Direct Comparison of Hyperspectral Stimulated Raman Scattering and Coherent Anti-Stokes Raman Scattering Microscopy for Chemical Imaging
Summary
This paper directly compares the resolution, sensitivity, and imaging contrasts of stimulated Raman scattering (SRS) and coherent anti-Stokes Raman scattering (CARS) integrated into the same microscope platform. The results show that CARS has a better spatial resolution, SRS gives better contrasts and spectral resolution, and both methods have similar sensitivity.
Abstract
Stimulated Raman scattering (SRS) and coherent anti-Stokes Raman scattering (CARS) microscopy are the most widely used coherent Raman scattering imaging technologies. Hyperspectral SRS and CARS imaging offer Raman spectral information at every pixel, which enables better separation of different chemical compositions. Although both techniques require two excitation lasers, their signal detection schemes and spectral properties are quite different. The goal of this protocol is to perform both hyperspectral SRS and CARS imaging on a single platform and compare the two microscopy techniques for imaging different biological samples. The spectral focusing method is employed to acquire spectral information using femtosecond lasers. By using standard chemical samples, the sensitivity, spatial resolution, and spectral resolution of SRS and CARS in the same excitation conditions (i.e., power at the sample, pixel dwell time, objective lens, pulse energy) are compared. The imaging contrasts of CARS and SRS for biological samples are juxtaposed and compared. The direct comparison of CARS and SRS performances would allow for optimal selection of the modality for chemical imaging.
Introduction
The Raman scattering phenomenon was first observed in 1928 by C. V. Raman1. When an incident photon is interacting with a sample, an inelastic scattering event can spontaneously occur, in which the energy change of the photon matches a vibrational transition of the analyzed chemical species. This process does not require the use of a chemical tag, making it a versatile, label-free tool for chemical analysis while minimizing sample perturbation. Despite its advantages, spontaneous Raman scattering suffers from a low scattering cross-section (typically 1011 lower than the infrared [IR] absorption cross-section), which necessitates long acquisition times for analysis2. Thus, the quest for increasing the sensitivity of the Raman scattering process is essential in pushing Raman technologies for real-time imaging.
One effective way to greatly enhance the sensitivity of Raman scattering is through coherent Raman scattering (CRS) processes, for which two laser pulses are typically used to excite molecular vibrational transitions3,4. When the photon energy difference between the two lasers matches the vibrational modes of sample molecules, strong Raman signals will be generated. The two most commonly used CRS processes for imaging are coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS)5. Over the past two decades, technological developments have advanced CARS and SRS microscopy techniques to become powerful tools for label-free quantification and elucidation of chemical changes in biological samples.
Chemical imaging by CARS microscopy can be dated to 1982 when laser scanning was first applied to acquire CARS images, demonstrated by Duncan et al6. The modernization of CARS microscopy was greatly accelerated after the wide applications of laser scanning multiphoton fluorescence microscopy7. Early work from the Xie group using high repetition rate lasers has transitioned CARS to be a high-speed, label-free, chemical imaging platform for the characterization of molecules in biological samples8,9,10. One of the major issues for CARS imaging is the presence of a nonresonant background, which reduces the image contrast and distorts the Raman spectrum. Many efforts have been made to either reduce the nonresonant background11,12,13,14,15 or to extract resonant Raman signals from the CARS spectra16,17. Another advancement that has greatly advanced the field is hyperspectral CARS imaging, which allows for spectral mapping at each image pixel with improved chemical selectivity18,19,20,21.
Stimulated Raman scattering (SRS) is a younger imaging technology than CARS, although it was discovered earlier22. In 2007, SRS microscopy was reported using a low repetition rate laser source23. Soon, several groups demonstrated high-speed SRS imaging using high repetition rate lasers24,25,26. One of the major advantages of SRS microscopy over CARS is the absence of the nonresonant background27, although other backgrounds such as cross-phase modulation (XPM), transient absorption (TA), two-photon absorption (TPA), and photothermal (PT) effect, may occur with SRS28. In addition, the SRS signal and sample concentration have linear relationships, unlike CARS, which has a quadratic signal-concentration dependence29. This simplifies chemical quantification and spectral unmixing. Multicolor and hyperspectral SRS has evolved in different forms30,31,32,33,34,35,36, with spectral focusing being one of the most popular approaches for chemical imaging37,38.
Both CARS and SRS require the focusing of the pump and Stokes laser beams onto the sample to match the vibrational transition of the molecules for signal excitation. CARS and SRS microscopes also share a lot in common. However, the physics underlying these two processes, and signal detections involved in these microscopy technologies have disparities3,39. CARS is a parametric process that does not have net photon-molecule energy coupling3. SRS, however, is a nonparametric process, and contributes to energy transfer between photons and molecular systems27. In CARS, a new signal at anti-Stokes frequency is generated, while SRS manifests as the energy transfer between the pump and Stokes laser beams.
CARS signal satisfies Eq (1)28.
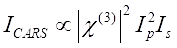 (1)
(1)
Meanwhile, SRS signal can be written as Eq (2)28.
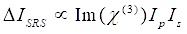 (2)
(2)
Here, Ip, Is, ICARS, and ΔISRS are the intensities of the pump beam, Stokes beam, CARS signal, and SRS signals, respectively. χ(3) is the third-order nonlinear optical susceptibility of the sample, and is a complex value composed of real and imaginary parts.
These equations express the spectral profiles and signal-concentration dependence of CARS and SRS. Differences in physics result in disparate detection schemes for these two microscopy technologies. Signal detection in CARS usually involves spectral separation of newly generated photons and detection using a photomultiplier tube (PMT) or charge-coupled device (CCD); for SRS, the energy exchange between the pump and Stokes beams is usually measured by high-speed intensity modulation using an optical modulator and demodulation using a photodiode (PD) paired with a lock-in amplifier.
Although many technological developments and applications have been published in recent years in both CARS and SRS fields, no systematic comparisons of the two CRS techniques have been performed on the same platform, especially for hyperspectral CARS and SRS microscopy. Direct comparisons in sensitivity, spatial resolution, spectral resolution, and chemical separation capabilities would allow biologists to select the best modality for chemical quantification. In this protocol, detailed steps to construct a multimodal imaging platform with both hyperspectral CARS and SRS modalities based on a femtosecond laser system and spectral focusing are provided. The two techniques have been compared in the forward direction for spectral resolution, detection sensitivity, spatial resolution, and imaging contrasts of cells.
Protocol
1. Instrumental setup for hyperspectral CRS imaging
NOTE: The generation of CRS signal requires the use of high-power (i.e., class 3B or class 4) lasers. Safety protocols must be addressed and proper personal protective equipment (PPE) must be worn at all times when working at such high peak powers. Consult proper documentation before experimentation. This protocol focuses on designing the beam path, chirping the femtosecond pulses, and optimizing imaging conditions. A general optical layout of this hyperspectral CRS microscope is shown in Figure 1. The configuration shown here is one of many existing configurations for CRS microscopy. The CRS microscopy system used in this protocol is built upon a dual-output femtosecond laser source and a laser scanning microscope.
- Ensure that the laser source provides two femtosecond pulse trains (120 fs width) with a repetition rate of 80 MHz, including a fixed wavelength at 1,045 nm used as the Stokes beam, and a tunable wavelength from 680 to 1,300 nm used as the pump beam. Synchronize the output pulses with an optical delay difference. Use a microscope frame to construct the imaging platform.
- Designing the beam path
- To control the laser power at the sample, use a half-wave plate and polarization beam splitter (PBS) combination for each laser beam.
- Install an acousto-optic modulator (AOM) in the Stokes laser beam path. Focus the beam with a 150 mm focal length lens into the AOM and recollimate the 0th order output with a 400 mm focal length lens.
- Use the same lens pair (150 mm and 400 mm focal lengths) to expand the pump beam to match the laser beam size with the Stokes.
- Place the 400 mm focal length lenses in both the pump and Stokes beam paths on separate one-dimensional translation stages for fine-tuning the beam divergence and optimizing the beam size before entering the microscope.
- Direct the pump beam with a right-angle reflecting mirror mounted on a motorized translation stage for optical delay tuning. If the Stokes beam needs optical delay, place these components instead into its beam path.
- Allow both beams to be combined at a dichroic mirror with a cutoff wavelength at ~1,000 nm (between the pump and Stokes wavelengths), so that the Stokes beam will transmit through the dichroic mirror while the pump beam is reflected by the dichroic mirror. Send the collinearly combined laser beams to the microscope.
- To chirp the pump and Stokes beams, place glass rods in their beam paths. See step 1.5 for details.
- To confirm proper alignment and beam size, use iris diaphragms after the dichroic mirror and before the microscope. Specifically, install one at a position close to and the other at a distance from the dichroic mirror to confirm good alignment and beam overlap. Use an IR-viewing card or IR viewer to visualize the beam during alignment.
- Use a fast PD and an oscilloscope to approximately measure the optical delay between the pump and Stokes pulses. Trigger the oscilloscope by sampling the laser pulse train.
- Block the pump beam and sample the Stokes beam. Zoom in on one of the pulses and place a vertical cursor on it to mark its temporal location on the oscilloscope.
- Unblock the pump beam and block the Stokes beam. Translate the delay stage until the sample pump pulses temporally align with the marked position.
- The laser scanning microscope
- For an upright microscope configuration, send the combined laser beams through a periscope to climb to an appropriate level before reaching the 2D galvo scanning mirrors.
- Measure the laser beam size before the microscope, and set up the proper lens pair after the galvo mirrors to expand the laser beam to best match the size of the entrance pupil of the objective lens.
- Construct a 4-f system using the two lenses, with the back aperture of the objective lens and the center of the two galvo mirrors being conjugate planes. Alternatively, use two separate 1D galvo mirrors with two 4-f lens systems for laser scanning.
- After the condenser, design a 2 in flip mirror to reflect the laser beams for signal collection. Position a lens with a 2 in diameter in the transmitted beam path to fully collect and focus transmission signals to the detectors.
- Direct the CARS signals to the PMT with a dichroic mirror having a cutoff at 776 nm, and allow the transmitted SRS signals to be detected by the PD. Use a bandpass filter (655/30 nm) before the PMT to reject residual excitation laser pulses. Use a short-pass filter (980 nm short pass) before the PD to block the Stokes beam from entering the detector.
- For CARS signal detection, connect a preamplifier and a current-voltage converter after the PMT and before sending the signal to the data acquisition system. Adjust the PMT voltage to optimize the signal and image contrast.
- Use a function generator to modulate the AOM at 1-10 MHz, and use the same frequency as the reference for the lock-in demodulation. Use a lock-in amplifier to extract SRS signals before data acquisition.
- Data acquisition and display
- Perform data acquisition using a digital data acquisition (DAQ) card in conjunction with a terminal block.
- Use the analog outputs from the DAQ to control the galvo mirrors and the analog inputs for signal acquisition.
- Use Lab-written software based on LabVIEW having a simultaneous multichannel display for real-time viewing and saving images (see Supplemental File).
- Chirping the femtosecond source and measuring the spectral resolution
NOTE: To achieve a good spectral resolution using spectral focusing, glass rods are used to introduce dispersions and chirp laser pulses from femtosecond to picosecond. To achieve the best spectral resolution, the chirp rate of the pump beam needs to equal that of the Stokes beam. For this laser system, the best spectral resolution can be achieved by chirping the ~120 fs output laser pulses to 3.4 ps for the pump and 1.8 ps for the Stokes. This chirping is achieved by using a 4+1 combination (four in the combined beam, one only in the Stokes beam) of 150 mm glass rod (SF-57) combination, as described below, and should achieve a 15 cm-1 spectral resolution. The pulse duration can be measured using an autocorrelator.- Insert one 150 mm glass rod into only the Stokes beam path.
- Insert two 150 mm glass rods into the combined pump/Stokes beam path after the dichroic beam splitter. To increase the chirping, let the combined laser beams double-pass the two glass rods by placing a dielectric mirror at one end of the rods.
- To measure the spectral resolution, prepare a standard chemical (e.g., dimethyl sulfoxide [DMSO]) sample pressed between two glass coverslips, and scan the delay stage until the maximum signal is achieved.
- Move the optical delay 1,000 µm in the red-shift direction. Then, run 200 frames at 10 µm/step toward the blueshift direction to collect the hyperspectral image stack.
- To convert the frame numbers to wavenumbers, perform a linear regression using the symmetric (2,913 cm-1) and asymmetric (2,994 cm-1) C-H stretching from DMSO and their corresponding frame numbers40.
- Use the XPM signal for measuring the spectral focusing intensity profile. Half-close the diaphragm on the condenser and move the focus to a blank coverslip. Collect the same number of steps as for hyperspectral SRS. To measure the CARS nonresonant background, focus on the glass coverslip and collect the same number of steps for the hyperspectral CARS measurements.
- Optimizing the signal-to-noise ratio (SNR) of images
- Prepare a chemical sample for system alignment. Follow the procedure described in step 3.1 for sample preparation.
NOTE: DMSO is a good choice because it is a common lab chemical with strong Raman signals and well-separated C-H symmetric and asymmetric peaks. - Place the sample on the microscope stage, and add water or immersion oil if needed for the objective lens or condenser. Properly move the edge of the DMSO droplet in the field of view and adjust the objective lens for the best focus. Center the condenser using the Köhler illumination method41. Fully open the diaphragm on the condenser.
- Tune the pump beam wavelength to 800 nm (1,045 nm Stokes) to target the 2,913 cm-1 CH3 peak. Set the power of both the pump and Stokes beam to ~30 mW before the microscope by adjusting the half-wave plate (~10 mW power at the sample plane).
- For SRS, set the lock-in amplifier gain to ~10 with a time constant of 7 µs (when using a 10 µs pixel dwell time). Ensure that the time constant is smaller than the pixel dwell time. Use Demod R for the AUX output for SRS signals.
- For CARS, send the PMT output to the preamplifier and current-voltage converter. Use the DAQ to acquire the output from the converter.
- Set the image acquisition parameters in the acquisition software. Use a pixel number of 200 x 200 with a scanning size of ~100 x 100 µm2. Make sure the image contains both the DMSO droplet and an empty area.
- Scan the sample and check the image on the computer screen. Scan the motorized delay stage in the Stokes/pump beam while monitoring the real-time images. Scan over the delay until the signal is maximized.
- Move the DMSO droplet to cover the whole field of view and check if the DC signal maximum is centered in the image (signal is pump beam-dependent). Adjust either the position of the pump beam via a mirror or the voltage offset in the imaging software.
- After DC optimization, adjust the Stokes beam mirrors until the AC signal is maximized by adjusting the threshold value to display ~50% saturation. Check whether the saturation is centered in the image. If not, fine-tune the mirrors only in the Stokes beam. Monitor the signal during alignment as real-time feedback on the quality of the alignment.
- To determine the SNR, select a small region of the DMSO image and measure the mean value. For the noise, select a small area in the empty region of the image and determine both the average mean value and the standard deviation. Subtract the noise mean value from the signal mean value and divide the results by the standard deviation of the empty region.
- If the calculated SNR is not high enough (typically 800-1,000 for SRS and >10,000 for CARS at a PMT voltage of 0.4 V with this power combination), check and reoptimize the beam overlap, beam sizes, and delay stage position, fine-tune the AOM, and change the function generator modulation frequency until the expected SNR is obtained.
- Prepare a chemical sample for system alignment. Follow the procedure described in step 3.1 for sample preparation.
2. Image analysis and data processing
- SNR analysis
- Open the ImageJ software. To import the saved DMSO sample .txt file, click File | Import | Text Image | Open.
- Once the image is imported, press CTRL+shift+C to bring up the brightness and contrast function (B&C). To find the maximum sample signal, press the auto button in the B&C until a region of the DMSO sample appears saturated.
- Click the oval selection tool on the ImageJ interface and highlight a small area of the saturated DMSO region. Once highlighted, press M to measure the mean and standard deviation of the selected area.
- For measuring the background, adjust the bars in the B&C function until the signal of the empty region can be observed. Click the oval selection and highlight a region of the background the same size as for step 2.1.3. Make sure the region selected does not contain DMSO. Press M to measure the statistics of the selected area.
- Calculate the SNR according to step 1.6.10.
- Processing hyperspectral CRS images
- Import the .txt file according to step 2.1.1. Once imported, click on Image | Stacks | Tools | Montage to Stack… to convert the file into an image stack.
- Scroll through the montage until the first DMSO peak is visible. Select a region on the DMSO and click on Image | Stack | Plot Z-axis Profile to plot the intensity versus frame number spectrum. To extract the raw spectral data, click on list and copy the profile data.
- To convert the recovered spectrum into wavenumber units, perform a linear regression as outlined in step 1.5.5.
- Fitting for measuring spectral resolution
NOTE: Lorentzian functions are used to fit the SRS and CARS spectra28.- Open the fitting software, then copy and paste the linear regression data into the program. For fitting the SRS data, highlight the data and then plot the data as a scatter plot.
- Pull up the scatter plot. Click on Analysis | Peaks and Baseline | Multiple Peak Fit | Open Dialog to bring up the peak analyzer. When pulled up, check that the input is the current plot and change the peak function to Lorentzian (Lorentz).
- Double-click on each of the two DMSO peaks on the graph to highlight the regions to be fitted. Next, click on Open NLfit to bring up the fitting window. Click on the Fit until converged button, and then OK, to see a tabulated summary of the fitting coefficients (see Eq (3)).
NOTE: The below equation shows the Lorentzian function format in the software. A1/2 are the amplitudes of the fitting peaks, w1/2 are the widths of the fitted peaks, and the x01/02 values are the centers of the fitted peaks. The independent variable is x and the dependent variable is y.
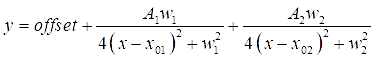 (3)
(3) - For CARS spectral fitting, click on Analysis | Fitting | Nonlinear curve fit | Open Dialog. Select category: new to define a new function for CARS. Use a two-peak CARS fitting function defined below (see Eq (4)) for CARS spectral fitting.
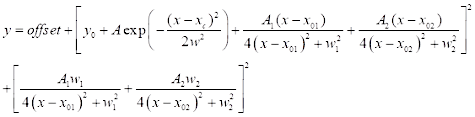 (4)
(4)
- Determining the spatial resolution
NOTE: Before this step, it is important to know the conversion between the pixel size at a specific magnification, pixel number, and the step size in µm. This can be performed by using a sample of a known diameter that is larger than the expected imaging resolution, measuring its line profile, and fitting a Gaussian function to determine the full width at half maximum (FWHM) value. Resolution targets or uniform samples such as polymeric beads can be used.- Acquire an image of cells or polymer particles less than 200 nm in diameter.
- Use ImageJ to draw a line across the smallest particle in the image.
- Press K to plot the intensity profile.
- Click list from the pop-up window and copy the information to fitting software.
- Plot the profile in fitting software and use Gaussian fitting (click on Analysis | Fitting | Nonlinear curve fit | Open Dialog | Category: Basic Functions; Function: Gauss).
- Read the peak width after fitting. Use the pixel to size conversion to obtain the actual resolution of the microscope.
3. Preparation of samples for hyperspectral CRS imaging
- Preparation of imaging slides and chemical samples
- Place a piece of double-sided tape onto a coverslip, and cut out a small rectangular shape of the tape from the middle of the placed tape to create an open area for the sample to be placed.
- Pipette 1-2 µL of pure DMSO and dispense the droplet at the center of the vacancy.
- Carefully place the top coverslip and gently press the edges of the coverslips to seal the chamber while making sure the DMSO sample does not contact the edges of the tape.
- For sensitivity experiments, prepare serial dilutions of DMSO in deuterium oxide (D2O) to give a concentration range of 50%-0%. Take 1-2 µL of each solution and prepare pressed samples as described above.
- Cell preparation
- Seed the cells in a 35 mm glass-bottom dish (or larger) in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
- Incubate the cells in an incubation chamber at 37 °C with a 5% CO2 atmosphere overnight or longer until ~50%-80% confluency is achieved.
- Image the live cells directly or fix the cells with a 10% formalin solution for imaging.
Representative Results
Comparisons of the spectral resolution
Figure 2 compares the spectral resolution of hyperspectral SRS (Figure 2A) and CARS (Figure 2B) microscopy using a DMSO sample. For the SRS spectrum, two Lorentzian functions (see protocol step 2.3) were applied to fit the spectrum, and a resolution of 14.6 cm-1 was obtained using the 2,913 cm-1 peak. For CARS, a two-peak-fitting function with a Gaussian background (see protocol step 2.3) was utilized for fitting, which gave the spectral resolution of 17.1 cm-1. These results show that, in the same measurement condition, SRS has a better spectral resolution than CARS. The reduced spectral resolution in CARS is mainly contributed by the involvement of the nonresonant background. In addition, it was found that the symmetric (2,913 cm-1) and asymmetric (2,995 cm-1) peak ratios were very different for SRS and CARS. This is due to the different signal correlations with the third-order nonlinear optical susceptibility, as described in equations (1) and (2). With the quadratic dependence of CARS, the intensity difference between the two peaks is amplified. The symmetric line shapes of the SRS peaks and the asymmetric line shapes of the CARS peaks can be observed in the spectrum. The asymmetry in the CARS signal is mainly due to the presence of the nonresonant background interference. The CARS spectral peaks appear slightly red-shifted (1-2 cm-1) to the SRS peaks. This also arises from the nonresonant background interference with resonant peaks.
Comparisons of the detection sensitivity
Figure 3 compares the detection sensitivity of hyperspectral SRS and CARS microscopy. The SNR of the DMSO SRS signals (2,913 cm-1) as the function of DMSO concentration in D2O at high concentrations are plotted first (1%-50%, Figure 3A). The results show a linear relationship, satisfying equation (2). Figure 3B plots the DMSO spectra at 0.1% and 0.01% concentrations, in which the 2,913 cm-1 peak can be resolved in the former but not in the latter, indicating the detection limit is between 0.1% and 0.01% DMSO. Using the limit of blank criteria, we estimated that the SRS detection limit is 0.021% DMSO. Figure 3C plots the CARS SNR as the function of DMSO concentration (1%-50%), showing a quadratic dependence in agreement with equation (1). The phase-retrieved CARS spectra are shown in Figure 3D for the 0.1% and 0.01% DMSO. To achieve these spectra, a spectral phase-retrieval method based on Kramers-Kronig relations was used and additional background removal was performed16. Similar to the SRS spectra, the DMSO 2,913 cm-1 peak can be clearly resolved for the 0.1% DMSO but not the 0.01%, indicating a detection limit between these two concentrations. Using the limit of blank criteria, we estimated that the SRS detection limit is 0.015% DMSO. The 0.02% DMSO corresponds to 2.8 mM. Therefore, the detection limit of the hyperspectral CRS microscope used here is ~2.1-2.8 mM DMSO.
Comparisons of the spatial resolution
Figure 4 compares the resolution of a small cellular feature detected in SRS (Figure 4A) and CARS (Figure 4B) images. The intensity profiles from the same line are displayed and fit using a Gaussian function to determine FWHM values for resolution comparison. The SRS signal gave a resolution of 398.6 nm (Figure 4C), while the CARS signal gave a resolution of 330.3 nm (Figure 4D). The resolution of CARS was ~1.2x better than that of the SRS. The reason for the resolution difference also lies in equations (1) and (2). Both pump and Stokes beams have a Gaussian point spread function at the focus. The signal of CARS is then proportional to the multiplication of three Gaussian functions, which roughly reduces the width by a factor of √3. Similarly, for SRS, the width is reduced by a factor of √2. Therefore, the resolution of CARS was √3/√2 = 1.2 times better than that of the SRS.
Comparisons of the images of cells
Figure 5 compares SRS and CARS images from MIA PaCa-2 cells at different optical delay positions. Figure 5A shows the SRS images at the optical delay that gave the strongest signal. In this image, lipid droplets (LDs), endoplasmic reticulum (ER), and nucleus (NU) can be detected, with LDs having the strongest signals shown as bright dots. Figure 5B shows the CARS channel image at the same optical delay, having much-reduced contrasts for LDs. The major reasons for this contrast difference are the presence of the nonresonant background and the red-shift of the same Raman peak in CARS spectra. At this optical delay, the generated signal has a large contribution from the nonresonant background of water. To enhance the lipid contrast in CARS, the optical delay was tuned to a red-shifted value. The red-shift improved the lipid contrasts by concentrating more energy to the 2,850 cm-1 for both SRS (Figure 5C) and CARS (Figure 5D), although the overall signal level was reduced. For CARS, a similar contrast of LDs as SRS was achieved by a ~98 cm-1 red-shift in spectral focusing (Figure 5D), though a background higher than that in the SRS image was still observed. At this optical delay, the SRS image shows much less protein and nucleic acid contents but strong lipid contents in LDs, ER, and cell membranes (Figure 5C).
CARS is a parametric process while SRS is nonparametric. Such a difference also contributes to contrast differences in the two modalities. The parametric CARS signals are determined by the interference of CARS signals from different layers close to the laser focus, which might show negative contrasts as indicated by arrows in Figure 5B and Figure 5D (also in Figure 4B). Such signal-interference-induced negative contrasts are absent in the SRS images. The negative contrast in CARS might provide information about the axial position of the target of interest.
The SRS signals have a linear relationship with the molecular concentration, while the CARS signals satisfy a near-quadratic concentration-dependence. Therefore, the CH2-rich LDs show a much stronger signal than the ER and the cell membranes in the CARS image than in the SRS image (Figure 5E,F). The SRS spectra can be extracted from hyperspectral images. Figure 5G shows the typical SRS spectra from LDs, ER, cytosol (CY), and NU. Both the intensity and spectral shape are different for different cellular compartments. LD shows a much stronger signal at 2,850 cm-1 than other organelles. As for CARS, similar spectra, though different in shapes, can be obtained. The raw CARS spectra show a small red-shift compared to the corresponding SRS spectra. Spectral phase-retrieval can be further used to extract the Raman responses using the CARS spectra.
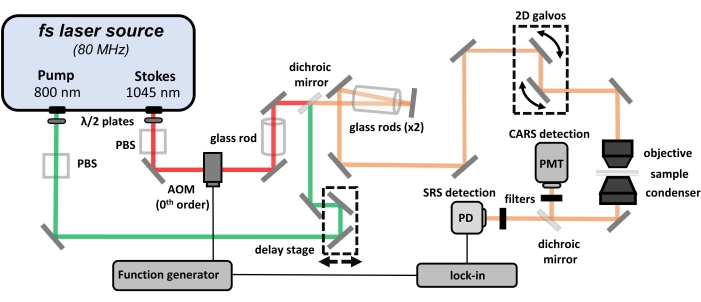
Figure 1: A schematic of the hyperspectral CARS/SRS microscope. Abbreviations: CARS = coherent anti-Stokes Raman scattering; SRS = stimulated Raman scattering; PBS = polarization beam splitter; PD = photodiode; PMT = photomultiplier tube; AOM = acousto-optic modulator. Please click here to view a larger version of this figure.
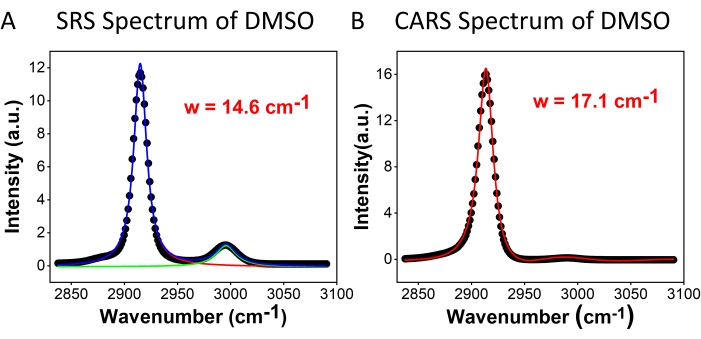
Figure 2: DMSO spectra. (A) SRS and (B) CARS spectra of DMSO. Dots are experimental data; curves are spectral fitting results. Abbreviations: CARS = coherent anti-Stokes Raman scattering; SRS = stimulated Raman scattering; DMSO = dimethyl sulfoxide; w = spectral resolution. Please click here to view a larger version of this figure.
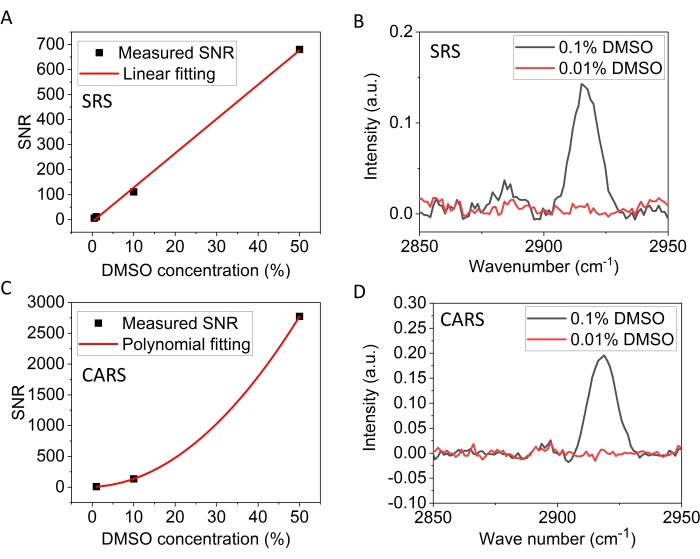
Figure 3: Signal-to-noise ratios and spectra of DMSO. (A) Signal-to-noise ratio of the DMSO symmetric peak at 2,913 cm-1 as a function of concentration in D2O measured by SRS. The dots are experimental data; the line is the linear fitting result. (B) The SRS spectra of 0.1% and 0.01% DMSO in D2O. (C) Signal-to-noise ratio of the DMSO symmetric peak at 2,913 cm-1 as a function of concentration in D2O measured by CARS. The dots are experimental data; the curve is the second-degree polynomial fitting result. (D) The CARS spectra of 0.1% and 0.01% DMSO in D2O. Abbreviations: CARS = coherent anti-Stokes Raman scattering; SRS = stimulated Raman scattering; DMSO = dimethyl sulfoxide; SNR = signal-to-noise ratio; D2O = deuterium oxide. Please click here to view a larger version of this figure.
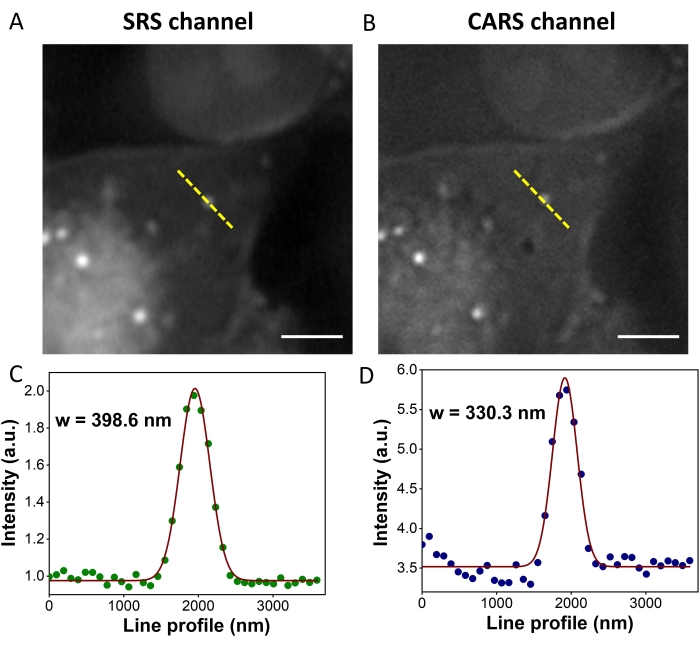
Figure 4: SRS and CARS images and intensity profiles of an MIA PaCa-2 cell. (A) An SRS image of an MIA PaCa-2 cell. (B) A CARS image of an MIA PaCa-2 cell at the same field-of-view as panel A. (C) Intensity profile of SRS along the yellow line in panel A. (D) Intensity profile of CARS along the yellow line in panel B. Dots are experimental data; curves are Gaussian function fitting results. Scale bars = 5 µm. Abbreviations: CARS = coherent anti-Stokes Raman scattering; SRS = stimulated Raman scattering; w = resolution. Please click here to view a larger version of this figure.
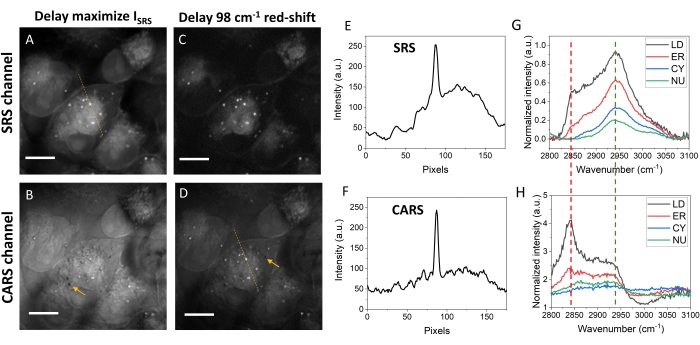
Figure 5: Images and intensity profile of MIA PaCa-2 cells. (A) An SRS image of MIA PaCa-2 cells at the optimized time delay for SRS intensity. (B) A CARS image at the same delay as in panel A. (C) An SRS image at the 98 cm-1 red-shifted delays as in panel A. (D) A CARS image at the same optical delay as in panel C. (E,F) The SRS and CARS intensity profiles plotted along the dotted lines in panels A and D. (G) Typical SRS spectra from the lipid droplets, endoplasmic reticulum, cytosol, and nucleus. (H) Typical CARS spectra of the four cellular compositions. The green and red dotted lines are delay positions for panels A/B and C/D, respectively. Scale bars = 10 µm. Abbreviations: CARS = coherent anti-Stokes Raman scattering; SRS = stimulated Raman scattering; LD = lipid droplets; ER = endoplasmic reticulum; CY = cytosol; NU = nucleus. Please click here to view a larger version of this figure.
Supplemental File: Lab-written software based on LabVIEW having a simultaneous multichannel display for real-time viewing and saving images. Please click here to download this File.
Discussion
The protocol presented here describes the construction of a multimodal CRS microscope and the direct comparison between CARS and SRS imaging. For the microscope construction, the critical steps are spatial and temporal beam overlapping and beam size optimization. It is recommended to use a standard sample such as DMSO before the biological imaging for optimizing SNR and calibrating Raman shifts. Direct comparison between CARS and SRS images reveals that CARS has a better spatial resolution, while SRS gives better spectral resolution and less convoluted chemical contrasts. Both CARS and SRS have similar detection limits.
CARS and SRS imaging use high-energy pulse lasers for excitation. This allows the platform to integrate other nonlinear optical imaging modalities such as multiphoton excitation fluorescence, harmonic generation, and transient absorption for additional chemical contrasts28,39.
CARS and SRS have been extensively used to study lipid composition with high chemical selectivity. However, the technologies are not limited to quantifying lipids. SRS has been applied to map drug distribution42, protein synthesis43, and DNA44. CARS and SRS have also been applied to image pharmaceutical ingredients and excipients in tablets45,46,47,48. Hyperspectral CARS and SRS have found applications in cancer diagnosis49, cardiovascular disease evaluation50, and neural imaging51. They can also be applied for COVID-19 studies52. Broadband CARS, which can cover spectral windows as broad as 3,000 cm-1, can elucidate rich chemical structures in biological samples53. However, due to the slow readout rate of the CCD, the pixel dwell time is on the level of milliseconds, much slower than the microsecond pixel dwell time for SRS microscopy34. Hyperspectral SRS microscopy currently has a typical bandwidth of 200-300 cm-1, limited by the laser bandwidth and the lack of lock-in-integrated array detectors34. Fourier transform SRS microscopy is an alternative way to potentially broaden the SRS spectral coverage35.
Although CARS and SRS provide rich chemical information without the need for labeling, the chemical selectivity lies in chemical bonds, making it difficult to distinguish specific proteins. Raman tags have shown the potential to enhance the chemical selectivity of CARS and SRS54,55. However, coherent Raman imaging still has much lower sensitivity compared to fluorescence detection. Surface enhancement was used for spontaneous Raman scattering spectroscopy to improve the signal levels56. It was also applied to CARS and SRS for signal amplification57,58,59. Although the enhancement factor is not as high as spontaneous Raman scattering, surface-enhanced CARS and SRS microscopy still show the potential to detect single molecules59,60. Nevertheless, the use of metal particles or surfaces deprives the advantage of the label-free approach. Improving the sensitivity of coherent Raman microscopy without using metal surfaces would greatly expand the application of the technology in biological science.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research was supported by the Purdue University Department of Chemistry startup fund.
Materials
| 2D galvo scanner set | Thorlabs | GVS002 | |
| Acousto-optic modulator | Isomet | M1205-P80L-0.5 | |
| AOM driver | Isomet | 532B-2 | |
| Data acquisition card | National Instruments | PCle 6363 | Custom ordered filter (980 sp) |
| Delay stage | Zaber | X-LSM050A | |
| Deuterium oxide | Millipore Sigma | 151882-100G | |
| Dichroic mirror for beam combination | Thorlabs | DMLP1000 | |
| Dichroic mirror for signal separation | Semrock | FF776-Di01-25×36 | |
| DMSO | MiliporeSigma | 200-664-3 | |
| MIA PaCa 2 Cells | ATCC | CRL-1420 | |
| Femtosecond laser system | Spectral Physics | InSightX3+ | |
| Filter for CARS | Chroma | AT655/30m | |
| Filter for SRS | Chroma | ET980sp | |
| Function generator | Rigol | DG1022Z | |
| Glass rods | Lattice Electro Optics | SF-57 | |
| Half-wave plate | Newport | 10RP02-51; 10RP02-46 | |
| LabVIEW 2020 | National Instruments | This is the image acquisition software | |
| Lock-in amplifier | Zurich Instrument | HF2LI | |
| Microscope housing | Olympus | BX51W1 | |
| Objective lens | Olympus | UPLSAPO60XW | |
| Origin Pro 2019b | OriginLab Corporation | This is the spectral fitting software | |
| Oscilloscope | Tektronix | TBS2204B | |
| Photodiode | Hamamatsu | S3994-01 | |
| PMT detector | Hamamatsu | H7422P-40 | |
| PMT voltage amplifier | Advanced Research Instrument Corp. | PMT4V3 | |
| Polarizing beamsplitter cube | Thorlabs | PBS255 | |
| Terminal block | National Instruments | BNC-2110 |
References
- Raman, C. V. A change of wave-length in light scattering. Nature. 121 (3051), 619 (1928).
- Li, S., Li, Y., Yi, R., Liu, L., Qu, J. Coherent anti-Stokes Raman scattering microscopy and its applications. Frontiers in Physics. 8, 515 (2020).
- Evans, C. L., Xie, X. S. Coherent anti-Stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annual Review of Analytical Chemistry. 1 (1), 883-909 (2008).
- Min, W., Freudiger, C. W., Lu, S., Xie, X. S. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annual Review of Physical Chemistry. 62, 507-530 (2011).
- Suhalim, J. L., Boik, J. C., Tromberg, B. J., Potma, E. O. The need for speed. Journal of Biophotonics. 5 (5-6), 387-395 (2012).
- Duncan, M. D., Reintjes, J., Manuccia, T. J. Scanning coherent anti-Stokes Raman microscope. Optics Letters. 7 (8), 350-352 (1982).
- Denk, W., Strickler, J. H., Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science. 248 (4951), 73-76 (1990).
- Zumbusch, A., Holtom, G. R., Xie, X. S. Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Physical Review Letters. 82 (20), 4142-4145 (1999).
- Cheng, J. -. X., Xie, X. S. Coherent anti-Stokes Raman scattering microscopy: instrumentation, theory, and applications. The Journal of Physical Chemistry B. 108 (3), 827-840 (2004).
- Evans, C. L., et al. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proceedings of the National Academy of Sciences of the United States of America. 102 (46), 16807 (2005).
- Cheng, J. -. X., Volkmer, A., Book, L. D., Xie, X. S. An epi-detected coherent anti-Stokes Raman scattering (E-CARS) microscope with high spectral resolution and high sensitivity. The Journal of Physical Chemistry B. 105 (7), 1277-1280 (2001).
- Volkmer, A., Book, L. D., Xie, X. S. Time-resolved coherent anti-Stokes Raman scattering microscopy: Imaging based on Raman free induction decay. Applied Physics Letters. 80 (9), 1505-1507 (2002).
- Marks, D. L., Boppart, S. A. Nonlinear interferometric vibrational imaging. Physical Review Letters. 92 (12), 123905 (2004).
- Ganikhanov, F., Evans, C. L., Saar, B. G., Xie, X. S. High-sensitivity vibrational imaging with frequency modulation coherent anti-Stokes Raman scattering (FM CARS) microscopy. Optics Letters. 31 (12), 1872-1874 (2006).
- Potma, E. O., Evans, C. L., Xie, X. S. Heterodyne coherent anti-Stokes Raman scattering (CARS) imaging. Optics Letters. 31 (2), 241-243 (2006).
- Liu, Y., Lee, Y. J., Cicerone, M. T. Broadband CARS spectral phase retrieval using a time-domain Kramers-Kronig transform. Optics Letters. 34 (9), 1363-1365 (2009).
- Masia, F., Karuna, A., Borri, P., Langbein, W. Hyperspectral image analysis for CARS, SRS, and Raman data. Journal of Raman Spectroscopy. 46 (8), 727-734 (2015).
- Knutsen, K. P., Johnson, J. C., Miller, A. E., Petersen, P. B., Saykally, R. J. High spectral resolution multiplex CARS spectroscopy using chirped pulses. Chemical Physics Letters. 387 (4-6), 436-441 (2004).
- Okuno, M., Kano, H., Leproux, P., Couderc, V., Hamaguchi, H. -. o. Ultrabroadband multiplex CARS microspectroscopy and imaging using a subnanosecond supercontinuum light source in the deep near infrared. Optics Letters. 33 (9), 923-925 (2008).
- Masia, F., Glen, A., Stephens, P., Borri, P., Langbein, W. Quantitative chemical imaging and unsupervised analysis using hyperspectral coherent anti-Stokes Raman scattering microscopy. Analytical Chemistry. 85 (22), 10820-10828 (2013).
- Pegoraro, A. F., Slepkov, A. D., Ridsdale, A., Moffatt, D. J., Stolow, A. Hyperspectral multimodal CARS microscopy in the fingerprint region. Journal of Biophotonics. 7 (1-2), 49-58 (2014).
- Eckhardt, G., et al. Stimulated Raman scattering from organic liquids. Physical Review Letters. 9 (11), 455-457 (1962).
- Ploetz, E., Laimgruber, S., Berner, S., Zinth, W., Gilch, P. Femtosecond stimulated Raman microscopy. Applied Physics B. 87 (3), 389-393 (2007).
- Freudiger, C. W., et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 322 (5909), 1857-1861 (2008).
- Nandakumar, P., Kovalev, A., Volkmer, A. Vibrational imaging based on stimulated Raman scattering microscopy. New Journal of Physics. 11 (3), 033026 (2009).
- Slipchenko, M. N., Le, T. T., Chen, H., Cheng, J. -. X. High-speed vibrational imaging and spectral analysis of lipid bodies by compound Raman microscopy. The Journal of Physical Chemistry B. 113 (21), 7681-7686 (2009).
- Min, W., Freudiger, C. W., Lu, S., Xie, X. S. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annual Review of Physical Chemistry. 62 (1), 507-530 (2011).
- Zhang, C., Zhang, D., Cheng, J. -. X. Coherent Raman scattering microscopy in biology and medicine. Annual Review of Biomedical Engineering. 17 (1), 415-445 (2015).
- Prince, R. C., Frontiera, R. R., Potma, E. O. Stimulated Raman scattering: from bulk to nano. Chemical Reviews. 117 (7), 5070-5094 (2017).
- Lu, F. -. K., et al. Multicolor stimulated Raman scattering microscopy. Molecular Physics. 110 (15-16), 1927-1932 (2012).
- Ozeki, Y., et al. High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nature Photonics. 6 (12), 845-851 (2012).
- Wang, P., et al. Label-free quantitative imaging of cholesterol in intact tissues by hyperspectral stimulated raman scattering microscopy. Angewandte Chemie International Edition. 125 (49), 13280-13284 (2013).
- Freudiger, C. W., et al. Stimulated Raman scattering microscopy with a robust fibre laser source. Nature Photonics. 8 (2), 153-159 (2014).
- Liao, C. -. S., et al. Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light: Science & Applications. 4 (3), 265 (2015).
- Liao, C. -. S., et al. Spectrometer-free vibrational imaging by retrieving stimulated Raman signal from highly scattered photons. Science Advances. 1 (9), 1500738 (2015).
- He, R., et al. Dual-phase stimulated Raman scattering microscopy for real-time two-color imaging. Optica. 4 (1), 44-47 (2017).
- Andresen, E. R., Berto, P., Rigneault, H. Stimulated Raman scattering microscopy by spectral focusing and fiber-generated soliton as Stokes pulse. Optics Letters. 36 (13), 2387-2389 (2011).
- Fu, D., Holtom, G., Freudiger, C., Zhang, X., Xie, X. S. Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. The Journal of Physical Chemistry B. 117 (16), 4634-4640 (2013).
- Zhang, C., Aldana-Mendoza, J. A. Coherent Raman scattering microscopy for chemical imaging of biological systems. Journal of Physics: Photonics. , (2021).
- Martens, W. N., Frost, R. L., Kristof, J., Theo Kloprogge, J. Raman spectroscopy of dimethyl sulphoxide and deuterated dimethyl sulphoxide at 298 and 77 k. Journal of Raman Spectroscopy. 33 (2), 84-91 (2002).
- Gill, G. W., Gill, G. W. . Cytopreparation: Principles & Practice. , 309-323 (2013).
- Fu, D., et al. Imaging the intracellular distribution of tyrosine kinase inhibitors in living cells with quantitative hyperspectral stimulated Raman scattering. Nature Chemistry. 6 (7), 614-622 (2014).
- Wei, L., Yu, Y., Shen, Y., Wang, M. C., Min, W. Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. Proceedings of the National Academy of Sciences. 110 (28), 11226-11231 (2013).
- Lu, F. -. K., et al. Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proceedings of the National Academy of Sciences. 112 (37), 11624-11629 (2015).
- Slipchenko, M. N., et al. Vibrational imaging of tablets by epi-detected stimulated Raman scattering microscopy. Analyst. 135 (10), 2613-2619 (2010).
- Slipchenko, M. N., Zhou, B., Pinal, R., Teresa Carvajal, M., Cheng, J. -. X. RAMAN-chemical imaging of solid dosage forms based on stimulated Raman scattering. American Pharmaceutical Review. 15 (3), 66 (2012).
- Sarri, B., et al. Discriminating polymorph distributions in pharmaceutical tablets using stimulated Raman scattering microscopy. Journal of Raman Spectroscopy. 50 (12), 1896-1904 (2019).
- Fussell, A. L., Kleinebudde, P., Herek, J., Strachan, C. J., Offerhaus, H. L. Coherent anti-Stokes Raman scattering (CARS) microscopy visualizes pharmaceutical tablets during dissolution. JoVE (Journal of Visualized Experiments). (89), e51847 (2014).
- Freudiger, C. W., et al. Multicolored stain-free histopathology with coherent Raman imaging). Laboratory Investigation. 92 (10), 1492-1502 (2012).
- Lim, R. S., et al. Multimodal CARS microscopy determination of the impact of diet on macrophage infiltration and lipid accumulation on plaque formation in ApoE-deficient mice [S]. Journal of Lipid Research. 51 (7), 1729-1737 (2010).
- Ji, M., et al. label-free detection of brain tumors with stimulated Raman scattering microscopy. Science Translational Medicine. 5 (201), (2013).
- Tabish, T. A., Narayan, R. J., Edirisinghe, M. Rapid and label-free detection of COVID-19 using coherent anti-Stokes Raman scattering microscopy. Mrs Communications. 10 (4), 566-572 (2020).
- Camp, C. H., et al. High-speed coherent Raman fingerprint imaging of biological tissues. Nature Photonics. 8 (8), 627-634 (2014).
- Wei, L., et al. Live-cell bioorthogonal chemical imaging: stimulated Raman scattering microscopy of vibrational probes. Accounts of Chemical Research. 49 (8), 1494-1502 (2016).
- Hu, F., Shi, L., Min, W. Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nature Methods. 16 (9), 830-842 (2019).
- Nie, S., Emory, S. R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 275 (5303), 1102-1106 (1997).
- Steuwe, C., Kaminski, C. F., Baumberg, J. J., Mahajan, S. Surface enhanced coherent anti-Stokes Raman scattering on nanostructured gold surfaces. Nano Letters. 11 (12), 5339-5343 (2011).
- Fast, A., Kenison, J. P., Syme, C. D., Potma, E. O. Surface-enhanced coherent anti-Stokes Raman imaging of lipids. Applied Optics. 55 (22), 5994-6000 (2016).
- Zong, C., et al. Plasmon-enhanced stimulated Raman scattering microscopy with single-molecule detection sensitivity. Nature Communications. 10 (1), 1-11 (2019).
- Yampolsky, S., et al. Seeing a single molecule vibrate through time-resolved coherent anti-Stokes Raman scattering. Nature Photonics. 8 (8), 650-656 (2014).

