Mycobacterium tuberculosis Extracellular Vesicle Enrichment through Size Exclusion Chromatography
Summary
This protocol describes size exclusion chromatography, a facile and reproducible technique for enriching Mycobacterium tuberculosis extracellular vesicles from culture supernatants.
Abstract
The role of extracellular vesicles (EVs) in the context of bacterial infection has emerged as a new avenue for understanding microbial physiology. Specifically, Mycobacterium tuberculosis (Mtb) EVs play a role in the host-pathogen interaction and response to environmental stress. Mtb EVs are also highly antigenic and show potential as vaccine components. The most common method for purifying Mtb EVs is density gradient ultracentrifugation. This process has several limitations, including low throughput, low yield, reliance on expensive equipment, technical challenges, and it can negatively impact the resulting preparation. Size exclusion chromatography (SEC) is a gentler alternative method that combats many of the limitations of ultracentrifugation. This protocol demonstrates that SEC is effective for Mtb EV enrichment and produces high-quality Mtb EV preparations of increased yield in a rapid and scalable manner. Additionally, a comparison to density gradient ultracentrifugation by quantification and qualification procedures demonstrates the benefits of SEC. While the evaluation of EV quantity (nanoparticle tracking analysis), phenotype (transmission electron microscopy), and content (Western blotting) is tailored to Mtb EVs, the workflow provided can be applied to other mycobacteria.
Introduction
Extracellular vesicle (EV) release by pathogens may be the key to unlocking new technologies to control infectious diseases1. Mycobacterium tuberculosis (Mtb) is a pathogen of high consequence, infecting approximately one-third of the world's population and claiming the lives of millions of people each year2. EV production by Mtb is well documented yet elusive in the biogenesis and varied roles (i.e., immunostimulatory, immunosuppressive, iron and nutrient acquisition) of these EVs in the context of infection3,4,5. Efforts to understand the composition of Mtb EVs revealed 50-150 nm lipid membrane-enclosed spheres derived from the plasma membrane containing lipids and proteins of immunological significance3,6. Investigation of the role of Mtb EVs in bacterial physiology has revealed the importance of bacterial EV modulation in response to environmental stress for survival5. Host-pathogen interaction studies have been more complicated to interpret, but evidence indicates that Mtb EVs can influence the immune response of the host and may potentially serve as an effective vaccination component3,4,7.
Most studies of Mtb EVs thus far have relied on density gradient ultracentrifugation for vesicle enrichment8. This has been effective for small-scale studies; however, this technique has several technical and logistical challenges. Alternate workflows couple multistep centrifugation, for the removal of whole cells and large debris, with a final ultracentrifugation step to pellet EVs. This methodology can vary in efficiency, and often results in low yield and co-purification of soluble non-vesicle associated biomolecules while also impacting vesicle integrity9. Additionally, this process is time-consuming, manually intensive, and very limited in throughput due to equipment constraints.
The present protocol describes an alternative technique to density gradient ultracentrifugation: size exclusion chromatography (SEC). This method has been demonstrated for environmental mycobacteria, and in the current work, it has been extrapolated to Mtb10. A commercially available column and automatic fraction collector can improve consistency in vesical preparation and reduce the necessity for specific, expensive equipment. It is also possible to complete this protocol in a fraction of the time compared to density gradient ultracentrifugation, increasing the throughput. This technique is less technically challenging, making it easier to master, and can increase inter/intra-laboratory reproducibility. Finally, SEC has high separation efficiency and is gentle, preserving the integrity of the vesicles.
Protocol
The Colorado State University Institutional Biosafety Committee approved the present study (19-046B). Cultivation of Mycobacterium tuberculosis and harvesting of EV-rich culture supernatants were performed by trained personnel in a high-containment laboratory. The materials were moved out of the high-containment area after a valid inactivation method was performed, confirmed, and approved by institutional biosafety policies. While replicating the protocol, if validated inactivation or sterile filtration method is not feasible, the following procedures need to be performed in a high-containment laboratory.
1. Preparation of crude Mtb EV concentrate
NOTE: For detailed procedures on the cultivation of Mtb and preparation of culture filtrate protein (CFP), see References11,12. It is recommended that bacterial culture media is free from growth supplements with EV-containing or proteinaceous components, such as Oleic Albumin Dextrose Catalase (OADC), and detergents such as Tween. It is also recommended that the bacterial culture's quality and harvested CFP be screened to ensure limited cell death and lysis13,14.
- Prepare a 100 kDa molecular weight cut-off (MWCO) centrifugal filter (see Table of Materials) by adding the full volume capacity of phosphate-buffered saline (1x PBS). Centrifuge for 5 min at 2,800 x g at 4 °C.
NOTE: If using a filter with no dead stop volume, care must be taken to ensure the sample volume does not reduce below the filter level, resulting in complete filter drying during centrifugation. - Discard the flow-through and any remaining PBS before filling the sample chamber with Mtb CFP to its maximum capacity. Centrifuge at 2,800 x g at 4 °C until the volume has reduced to the minimum volume of the ultrafiltration device. Repeat as necessary, adding more CFP to the unit until the entire sample has been reduced sufficiently.
NOTE: Retain a portion of the 100 kDa flow-through (100F) material for downstream qualification if desired. Store at 4 °C. - Add 1x PBS to the filter unit containing the concentrate up to the total device capacity. Reduce the volume as described in 1.2. Repeat this step five times to ensure complete washing and buffer exchange.
- Recover the 100 kDa concentrated CFP retentate (100R) according to the ultrafiltration device specifications (see Table of Materials). Once the 100R is recovered, wash the filter with a minimal volume of 1x PBS at least three times, and pool the wash with the 100R to maximize the recovery.
- Quantitate the 100R material with a bicinchoninic assay (BCA) following the manufacturer's instructions (see Table of Materials). To perform the assay, use several dilutions of samples 1:2, 1:5, and 1:10 in PBS. Test the sample in triplicate.
NOTE: Retain a portion of the 100R material for downstream qualification if desired. Store at 4 °C.
2. Size exclusion chromatography for the enrichment of Mtb EVs from CFP
NOTE: The following procedure is specific for using 3 mg of 100R Mtb CFP with SEC column and automatic fraction collector (AFC, see Table of Materials). It can be adapted for other starting concentrations and column types by following the manufacturer's specifications. Additionally, the users are recommended to read and understand the automatic fraction collector user manual.
- Allow the SEC column to equilibrate to room temperature.
- Turn on the AFC using the power switch on the back of the tower unit and adjust the settings for the load cell, if necessary, by pressing SETUP on the main menu touchscreen. The load cell must only be calibrated upon first use, after every software update, and if inconsistency in fraction collection volumes is noticed.
- From the SETUP screen, align the carousel by selecting CAROUSEL > CALIBRATE. Insert the carousel with the 13 small holes facing up into the AFC tower, and adjust the carousel so that the fluid nozzle is directly above the flush position by pressing the – and/or + buttons.
- Remove the caps from the equilibrated SEC column. Slide the SEC column into the appropriate column mount and carefully install it on the AFC tower. Ensure the radio frequency identification (RFID) tag on the column faces the AFC, and check that the connection between the column and the valve is secure. Place the waste outlet tubing in a collection container.
- From the SETUP screen, select Collection Schedule and set the count to 13 and the size to 0.5 mL by pressing the – and/or + buttons. Leave the "Buffer Volume" setting as the default of 2.7 mL, and then close the "Collection Schedule" window by pressing X.
- Select START COLLECTION from the home screen and confirm the collection parameters by selecting YES. Load 13 labeled 1.7 mL microcentrifuge tubes with the lids open and pointing toward the carousel center.
- Advance the AFC by selecting OK, then mount the column reservoir and advance again by pressing OK. Select the option to flush the column by pressing YES, and add one column volume of 0.2 µm filtered PBS to remove any storage buffer. Once the flush is complete, advance the AFC by pressing OK.
- Place the carousel cover over the AFC and press OK. Prepare one column volume of 0.2 µm filtered PBS. Use a pipette to remove any excess buffer from the column.
- Bring 3 mg of 100R sample as quantified in step 1.5 to 500 µL with 1x PBS. Add the 3 mg sample to the top of the column. Advance the AFC and allow the sample to run into the frit. Once the sample has fully entered the column, add the PBS prepared in step 2.6 to the reservoir.
- Monitor the run of the AFC while it collects first the void volume and then the specified fractions. After the run completes and the carousel returns to waste position, remove the cover, and remove the fraction tubes from the carousel. Store the fractions at 4 °C prior to quantification (step 3) and qualification (step 4).
- If the column is to be reused, select the option to clean the column by pressing YES; otherwise, press NO. Follow the instructions on the AFC.
NOTE: Once the column is washed and the storage buffer has run through, it can be removed, capped, and stored at 4 °C.
3. Quantification of the Mtb EVs
- Measure the protein concentration for each fraction using BCA and micro BCA12.
- For 0.5 mL fractions collected from 3 mg of 100R Mtb CFP, use the micro BCA with a 1:3 dilution for fractions numbered 1-7 of each sample in triplicate.
- For 0.5 mL fractions numbered 5-13, use the BCA with no dilution for each sample in triplicate.
NOTE: Overlapping the assays for fractions 5-7 will help ensure all fractions have a readable output.
- Quantitate the particle concentration for each fraction using nanoparticle tracking analysis (NTA).
- Dilute the samples in 1 mL of 1x PBS using the following suggested starting ratios; for fractions 1-3, use 1:100, and then for the remaining fractions, use a 1:10 dilution.
- Vortex the diluted solutions and then draw the sample into a 1 mL disposable syringe. Set the syringe in an automatic syringe pump if available.
- Set the syringe pump to 30 µL/min and use the video capture settings with a screen gain of 10-12 and a camera level of 10-12. Adjust the focus for each sample.
- Collect a minimum of three videos at 30 s each using a constant flow.
- Perform the analysis with the software detection threshold set to five.
NOTE: It may not be possible to obtain NTA data for the latest fractions (>7) without sacrificing significant sample volume.
4. Qualification of Mtb EVs
- Visualize the general protein profile of each fraction using a silver-stained protein gel.
NOTE: Detailed procedures for SDS-PAGE and silver stain can be found in Reference15.- Add SDS sample buffer to 10 µL of each fraction and 5 µg of CFP, 100R, and 100F. Boil for 5 min at 100 °C, then load on a 4%-12% Bis-Tris Gel with a molecular weight ladder for polyacrylamide gel electrophoresis (PAGE) (see Table of Materials).
- Run the gel using 1x 2-[N-morpholino]ethanesulfonic acid (MES) running buffer (see Table of Materials) and a 200 V current for 35 min.
- Remove the gel from the cassette and proceed with silver staining15.
- Evaluate the presence or absence of EV markers in each fraction by Western blot.
NOTE: Detailed procedures for Western blotting can be found in Reference15.- Run an SDS-PAGE gel as described in steps 4.1.1-4.1.2.
- Remove the gel from the cassette and transfer the resolved proteins to a 0.2 µm nitrocellulose membrane (see Table of Materials) by applying a 50 V current for a minimum of 1 h.
- Perform Western blot analysis to evaluate proteins of interest. Primary antibodies against LpqH, lipoarabinomannan (LAM), and GroES (see Table of Materials) are recommended.
- Pool fractions based on the presence or absence of markers as applicable for the downstream application.
- Confirm the presence of intact vesicles by transmission electron microscopy (TEM).
- Fix 15 µL of the vesicle sample by adding 15 µL of 4% EM grade paraformaldehyde and store at 4 °C overnight.
- Prepare a 200 mesh formvar-carbon coated copper TEM grid by cleaning the surface and making a hydrophilic substrate.
NOTE: Plasma cleaning with 95% argon, 5% oxygen, and 30% plasma power for 1 min is recommended. - Drop 10 µL of the fixed sample onto the grid and allow the sample to adhere for 10 min, then blot away the excess liquid with filter paper.
- Float the grid on a drop of ultrapure water for 30 s, then blot away excess liquid.
- Float the grid on a d% EM grade uranyl acetate drop for 2 min, then blot away the excess liquid.
- Allow the grid to air dry completely.
- Image using a TEM (see Table of Materials) at 100 kV or similar.
NOTE: Other EV quantitation and characterization methods should be used based on downstream applications. The Minimal Information for Studies of Extracellular Vesicles18 should be consulted for guidelines to ensure the rigor and reproducibility of all EV studies.
Representative Results
Culture filtrate protein (CFP) from Mycobacterium tuberculosis (Mtb) was concentrated, quantified, and then 3 mg of material was applied to a size exclusion chromatography (SEC) column. The protein and particle concentrations were enumerated by BCA and NTA, respectively. Expected ranges for protein and particle recovery plus the exact values obtained for these results are reported in Table 1. Values much higher than these ranges may indicate contamination or column integrity issues. Values significantly lower point to problems in the ultrafiltration steps, and therefore the flow-through needs to be compared to the starting CFP and 100R to determine if a successful concentration occurred. A non-specific protein silver-stain shows that later fractions contain more protein and resemble the 100R material (Figure 1).
Western blots of the fractions demonstrate that lipoarabinomannan (LAM) is present across the fractions, with later fractions showing lower intensity banding (Figure 2A, Supplementary Figure 1). The 19 kDa lipoprotein LpqH, known to be present in Mtb EVs3,19, is enriched in the earliest fractions from the SEC (Figure 2B, Supplementary Figure 1). The 10 kDa chaperonin protein GroES is absent in the earliest fractions from the SEC (Figure 2C, Supplementary Figure 1); this finding aligns with previously published studies regarding GroES as a contaminant during Mtb EV enrichment12 and serves as a negative control for Mtb EV presence. Transmission Electron Microscopy (TEM) confirms the presence of closed, membrane-bound vesicles (Figure 3A). A comparison of Mtb EVs separated by density gradient ultrafiltration and this SEC method is reported in Table 2. SEC provides higher protein and particle recovery for three technical replicates from the same CFP. These results have been consistent across multiple batches of CFP (data not shown). Both methods result in closed, membrane-bound vesicles in the expected size range, as demonstrated by TEM (Figure 3) and NTA (Figure 4).
Altogether, these data demonstrate enrichment of Mtb EVs in the early SEC fractions. The highest NTA values occur in fractions 1-3, and the protein content increases as the fraction number rises, indicating the separation of soluble proteins from the EVs (Table 1). Because fraction 4 contains evidence of GroES (Figure 2C, Supplementary Figure 1), this fraction was not included in the pooled material for TEM. Depending on the downstream application, the inclusion or exclusion of specific fractions for the pooling strategy must be considered.
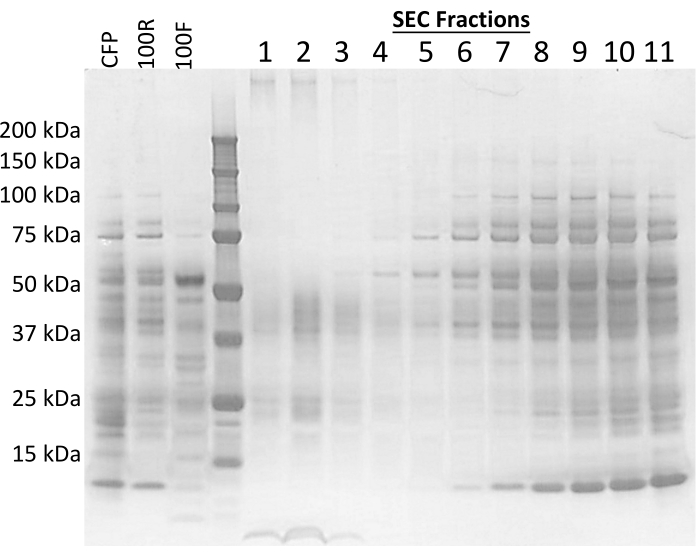
Figure 1: Silver stain by fraction. An SDS-PAGE gel stained with silver demonstrates the overall protein profile for each fraction. CFP = 5 µg of Mtb CFP, 100R = 5 µg of 100R, 100F = 5 µg of 100F, 1-11 = 10 µL of SEC fractions 1-11. Please click here to view a larger version of this figure.
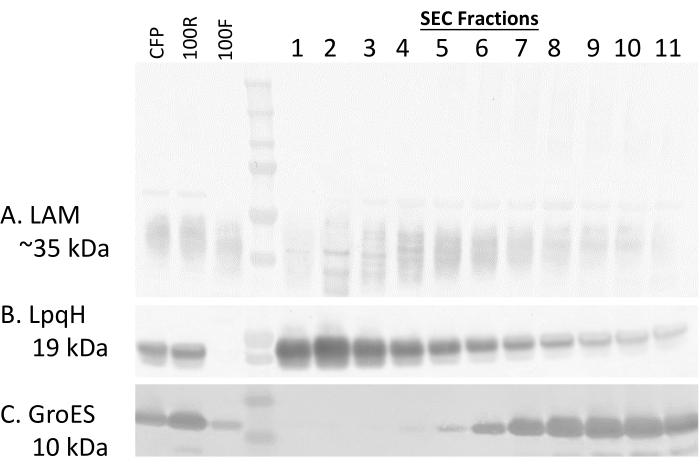
Figure 2: Western blots by fraction. Western blots detecting (A) LAM, (B) LpqH, and (C) GroES, demonstrating protein marker changes across the fractions. CFP = 5 µg of Mtb CFP, 100R = 5 µg of 100R, 100F = 5 µg of 100F, 1-11 = 10 µL of SEC fractions 1-11. Please click here to view a larger version of this figure.
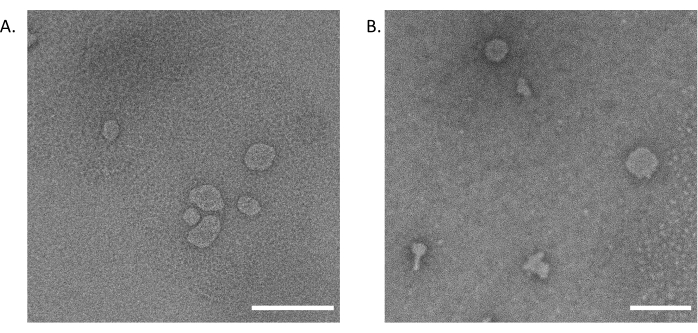
Figure 3: Transmission Electron Microscopy (TEM) images. (A) SEC fractions 1-3 pooled prior to fixation. (B) The density gradient ultracentrifugation of Mtb EVs was negatively stained for TEM imaging. Scale bars = 200 nm. Please click here to view a larger version of this figure.
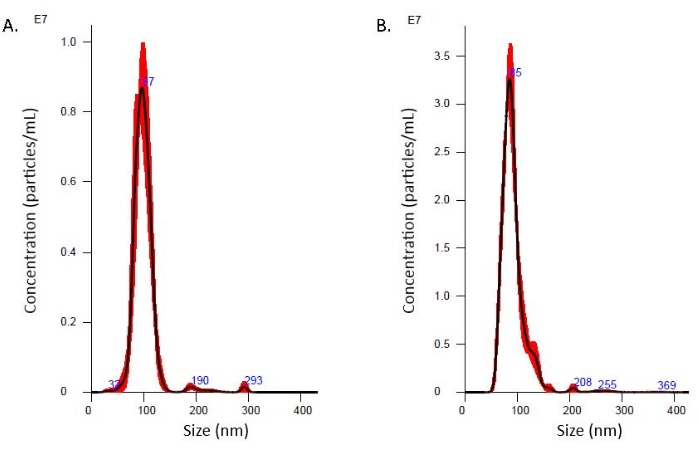
Figure 4: Nanoparticle Tracking Analysis (NTA) distribution. (A) SEC fractions 1-3. (B) The density gradient ultracentrifugation Mtb EVs were analyzed with nanoparticle tracking analysis. The size distribution is displayed. Please click here to view a larger version of this figure.
| Fraction | Protein Recovery Range (µg/µL) | Particle Recovery Range (per µL) | Result Protein Recovery (µg/µL) | Result Particle Recovery (per µL) |
| 1 | 0.01-0.03 | 1E8 – 3E8 | 0.013 | 1.60E+08 |
| 2 | 0.02-0.04 | 2E8 – 4E8 | 0.026 | 2.10E+08 |
| 3 | 0.02-0.04 | 2E7 – 6E7 | 0.024 | 5.20E+07 |
| 4 | 0.03-0.05 | 7E6 – 2E7 | 0.032 | 1.30E+07 |
| 5 | 0.05-0.09 | 1E6 – 5E6 | 0.053 | 4.20E+06 |
| 6 | 0.09-0.2 | 1E6 – 5E6 | 0.099 | 5.20E+06 |
| 7 | 0.1-0.3 | 1E6 – 3E6 | 0.234 | 2.70E+06 |
| 8 | 0.3-0.5 | 1E6 – 4E6 | 0.398 | 4.20E+06 |
| 9 | 0.4-0.6 | 1E6 – 3E6 | 0.543 | 4.10E+06 |
| 10 | 0.5-0.7 | 1E6 – 3E6 | 0.661 | 1.70E+06 |
| 11 | 0.6-0.8 | 1E6 – 3E6 | 0.736 | 1.40E+06 |
Table 1: Protein and particle recovery by fraction. Expected protein and particle recovery range per fraction based on 3 mg of starting material. The exact values for obtaining the material used in this study are included in the two rightmost columns.
| Enrichment Method | Total Protein Recovered (µg) | Total Particles Recovered |
| Density gradient ultracentrifugation | 3.14 | 1.82E+10 |
| 3.88 | 1.93E+10 | |
| 3.20 | 1.65E+10 | |
| Size exclusion chromatography F1-3 | 12.96 | 4.05E+10 |
| 14.14 | 4.15E+10 | |
| 14.74 | 4.35E+10 |
Table 2: Method comparison of protein and particle recovery. Protein yield measured with BCA and total particle count measured by NTA for Mtb EVs originating from 3 mg of the same 100R starting material, enriched using the referenced density gradient ultracentrifugation method8 or this SEC method (triplicate).
Supplementary Figure 1: Original Western blots (uncropped) from Figure 2. Please click here to download this File.
Discussion
Mycobacterium tuberculosis extracellular vesicles are highly antigenic reservoirs, which present them as an attractive avenue for developing diagnostic tools and future vaccines4,19,20. Historically, density gradient ultracentrifugation has been used to separate Mtb EVs from other soluble, secreted material8. While this process is effective, it is also time-consuming, technically challenging, and may impact the integrity of the resulting EV preparations9,10. The presented protocol offers an alternative method for Mtb EV preparations through size exclusion chromatography (SEC).
There are several critical points for success in this method. The initial Mtb CFP preparation will influence the quality and yield of vesicles following SEC. It is recommended that CFP be harvested at or before the mid-log phase of growth to limit the level of cellular lysis at the time of harvest. Late-log and stationary growth phase cultures can contain higher levels of cell lysis and can result in the identification of intracellular proteins and artifactual EV-like structures, spontaneously generated from the membrane fragments of the lysed cells, as a significant contributor to the collected CFP12,13,14. This should be evaluated prior to beginning this protocol as membrane vesicles from lysed cells will co-purify with secreted Mtb EVs and potential skew downstream analyses. Care must be taken during ultrafiltration to ensure that the filter membrane remains intact and wet. Damage to the filter can cause desired material to flow through. Including the original CFP, 100R, and 100F on downstream quality evaluations will assist in the evaluation of ultrafiltration integrity.
Proper setup and washing of the SEC column are necessary for the highest quality EV preparation. Always follow the user manuals for setup and takedown. While fractions are being collected, ensure the AFC is not bumped or moved as this can interrupt the process and lead to skipped or inconsistent fractions. The antibodies used for qualification will depend on the downstream application of the enriched vesicles, and should include a marker expected to be in Mtb EVs (LpqH) and a marker found in CFP but not enriched in Mtb EVs (GroES). One limitation to the methods presented here is potential variation in appropriate protein controls. This protocol has been developed using standard culturing methods. Work performed with Mycobacterium avium suggested that levels of chaperonins like GroES change in CFP and EVs based on the medium used for growth21. As more information emerges regarding mycobacterial EV composition, adjusting the specific negative control marker may be necessary.
Finally, all quantification and qualification procedures and downstream applications should be performed quickly. If material storage is required, 4 °C is recommended over freezing to prevent vesicle integrity disruption during the freeze-thaw process. If freezing is performed, aliquot the material to avoid repeated freeze-thaw cycles. Enriched EVs should be re-evaluated both quantitatively and qualitatively if a significant amount of time passes between the initial evaluation and use.
This method effectively enriches Mtb EVs, and there is flexibility for adaptation to other mycobacteria. When adapting this procedure for other organisms, ensure the starting culture has limited cellular lysis. The amount of material loaded on the column should be adjusted, as the kinetics of EV release will vary. Do not exceed a maximum protein concentration of 7 g per 100 mL based on the manufacturer's specifications. It is also necessary to evaluate each fraction individually for the presence of EV-associated markers and contaminants. Protein concentration and NTA data may be misleading in method optimization: very low protein concentration does not mean the absence of vesicles in those fractions. Additionally, changing SEC columns and fraction collection parameters allow the user to scale the protocol based on input and downstream applications. Limitations of this method include the cost of the AFC and consumables, the dilute output, and, as mentioned earlier, the development and optimization of the faction pooling scheme. While density gradient ultracentrifugation has its advantages and maybe more applicable for certain experiments, enrichment of Mtb EVs by SEC is comparable in quality and superior in access and ease of use, as presented here.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to acknowledge support from the College of Veterinary Medicine and Biomedical Sciences Experiential Award and College Research Council Shared Research Program to NKG and funding by ATCC (award # 2016-0550-0002) to KMD. We would also like to acknowledge Anne Simpson for technical support and BEI Resources, NIAID, NIH for the following reagents: Monoclonal Anti-Mycobacterium tuberculosis LpqH (Gene Rv3763), IT-54 (produced in vitro), NR-13792, Monoclonal Anti-Mycobacterium tuberculosis GroES (Gene Rv3418c), Clone IT-3 (SA-12) (produced in vitro), NR-49223, and Monoclonal Anti-Mycobacterium tuberculosis LAM, Clone CS-35 (produced in vitro), NR-13811.
Materials
| 20x MES SDS Running Buffer | ThermoFisher Scientific | NP0002 | |
| 96 well plate | Corning | 15705-066 | |
| Automatic Fraction Collector | IZON Science | AFC-V1-USD | |
| BenchMark Pre-stained Protein Ladder | Invitrogen | 10748010 | |
| Benchtop centrifuge | Beckman Coulter | Allegra 6R | |
| Centricon Plus – 70 Centrifugal filter, 100 kDa cutoff | Millipore Sigma | UFC710008 | Ultrafiltration device used in step 1.1 |
| Electroblotting System | ThermoFisher Scientific | 09-528-135 | |
| EM Grade Paraformaldehyde | Electron Microscopy Sciences | 15714-S | |
| Formvar/Carbon 200 mesh Cu Grids | Electron Microscopy Sciences | FCF200H-Cu-TA | |
| Goat Anti-Mouse IgG H&L (Alkaline Phosphatase), whole molecule, 1 mL | AbCam | ab6790 | Secondary antibody |
| JEM-1400 Transmission Electron Microscope | JOEL | ||
| Micro BCA Protein Assay Kit | ThermoFisher Scientific | 23235 | |
| Microplate reader | BIOTEK | Epoch | |
| Monoclonal Anti-Mycobacterium tuberculosis GroES (Gene Rv3814c) | BEI Resources | NR-49223 | Primary antibody |
| Monoclonal Anti-Mycobacterium tuberculosis LpqH (Gene Rv3763) | BEI Resources | NR-13792 | Primary antibody |
| Monocolonal Anti-Mycobacterium tuberculosis LAM, Clone CS-35 | BEI Resources | NR-13811 | Primary antibody |
| NanoClean 1070 | Fischione Instruments | For plasma cleaning of the TEM grid | |
| Nanosight equipped with syringe pump and computer with NanoSight NTA software | Malvern Panalytical | NS300 | |
| Nitrocellulose membrane, Roll, 0.2 μm | BioRad | 1620112 | |
| NuPAGE 4-12% Bis-Tris Protein Gels | ThermoFisher Scientific | NP0323BOX | |
| Phosphate-buffered Saline, 1X without calcium and magnesium | Corning | 21-040-CV | |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | 23225 | |
| PowerPac Basic Power Supply | BioRad | 1645050 | |
| qEV Original 35 nm 5/pk | IZON Science | SP5-USD | SEC column |
| SDS sample buffer | Boster | AR1112 | In-house recipe used in this procedure, however this product is equivalent |
| SDS-PAGE gel chamber | ThermoFisher Scientific | EI0001 | |
| Sigmafast BCIP/NBT | Millipore Sigma | B5655 | |
| Silver Stain Plus Kit | BioRad | 1610449 | In-house protocol used in this procedure, however this kit is equivalent |
| Uranyl Acetate | Electron Microscopy Sciences | 22400 |
References
- Gill, S., Catchpole, R., Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiology Reviews. 43 (3), 273-303 (2019).
- World Health Organization. GLOBAL TUBERCULOSIS REPORT 2021. World Health Organization. , (2021).
- Prados-Rosales, R., et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. Journal of Clinical Investigation. 121 (4), 1471-1483 (2011).
- Prados-Rosales, R., et al. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of mycobacterium tuberculosis. mBio. 5 (5), 01921 (2014).
- Prados-Rosales, R., et al. Role for mycobacterium tuberculosis membrane vesicles in iron acquisition. Journal of Bacteriology. 196 (6), 1250-1256 (2014).
- Lee, J., et al. Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis. Proteomics. 15 (19), 3331-3337 (2015).
- Athman, J. J., et al. Mycobacterium tuberculosis Membrane Vesicles Inhibit T Cell Activation. The Journal of Immunology. 198 (5), 2028-2037 (2017).
- Prados-Rosales, R., Brown, L., Casadevall, A., Montalvo-Quirós, S., Luque-Garcia, J. L. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX. 1, 124-129 (2014).
- Cvjetkovic, A., Lötvall, J., Lässer, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. Journal of Extracellular Vesicles. 3 (1), 23111 (2014).
- Dauros Singorenko, P., et al. Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. Journal of Extracellular Vesicles. 6 (1), 1324731 (2017).
- Wallace, E., et al. Culturing mycobacteria. Methods in Molecular Biology. 2314, 1-58 (2021).
- Lucas, M., et al. Extraction and separation of mycobacterial proteins. Methods in Molecular Biology. 2314, 77-107 (2021).
- Walker, S. A., Kennedy, M. T., Zasadzinski, J. A. Encapsulation of bilayer vesicles by self-assembly. Nature. 387 (6628), 61-64 (1997).
- Edwards, D. A., et al. Spontaneous vesicle formation at lipid bilayer membranes. Biophysical Journal. 71 (3), 1208 (1996).
- . Production Manuals & SOPs. SP007: Running of polyacrylamide gels, SP011: Western blot, and SP0012: Silver staining protocols Available from: https://labs.vetmebiosci.colostate.edu/dobos/bei-resources/ (2022)
- Engers, H. D., et al. Results of a World Health Organization-sponsored workshop to characterize antigens recognized by mycobacterium-specific monoclonal antibodies. Infection and Immunity. 51 (2), 718-720 (1986).
- Chatterjee, D., Lowell, K., Rivoire, B., McNeil, M. R., Brennan, P. J. Lipoarabinomannan of Mycobacterium tuberculosis. Capping with mannosyl residues in some strains. Journal of Biological Chemistry. 267 (9), 6234-6239 (1992).
- Théry, C., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles. 7 (1), 1535750 (2018).
- Palacios, A., et al. Mycobacterium tuberculosis extracellular vesicle-associated lipoprotein LpqH as a potential biomarker to distinguish paratuberculosis infection or vaccination from tuberculosis infection. BMC Veterinary Research. 15 (1), 1-9 (2019).
- Ziegenbalg, A., et al. Immunogenicity of mycobacterial vesicles in humans: Identification of a new tuberculosis antibody biomarker. Tuberculosis. 93 (4), 448-455 (2013).
- Chiplunkar, S. S., Silva, C. A., Bermudez, L. E., Danelishvili, L. Characterization of membrane vesicles released by Mycobacterium avium in response to environment mimicking the macrophage phagosome. Future Microbiology. 14 (4), 293-313 (2019).

