested from a maximum of 2 whole-cerebellum tissues.
- Prepare additional culture dishes for additional cerebellums.
NOTE: See steps 2.1.18 and 2.1.19 for the use of the preplating culture dish.
2. Experimental day 0
- Isolation and culturing of mouse primary GCPs
NOTE: The GCP culture method was modified from a standard protocol, and the brief protocol was briefly described in our previous work6,11,12. For optimal GCP yield, use postnatal (P) day 6 or P7 pups for the isolation of GCP. Complete the following dissection steps 2.1.6-2.1.10 as quickly as possible for optimal cell viability.- Prewarm SFM, digestion blocking medium, culture medium, and Opti-MEM at 37 °C during dissection.
- On a clean bench, presoak all the dissection apparatus in 70% ethanol for disinfection.
- Fill a 60 mm cell culture dish with 2-3 mL of dissection buffer and chill it on ice.
- Prepare fresh digestion buffer as described in section 1.1.5 and keep it warm at 37 °C.
- Use 70% ethanol to wipe the head of the pup for disinfection.
- Decapitate the pup without anesthesia. Use forceps to hold the head and sterilized surgical scissors to cut from the back of the skull to decapitate the pup. Carefully remove the skin and skull to uncover the brain by using forceps.
- Use forceps to pinch off the cerebellum and quickly soak it in the pre-chilled dissection buffer prepared in step 2.1.3.
- Remove the meninges with the cerebellum submerged in the dissection buffer under a dissecting microscope. Blood vessel-enriched meninges should appear pinkish under the dissecting microscope.
- Remove all the meninges using forceps.
NOTE: A cerebellum without meninges should have a whitish appearance. - Remove any visible midbrain tissues and the choroid plexus from the cerebellum.
- Transfer the cerebellum to a 35 mm culture dish prefilled with warm digestion buffer prepared in step 2.1.4. Avoid transferring excess dissection buffer. Cut the cerebellum into fine pieces using microspring scissors as quickly as possible.
- Immediately incubate the minced cerebellum at 37 °C for 15 min in a CO2 incubator. Extend the incubation time to 20 min if more than 4 cerebellums are to be processed.
NOTE: After incubation, the tissue will clump together. - Immediately transfer the digested tissue to the bottom of a new sterile 15 mL centrifuge tube using a P1000 pipette tip prewet with digestion blocking medium (avoid transferring digestion buffer).
- Add an appropriate volume of digestion blocking medium (1 mL for 1 cerebellum) to terminate the digestion and pipette up and down gently 30 times using a P1000 micropipette to further dissociate the tissue into a single-cell suspension. Avoid air bubble formation.
- Gently pass the cell suspension through a 70 µm cell strainer into a new sterile centrifuge tube to remove cell clumps.
- Pass an additional 1 mL of fresh digestion blocking medium through the cell strainer to collect residual cells from the cell strainer.
- Centrifuge the filtrate at 200 × g for 5 min at room temperature (RT). Remove the supernatant and resuspend the pellet with 1 mL of SFM. Repeat this step twice. Avoid forming air bubbles.
- Resuspend the pellet in a final volume of 2 mL of GCP culture medium and transfer the cell suspension to the PDL-coated 60 mm preplating culture dish prepared in step 1.3. Incubate for 15 min at 37 °C in a CO2 incubator.
NOTE: Do not exceed 20 min. - After incubation, tap the culture dish from the side to dislodge the loosely adherent GCP cells. Collect the adherent GCP cells in culture medium by gentle pipetting using a P1000 pipette. Collect this GCP suspension into a new 15 mL centrifuge tube. Discard the 60 mm dish.
NOTE: Strongly adherent astroglia and fibroblast cells will remain attached on the 60 mm dish bottom and will be separated in this step. Omitting this step will compromise the purity of the GCP culture. - Count the cells and proceed to electroporation immediately.
- Day in vitro (DIV) 0: Electroporation for Hh receptor transgene overexpression: pEGFP-Smo
- For electroporation using a 2 mm gap cuvette, prepare the following plasmid-cell electroporation mixture for each reaction of electroporation: 1.2 × 106 cells and 10 µg of pEGFP-mSmo (adjust the DNA stock concentration to ~2-5 µg/µL in Tris-EDTA buffer or sterile dH2O) in 100 µL of Opti-MEM.
NOTE: Reduce the total cell number if the cells are insufficient (though this may reduce the electroporation efficiency). However, do not adjust the amount of plasmid nor the total volume of Opti-MEM used per cuvette. If the total cell number required for the experiment exceeds 1.5 × 106 cells, scale up the electroporation mixture accordingly and perform multiple electroporation reactions separately. The cuvette may be reused up to 5 times. - Prepare the post-electroporation cell seeding plate by adding 0.5 mL of culture medium into each well of the 24-well culture plate containing coated coverslips (from step 1.2) and keep it warm at 37 °C in a CO2 incubator.
- From step 2.1.20, pipette the required number of cells into a sterile 1.5 mL microcentrifuge tube and spin at 200 × g for 5 min at RT. Discard the supernatant and resuspend the cell pellet in 200 µL of Opti-MEM. Repeat this step twice with Opti-MEM to ensure no residual culture medium is present in the tube
NOTE: For every well/coverslip of a 24-well plate, electroporate and seed 1.2-1.3 × 106 cells/well to obtain ~70-75% cell confluency on the next day of culture. - Set the parameters of electroporation as shown in Table 1.
- Pipette the electroporation reaction gently to mix well and use a long P200 tip to transfer an exact volume of 100 µL of the mixture into the 2 mm gap cuvette. Avoid forming bubbles.
- Place the cuvette in the cuvette chamber.
- Press the Ω button of the electroporator (see the Table of Materials) and make a note of the impedance value, which should be ~30-35. To ensure the electric impedance value Ω falls within the range of 30-35, adhere to a precise volume of 100 µL.
- Press the start button to initiate the pulse.
- Record the values of the measured current and joules shown on the reading frame.
- Remove the cuvette from the cuvette chamber.
- Immediately add 100 µL of prewarmed culture medium into the cuvette and resuspend it by gentle pipetting up and down 2-3 times. Immediately transfer the cell suspension to the 24-well plate prepared in step 2.2.2.
NOTE: To minimize cell death, seed the cells immediately after electroporation. - Incubate the cells at 37 °C in a CO2 incubator. Leave the cells undisturbed for 3 h to avoid cell detachment.
- At 3 h post seeding, gently aspirate and discard half of the supernatant medium to remove floating dead cells and debris and replace with the same amount of prewarmed culture medium.
- Incubate the cells at 37 °C in the CO2 incubator and replenish half of the medium every other day.
- Observe the cells under the fluorescence microscope on the next day, i.e., DIV1 (Figure 1) for GFP signal to determine the efficiency of Smo overexpression.
- For the stimulation of the Hh signaling pathway on DIV 1 after replenishing half of the medium, add 0.2 µM Smoothened agonist (SAG) to the cells while adding an equal volume of DMSO to the control. Incubate for 24 h prior to cell fixation.
NOTE: Keep the volume of DMSO/SAG added as low as possible. Here, 0.5 µL DMSO or 0.5 µL of 0.2 mM SAG was added per 500 µL of medium per well, which is a dilution ratio of 1:1,000.
- For electroporation using a 2 mm gap cuvette, prepare the following plasmid-cell electroporation mixture for each reaction of electroporation: 1.2 × 106 cells and 10 µg of pEGFP-mSmo (adjust the DNA stock concentration to ~2-5 µg/µL in Tris-EDTA buffer or sterile dH2O) in 100 µL of Opti-MEM.
3. DIV 2: Visualization of primary cilia and investigation of the Hh signaling pathway
- Cell fixation
- At 24 h post treatment, remove the culture medium and rinse the cells 2-3 times with phosphate-buffered saline (PBS) using a disposable Pasteur pipette while avoid dislodging the cells.
- Fix the cells by adding ~400 µL of 4% paraformaldehyde (prepared in PBS) and incubate at RT for 10 min.
- Rinse and wash 2-3 times with PBS using a disposable Pasteur pipette.
- Store in PBS at 4 °C for up to 2 months or proceed to immunostaining.
- Immunocytochemical staining of GCPs and primary cilium marker
- In a 24-well plate, washthe cells twice in PBS with gentle shaking for 5 min each time.
- Add 0.5 mL of 100 mM ammonium chloride to the cell and incubate at RT for 10 min to quench the fixative.
- Rinse once and wash 2-3 times with PBS with gentle shaking for 10 min each time.
- Gently pipette 30 µL of blocking buffer (BB: 0.1% Triton X-100, 1% BSA, 2% heat-inactivated horse serum in PBS) onto a piece of parafilm to form a droplet without air bubbles.
- Gently transfer the coverslip from the well onto the BB droplet with the cell-seeded side facing downwards. Incubate the cells with BB in a humid chamber for 1 h at RT.
- Prepare the primary antibody mix in BB as shown in the Table of Materials. To probe the cells with primary antibody, repeat steps 3.2.4 and 3.2.5, replacing BB with the primary antibody mix. Incubate the cells with the primary antibody for 2 h at RT.
- Using forceps, transfer the coverslips back to the 24-well plate with the cell-seeded side facing upwards. Rinse the cells once and wash 3-4 times in PBS with gentle shaking for 10 min each time.
- Prepare the secondary antibody mix in BB as shown in the Table of Materials. To incubate the cells with the secondary antibody, repeat steps 3.2.4 and 3.2.5, replacing BB with the secondary antibody mix. Incubate in the dark for 1 h at RT.
- Repeat step 3.2.7 for post-secondary antibody incubation washing.
- Mount the coverslip on a clean microscopic glass slide using mounting medium.
- Air-dry the slide in the dark overnight and proceed to confocal imaging13.
Representative Results
Using the Opti-MEM (see the Table of Materials) as the universal reagent, this proposed electroporation methodology could achieve consistently high electroporation efficiency at ~80-90% (Figure 1). The electroporation efficiency of the Smo-EGFP vector was determined at DIV 2 post electroporation by quantification of the percentage of green fluorescence-positive cells in all paired box protein-6 (Pax6)-expressing GCP cells. The electroporation efficiency of DMSO- and SAG-treated groups appeared comparable (Figure 1 and Table 2).
In addition, immunostaining of the primary cilium marker, Arl13b, demonstrates that the ciliation rate of GCP at DIV 2 of culture was ~18% in both the vehicle- and SAG-treated groups (DMSO: 17.35% ± 0.59%; SAG: 18.24% ± 0.88%). The ciliation rate is illustrated as the percentage of Pax6-expressing GCPs bearing a primary cilium (Arl13b-positive) on the cell surface at DIV 2 post electroporation (Figure 2 and Table 3).
To decipher the primary cilium-dependent Hh signaling pathway, an agonist of Smo, SAG, was used to activate the Hh signaling pathway. Upon Hh pathway activation, the Smo receptor is enriched at the axoneme of the primary cilium14. Our results show significantly increased Smo-EGFP localization on the primary cilium axoneme of Pax6-expressing GCP cells at 24 h post SAG treatment (Figure 3, quantification data modified from previous work6), indicating a profound activation of the primary cilium-dependent Hh signaling pathway.
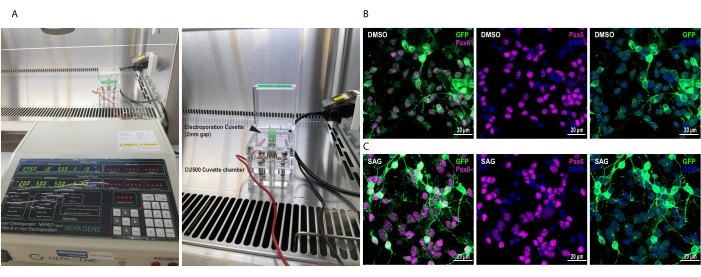
Figure 1: Electroporation setup and the electroporation efficiency of GCPs. (A) Electroporation setup. Right, black arrowhead denotes electroporation cuvette. (B, C) Representative images depict the electroporation efficiency of the Smo-EGFP vector determined by quantification of the percentage of GFP-positive cells in all Pax6-expressing GCP cells (Table 2). Representative images depict the green fluorescent signals on Pax6-expressing (violet) GCP cells on DIV 2 post electroporation after 24 h treatment with (B) DMSO and (C) SAG. Nuclei were labeled with DAPI (blue). Scale bars = 20 µm. Abbreviations: GCPs = granule cell precursors; GFP = green fluorescent protein; Pax6 = paired box protein-6; DIV = day in vitro; DMSO = dimethyl sulfoxide; SAG = Smoothened agonist; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
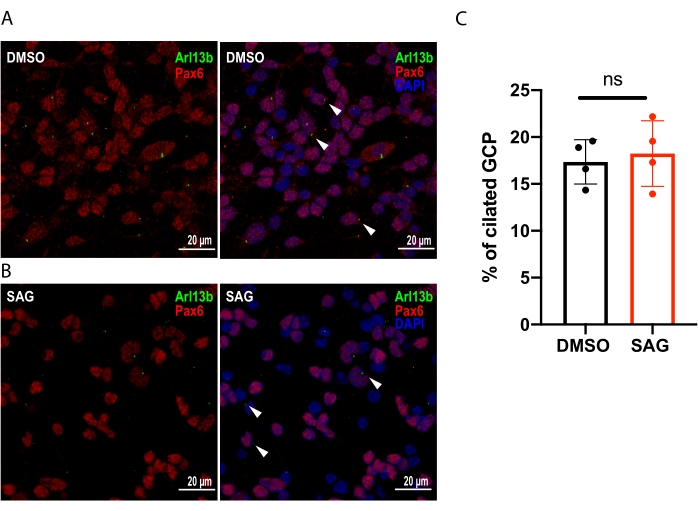
Figure 2: Percentage of ciliation on DIV 2 of GCP primary culture. (A, B) Representative images depict the percentage of ciliation on DIV 2 of GCP primary culture. Representative images depict the primary cilia (green) on Pax6-expressing (red) GCP cells on DIV 2 post electroporation after 24 h treatment with (A) DMSO and (B) SAG. Nuclei were labeled with DAPI (blue). The primary cilium (green) is denoted by white arrowheads. Scale bars = 20 µm. (C) Graph illustrates quantification data of 4 independent experiments. Statistical analysis, Unpaired Student's t-test. Error bars depict ±SEM. Abbreviations: GCP = granule cell precursor; Pax6 = paired box protein-6; DIV = day in vitro; DMSO = dimethyl sulfoxide; SAG = Smoothened agonist; DAPI = 4',6-diamidino-2-phenylindole; n.s. = Not significant; SEM = standard error of the mean; Arl13b = ADP ribosylation factor-like protein 13B. Please click here to view a larger version of this figure.
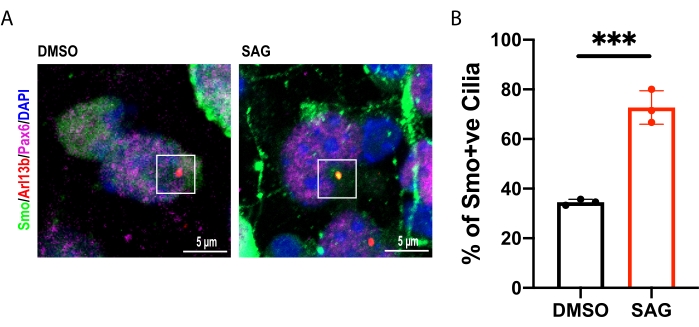
Figure 3: Increased Smo localization on the primary cilium of Pax6-expressing GCP cells upon SAG treatment. (A) Representative images depict the Smo-EGFP localization (green) on the primary cilium (red, white square box) on Pax6-expressing (violet) GCP cells at DIV 2 post electroporation after 24 h treatment with DMSO and SAG. Nuclei were labeled with DAPI (blue). Scale bars = 5 µm. (B) Graph illustrates quantification data of 4 independent experiments. Statistical analysis, unpaired Student's t-test. *** P ≤ 0.001. Error bars depict ± SEM. Total n for DMSO group = 97, total n for SAG group = 130. Figure 3B was modified from 6. Abbreviations: GCP = granule cell precursor; Smo = Smoothened; Pax6 = paired box protein-6; DIV = day in vitro; DMSO = dimethyl sulfoxide; SAG = Smoothened agonist; DAPI = 4',6-diamidino-2-phenylindole; SEM = standard error of the mean; Arl13b = ADP ribosylation factor-like protein 13B. Please click here to view a larger version of this figure.
| Poring Pulse Setting | Transfer Pulse Setting | |||
| Mouse Primary GCPs | Primary neurons | Mouse Primary GCPs | Primary neurons | |
| Voltage | 275 V | 275 V | 20 V | 20 V |
| Length | 1 ms | 0.5 ms | 50 ms | 50 ms |
| Interval | 50 ms | 50 ms | 50 ms | 50 ms |
| No. | 2 | 2 | 5 | 5 |
| D rate | 10% | 10% | 40% | 40% |
| Polarity | + | + | ± | ± |
Table 1: The electroporation parameters of mouse primary GCPs and primary neurons using Super Electroporator NEPA21 TYPE II. Abbreviation: GCP = granule cell precursor.
| Electroporation efficiency | Exp. 1 (n = 486) | Exp. 2 (n = 1314) | Exp. 3 (n = 704) | Exp. 4 (n = 476) | Average |
| DMSO-treated group | 90.57% ± 10.12% | 96.62% ± 3.09% | 98.89% ± 0.97% | 90.72% ± 11.31% | 94.02% ± 1.36% |
| SAG-treated group | 91.8% ± 8.69% | 79.97% ± 2.77% | 89.35% ± 5.67% | 88.59% ± 13.54% | 87.42% ± 1.71% |
| Average Electroporation efficiency | 91.31% ± 7.99% | 88.27% ± 10.81% | 94.12% ± 6.36% | 89.65% ± 11.21% | 90.84% ± 0.84% |
Table 2: Electroporation efficiency of Smo-EGFP vectordeterminedbyquantification of thepercentage of GFP-positive cells in all Pax6-expressing GCP cells. Data of four independent experiments (Exp.) are shown. (Total n = 2980). Abbreviations: Smo = Smoothened; GFP = green fluorescent protein; GCP = granule cell precursor; Pax6 = paired box protein-6.
| Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | average | |
| Ciliation rate – DMSO | 18.88% ± 3.61% | 19.58% ± 7.42% | 16.60% ± 1.48% | 14.35% ± 7.99% | 17.35% ± 0.59% |
| Ciliation rate – SAG | 13.93% ± 3.39% | 17.30% ± 2.15% | 22.19% ± 10.35% | 19.56% ± 1.15% | 18.24% ± 0.88% |
Table 3: The percentage of ciliation on DIV 2 of GCP primary culture. Data of four independent experiments (Exp.) are shown. (Total n for DMSO group = 1169, Total n for SAG group = 816). Abbreviations: GCP = granule cell precursor; DIV = day in vitro; DMSO = dimethyl sulfoxide; SAG = Smoothened agonist.
Discussion
Transfection of transgenes in primary GCP culture by electroporation method is typically associated with low cell viability and poor transfection efficiency9,10. This paper introduces a cost-effective and reproducible electroporation protocol that has demonstrated high efficiency and viability. In addition, we also demonstrate a straightforward method of studying the primary cilium-dependent Hh signaling pathway in primary GCP cells.
Other common electroporation methods often require costly cell-type-specific electroporation reagents that must be purchased from specific manufacturers. The method described here is deemed favorable as it uses a common and economical electroporation reagent for different cell types. Moreover, these data showed that the electroporation efficiency reached ~80-90%, which is highly efficient compared to other electroporation and transfection methods9,10.
To maintain higher cell viability, there are a few critical steps that one should take into consideration. The cerebellum dissection and dissociation procedures should be completed within the shortest possible time window of 1-2 h. Another critical step is to avoid bubble formation in the plasmid-cell electroporation mixture before pulses during electroporation. After pulses, prewarmed culture medium should be added immediately into the cuvettes and the cells seeded as quickly as possible. The cells must be undisturbed in the first 3 h post cell seeding. The aforementioned precautions will enhance cell viability up to approximately 70-80% on the second day of culture.
One notable limitation of studying the primary cilium in the primary culture platform is that the rate of ciliation in cultured cells is generally lower than that observed in vivo. Previous data 6 showed that the in vivo rate of ciliation on GCP at both E15.5 and P15 was approximately 60-80%. In contrast, the in vitro rate of ciliation in primary GCP culture was ~20%6. Nonetheless, this is a general phenomenon that is discernible across most (if not all) cell types when comparing the rate of ciliation between in vitro and in vivo studies.
Notably, this method is also applicable to other primary cultures such as neural progenitor cells and cortical and hippocampal neuron culture, which is achievable by modifying the electroporation parameter, i.e., poring pulse voltage, length, and number of pulses. To extend the application of this protocol to a broader field of study, the recommended electroporation parameters for primary neurons are provided in Table 1. In addition, the universal electroporation reagent, i.e., Opti-MEM used in this protocol also helps avoid additional tedious optimization effort compared to other electroporation protocols that require optimization with respect to reagent compatibility. This optimized, cost-effective electroporation protocol for the investigation of the primary cilium and Hh signaling in primary GCP cultures could be used as the reference procedure for other primary cilium-related studies using primary cultures.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This study was supported by HKBU Seed Fund and Tier-2 Start-up Grant (RG-SGT2/18-19/SCI/009), Research Grant Council-Collaborative Research Fund (CRF-C2103-20GF) to C.H.H. Hor.
Materials
| GCP Culture | |||
| B27 supplement | Life Technologies LTD | 17504044 | |
| Cell strainer, 70 µm | Corning | 352350 | |
| DNase I from bovine pancreas | Roche | 11284932001 | |
| Earle’s Balanced Salt Solution | Gibco, Life Technologies | 14155063 | |
| FBS, qualified | Thermo Scientific | SH30028.02 | |
| GlutamMAXTM-I ,100x | Gibco, Life Technologies | 35050061 | L-glutamine substitute |
| L-cysteine | Sigma Aldrich | C7352 | |
| Matrigel | BD Biosciences | 354277 | Basement membrane matrix |
| Neurobasal | Gibco, Life Technologies | 21103049 | |
| Papain,suspension | Worthington Biochemical Corporation | LS003126 | |
| Poly-D-lysine Hydrobromide | Sigma Aldrich | P6407 | |
| SAG | Cayman Chemical | 11914-1 | Smoothened agonist |
| IF staining | |||
| Bovine Serum Albumin | Sigma Aldrich | A7906 | |
| Paraformaldehyde | Sigma Aldrich | P6148 | |
| Triton X-100 | Sigma Aldrich | X100 | |
| Primary antibody mix | |||
| Anti-GFP-goat ab | Rockland | 600-101-215 | Dilution Factor: 1 : 1000 |
| Anti-Arl13b mouse monoclonal ab | NeuroMab | 75-287 | Dilution Factor: 1 : 1000 |
| Anti-Pax6 rabbit polyclonal ab | Covance | PRB-278P | Dilution Factor: 1 : 1000 |
| Secondary antibody mix | |||
| Alexa Fluor 488 donkey anti-goat IgG | Invitrogen | A-11055 | Dilution Factor: 1 : 1000 |
| Alexa Fluor 555 donkey anti-mouse IgG | Invitrogen | A-31570 | Dilution Factor: 1 : 1000 |
| Alexa Fluor 647 donkey anti-rabbit IgG | Invitrogen | A-31573 | Dilution Factor: 1 : 1000 |
| DAPI | Thermo Scientific | 62247 | Dilution Factor: 1 : 1000 |
| Electroporation | |||
| CU 500 cuvette chamber | Nepagene | CU500 | |
| EPA Electroporation cuvette (2 mm gap) | Nepagene | EC-002 | |
| Opti-MEM | Life Technologies LTD | 31985070 | reduced-serum medium for transfection |
| pEGFP-mSmo | Addgene | 25395 | |
| Super Electroporator NEPA21 TYPE II In Vitro and In Vivo Electroporation | Nepagene | NEPA21 | electroporator |
References
- Chang, C. H., et al. Atoh1 controls primary cilia formation to allow for SHH-triggered granule neuron progenitor proliferation. Developmental Cell. 48 (2), 184-199 (2019).
- Dahmane, N., Ruizi Altaba, A. Sonic Hedgehog regulates the growth and patterning of the cerebellum. Development. 126 (14), 3089-3100 (1999).
- Lewis, P. M., Gritti-Linde, A., Smeyne, R., Kottmann, A., Mcmahon, A. P. Sonic Hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Biologie du développement. 270 (2), 393-410 (2004).
- Wechsler-Reya, R. J., Scott, M. P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 22 (1), 103-114 (1999).
- Bangs, F., Anderson, K. V. Primary cilia and mammalian Hedgehog signaling. Cold Spring Harbor Perspectives in Biology. 9 (5), 028175 (2017).
- Hor, C. H. H., Lo, J. C. W., Cham, A. L. S., Leung, W. Y., Goh, E. L. K. Multifaceted functions of Rab23 on primary cilium- and Hedgehog signaling-mediated cerebellar granule cell proliferation. Journal of Neuroscience. 41 (32), 6850-6863 (2021).
- Han, Y. G., et al. Dual and opposing roles of primary cilia in medulloblastoma development. Nature Medicine. 15 (9), 1062-1065 (2009).
- Spassky, N., et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Biologie du développement. 317 (1), 246-259 (2008).
- Wang, T., Larcher, L., Ma, L., Veedu, R. N. Systematic screening of commonly used commercial transfection reagents towards efficient transfection of single-stranded oligonucleotides. Molecules. 23 (10), 2564 (2018).
- Chicaybam, L., et al. An efficient electroporation protocol for the genetic modification of mammalian cells. Frontiers in Bioengineering and Biotechnology. 4, 99 (2017).
- Lee, H. Y., Greene, L. A., Mason, C. A., Chiara Manzini, M. Isolation and culture of post-natal mouse cerebellar granule neuron progenitor cells and neurons. Journal of Visualized Experiments: JoVE. (23), e990 (2009).
- Mizoguchi, T., et al. Impaired cerebellar development in mice overexpressing VGF. Neurochemical Research. 44 (2), 374-387 (2019).
- Zhou, Y. Confocal imaging of nerve cells. Current Laboratory Methods in Neuroscience Research. , 235-247 (2014).
- Corbit, K. C., et al. Vertebrate Smoothened functions at the primary cilium. Nature. 437 (7061), 1018-1021 (2005).

