Isolation and Quantification of Epstein-Barr Virus from the P3HR1 Cell Line
Summary
This protocol permits the isolation of Epstein-Barr virus particles from the human P3HR1 cell line upon inducing the viral lytic cycle with phorbol 12-myristate 13-acetate. DNA is subsequently extracted from the viral preparation and subjected to real-time PCR to quantify the viral particle concentration.
Abstract
The Epstein-Barr virus (EBV), formally designated as Human herpesvirus 4 (HHV-4), is the first isolated human tumor virus. Nearly 90-95% of the world's adult population is infected by EBV. With the recent advancements in molecular biology and immunology, the application of both in vitro and in vivo experimental models has provided deep and meaningful insight into the pathogenesis of EBV in many diseases as well as into EBV-associated tumorigenesis. The aim of this visualized experiment paper is to provide an overview of the isolation of EBV viral particles from cells of the P3HR1 cell line, followed by quantification of the viral preparation. P3HR1 cells, originally isolated from a human Burkitt lymphoma, can produce a P3HR1 virus, which is a type 2 EBV strain. The EBV lytic cycle can be induced in these P3HR1 cells by treatment with phorbol 12-myristate 13-acetate (PMA), yielding EBV viral particles.
Using this protocol for the isolation of EBV particles, P3HR1 cells are cultured for 5 days at 37 °C and 5% CO2 in complete RPMI-1640 medium containing 35 ng/mL PMA. Subsequently, the culture medium is centrifuged at a speed of 120 x g for 8 min to pellet the cells. The virus-containing supernatant is then collected and spun down at a speed of 16,000 x g for 90 min to pellet the EBV particles. The viral pellet is then resuspended in a complete RPMI-1640 medium. This is followed by DNA extraction and quantitative real-time PCR to assess the concentration of EBV particles in the preparation.
Introduction
The Epstein-Barr virus (EBV) is the first human tumor virus to have been isolated1. EBV, formally referred to as Human herpesvirus 4 (HHV-4)2, is part of the gamma herpes virus subfamily of the herpes virus family and is the prototype of the Lymphocryptovirus genus. Nearly 90-95% of the world's adult population is infected by the virus3. In most cases, initial infection occurs within the first 3 years of life and is asymptomatic, however, if infection occurs later during adolescence, it may give rise to an illness referred to as infectious mononucleosis4. EBV is able to infect resting B cells inducing them to become proliferative B lymphoblasts in which the virus establishes and maintains a latently infected state5. EBV can reactivate at any time and thus lead to recurrent infections6.
Over the past 50 years, the association between some viruses and the development of human malignancies has become increasingly apparent, and today it is estimated that 15% to 20% of all human cancers are related to viral infections7. The herpes viruses, including EBV, are some of the best studied examples of these types of tumor viruses8. In fact, EBV can cause many types of human malignancies, such as Burkitt lymphoma (BL), Hodgkin lymphoma (HL), diffuse large B cell lymphoma, and lymphoproliferative diseases in immunocompromised hosts9,10. EBV has also been shown to be associated with the development of systemic autoimmune diseases. Some examples of these autoimmune disorders are rheumatoid arthritis (RA), polymyositis-dermatomyositis (PM-DM), systemic lupus erythematosus (SLE), mixed connective tissue disease (MCTD), and Sjögren's syndrome (SS)11. EBV is also associated with the development of inflammatory bowel disease (IBD)12.
Many of these diseases can be studied or modeled using cell culture, mice, or other organisms that are infected with EBV. That is why EBV particles are needed to infect cells or organisms, whether in in vitro or in vivo models13,14,15,16, hence the need to develop a technique that allows isolation of viral particles at a low cost. The protocol described here provides guidelines for an easy way to reliably isolate EBV particles from a relatively accessible cell line and to quantitate the particles using real-time PCR, which is cost-effective and readily available to most laboratories. This is in comparison to several other methods that have been described to isolate EBV from different cell lines17,18,19,20.
P3HR-1 is a BL cell line that grows in suspension and is latently infected with an EBV type 2 strain. This cell line is an EBV producer and can be induced to produce viral particles. The goal of this manuscript is to showcase a method that permits the isolation of EBV particles from the P3HR-1 cell line, followed by quantification of the viral stock that could later be used for both in vitro and in vivo EBV experimental models.
Protocol
NOTE: EBV should be considered a potentially biohazardous material, and thus should be handled under Biosafety Level 2 containment or higher. A lab coat as well as gloves should be worn. If there is potential for exposure to splashes, eye protection should also be considered. The following procedure should be conducted in a Biological Safety Cabinet.
1. Counting the P3HR1 cells
- Centrifuging and resuspending cells
- Transfer the cell suspension from a 100 mm culture plate (or T-25 flask) of an ongoing P3HR1 cell culture at 80% confluency to a 15 mL conical tube. The seeding density of P3HR-1 cells is 1 x 106 cells/mL. Maintain in complete RPMI culture medium (79% RPMI culture medium, 20% Fetal Bovine Serum, 1% Pen-Strep antibiotic) at 37 °C and 5% CO2, and passage every 3-4 days.
- Centrifuge for 8 min at 120 x g. After centrifugation, discard the supernatant, resuspend the cell pellet in 1 mL of complete RPMI culture medium, and mix well (this solution will be referred to as suspension A).
- Preparing the cell suspension for counting
- Prepare a diluted cell suspension B by mixing 2 µL of cell suspension A with 8 µL of culture medium. Add 10 µL of 0.4% trypan blue to suspension B to obtain suspension C. Mix the preparation well by gentle pipetting.
- Counting cells using a hemocytometer
- Place a glass coverslip on the hemocytometer and load 10 µL of suspension C. Place the hemocytometer under a light microscope and count the number of cells that are not stained blue in each of the four quadrants of the hemocytometer chamber using the 40x microscope objective (Figure 1).
- Trypan blue stains dead cells blue; do not count these cells, nor cells that are touching any of the top, bottom, right, or left borders of each hemocytometer quadrant.
- Calculate the concentration of cells per mL using the following formula:
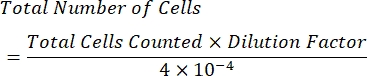
NOTE: The number of quadrants counted is four, and 10-4 mL is the volume of the squares on the hemocytometer. The four quadrants which should be counted are represented in green in Figure 1. Total Cells Counted in the formula is the sum of the numbers of cells in quadrants 1, 2, 3, and 4. Cells represented in blue in Figure 1 should be counted, while cells in red should not be counted since they are touching the top, right, bottom, or left borders of the quadrant.
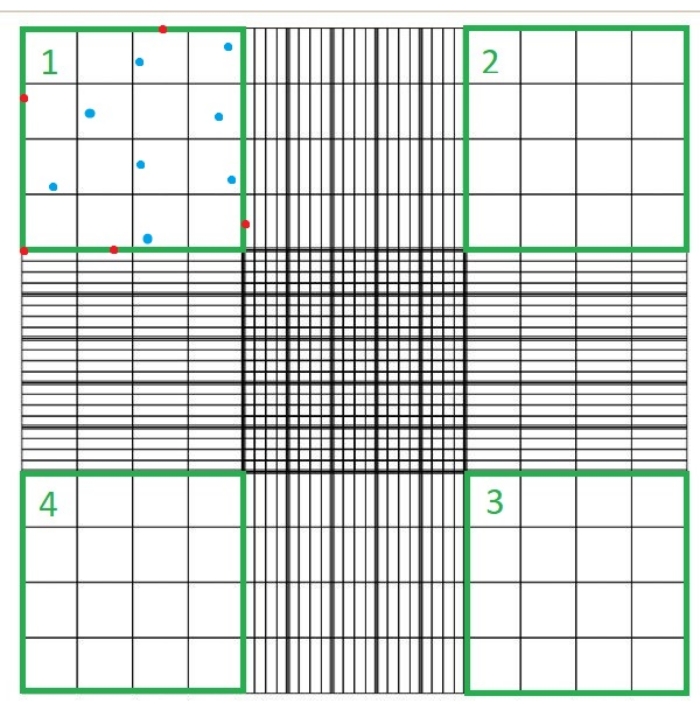
Figure 1: Cell counting using a hemocytometer chamber. Four quadrants are counted using a light microscope; these quadrants are represented in green. Total Cells Counted in the formula indicated in step 1.3.3 of the protocol described here is the sum of the number of cells in quadrants 1, 2, 3 and 4. Cells indicated in blue should be counted, while cells in red should not be counted since they are touching the top, right, bottom, or left borders of the quadrant Please click here to view a larger version of this figure.
2. Preparing the plate for culture
- Place a volume of cell suspension A in a 15 mL conical tube. The volume required is dependent on the cell count. Adjust the volume such that the number of cells is roughly 2.2 x 106 cells for a 100 mm culture plate.
- Add 5 mL of complete culture medium to the tube and then transfer the contents of the tube to a 100 mm culture plate. Add 80 µL of dimethyl sulfoxide (DMSO) to the culture plate.
NOTE: DMSO is light sensitive, hence it should be stored in a light-resistant container or covered with an opaque material like aluminum foil. - Add 350 µL of 1 mg/mL phorbol 12-myristate 13-acetate (PMA) to the tube. The final concentration of PMA should be 35 ng/mL, and that of DMSO should be 0.08%.
CAUTION: PMA is toxic, corrosive, and carcinogenic, hence it should be handled with extreme care. PMA is light sensitive, hence it should be stored in a light-resistant container or covered with an opaque material like aluminum foil. - Add 4.27 mL of culture medium so that the total volume is 10 mL. Mix the contents of the tube by tilting, then transfer the contents to a 100 mm culture plate. Leave the plate in a cell culture incubator for 5 days at 37 °C, 5% CO2.
3. Induction and isolation of Epstein-Barr virus particles
- After 5 days in the incubator, centrifuge the contents of the plate at 120 x g for 8 min to pellet the cells. Collect the cell-free, virus-containing supernatant and discard the cell pellet.
- Centrifuge the supernatant at 16,000 x g for 90 min at 4 °C to pellet the virus particles. Discard the supernatant and resuspend the virus pellet in 5 mL of culture medium.
NOTE: Phosphate-buffered saline (PBS) can be used to resuspend the virus pellet instead of culture medium. - Aliquot the viral suspension obtained into 20 tubes, each containing 250 µL of the viral suspension. Store the suspension at -80 °C.
4. DNA extraction from viral particles
CAUTION: Extreme care should be taken when handling phenol, as it is toxic and corrosive and has the ability to cause severe burns. Phenol is light sensitive and oxidizes upon contact with light or air. Store it in a light-resistant container or alternatively cover the phenol tube with an opaque material like aluminum foil.
- Separation of proteins from DNA
- Add 500 µL of TrisCl-saturated phenol to one of the 250 µL tubes. Add 100 µL of water (to increase the volume of the aqueous phase) and vortex well to obtain a pink emulsion. Centrifuge for 15 min at 9650 x g.
- DNA precipitation
- Collect the supernatant (transparent, aqueous phase) and transfer it to a new 1.5 mL microcentrifuge tube.
- Add an equivalent of 1/10 of the supernatant's volume of cold sodium acetate (3 M, pH 5.2) and mix by pipetting up and down. Add 1 µL of 20 mg/mL glycogen and mix by pipetting.
NOTE: This step is optional, but the addition of glycogen enhances DNA precipitation as well as makes it easier to visualize the DNA pellet. - Add three times the supernatant's volume of cold 100% ethanol. Store at -80 °C overnight.
NOTE: The procedure can be stopped here to be resumed at a later time. The sample can be stored at -80 °C for 1 h, or alternatively overnight. Overnight storage enhances DNA precipitation and is thus recommended.
- Isolating the viral DNA
- The following day, centrifuge at 9650 x g for 15 min at 4 °C and discard the supernatant to obtain a DNA pellet.
- Wash the pellet three times with 1 mL of cold 70% ethanol, centrifuge at 9650 x g for 15 min, and discard the supernatant.
- Air-dry the pellet for about 10 min. Resuspend the pellet in 10-50 µL of nuclease-free distilled water (depending on the size of the DNA pellet).
- Store the samples at -20 °C for later processing or at 4 °C overnight to ensure maximal DNA dissolution followed by storage at -20 °C. Alternatively, directly proceed with step 5.
5. Checking DNA concentration and purity
- After cleaning the pedestal of a microspectrophotometer with a delicate task wiper, load 1 µL of the DNA preparation.
- Note the concentration of the DNA in the sample. Check the ratios of absorbance at both
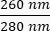 and
and 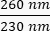 . DNA will absorb at 260 nm, proteins at 280 nm, and organic substances such as phenol will absorb at 230 nm. A ratio of 1.8-2 is considered sufficient for real-time PCR.
. DNA will absorb at 260 nm, proteins at 280 nm, and organic substances such as phenol will absorb at 230 nm. A ratio of 1.8-2 is considered sufficient for real-time PCR.
6. Quantification by real-time polymerase chain reaction
- Preparation of the PCR reaction mixes
- Place 5 µL of a SYBR green real-time PCR mix in 0.2 mL PCR tubes. Add to each tube 1 µL of 7.5 pmol/µL forward primer and 1 µL of 7.5 pmol/µL reverse primer. Forward primer sequence: 5'-CCCTAGTGGTTTCGGACACA-3'; reverse primer sequence: 5'-ACTTGCAAATGCTCTAGGCG-3'. The gene amplified to determine the EBV genome copy number is the Epstein-Barr virus small RNA 2 (EBER-2) gene.
- To one of the tubes, add 2 µL of nuclease-free water and 1 µL of viral DNA from step 4. The total volume of the reaction mixture is 10 µL.
NOTE: Depending on the concentration of DNA in the sample, a dilution step may be needed. The dilution factor F should be noted and will be integrated in the formula in step 6.3. - Prepare other tubes that will be used as standards with known EBV genome copy numbers (1000, 2000, 5000, 10,000, and 54,000 copies).
- Run the PCR mixtures starting with an initial step of activation at 95 °C for 5 min, then 40 cycles at 95 °C and 58 °C (annealing) for 15 s and 30 s, respectively.
- Generate a qPCR standard curve by plotting the Ct values of the standards against the log of the number of EBV genome copies per standard tube. Employing the standard curve plot equation, derive the number of EBV genome copies in the PCR tube containing the induced viral DNA. Then, use the following formula to calculate the concentration of the induced viral preparation:

Where X is the number of EBV genome copies derived from the standard curve and F is the dilution factor used for setting up the DNA utilized per PCR reaction.
7. Checking for biological activity/infectivity of the viral particles
- Transfer BC-3 cells from a 100 mm culture plate at 80% confluency to a 15 mL conical tube. The seeding density of P3HR-1 cells is 1 x 106 cells/mL. Maintain in complete RPMI culture medium (79% RPMI culture medium, 20% Fetal Bovine Serum, 1% Pen-Strep antibiotic) at 37 °C and 5% CO2, and passage every 3-4 days.
- Count the BC-3 cells the same way the P3HR-1 cells were counted by following step 1. To a 96-well plate, add 105 BC-3 cells to each well. Use three, six, nine, or 12 wells to ensure the significance of results and minimize errors.
- Add a volume of viral stock to each well, depending on the concentration of the viral preparation determined in step 6.3. The MOI can vary, and optimization of this protocol will reveal the best MOI for infection (an MOI range of 2-50 is recommended). Ensure the total volume of the mixture present in each well is 250 µL.
- To determine the required volume, the concentration of the viral stock should be known, and an MOI should be selected. For example, if the MOI is 2, then the number of viral particles added should be twice the number of cells. Calculate the volume V of the viral stock required using the formula:
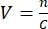 , with n being the number of viral particles needed, and C the known concentration of the viral stock
, with n being the number of viral particles needed, and C the known concentration of the viral stock
- To determine the required volume, the concentration of the viral stock should be known, and an MOI should be selected. For example, if the MOI is 2, then the number of viral particles added should be twice the number of cells. Calculate the volume V of the viral stock required using the formula:
- Incubate the contents of the 96-well plate for 5 days. After 5 days, transfer the contents of each well to a 1.5 mL tube.
- Centrifuge the tubes at 120 x g for 8 min to separate the cells from the virus-containing culture medium. Subject the cell pellets as well as the supernatants to DNA extraction followed by real-time PCR quantification to check for the presence of viral genomes in each fraction, thus assessing the infectivity of the viral particles.
Representative Results
The goal of this procedure is to isolate EBV particles in a suspension with known viral titer, that could subsequently be used to model EBV infection. Thus, it is of utmost importance to use optimal concentrations of the different reagents to obtain the highest EBV yield out of the procedure.
An optimization trial was performed to determine the concentrations of PMA and DMSO that would yield the highest number of EBV particles (Figure 2). A DMSO concentration of 0.8% and a PMA concentration of 35 ng/µL were optimal and resulted in high EBV concentrations. The concentration of viral particles obtained from the optimization protocol was 917,471 viral particles/µL.
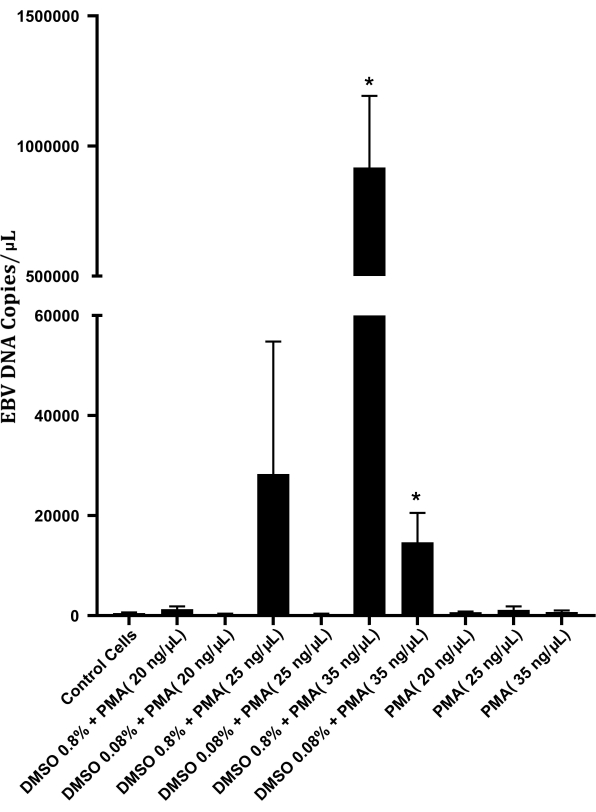
Figure 2: EBV DNA copies/mL obtained upon induction of P3HR-1 cells with phorbol 12-myristate 13-acetate (PMA). P3HR-1 cells (105 cells) were cultured alone or induced with different concentrations of PMA and DMSO using the protocol described here. The error bars indicate the standard deviation. A Student's t-test was performed comparing PMA/PMA and DMSO-treated cells to control cells; * indicates p < 0.05. Please click here to view a larger version of this figure.
For this protocol to be considered effective, step 6 should reveal a reasonable concentration of viral particles, and step 7 should show that these viral particles are indeed infectious. The BC-3 cell line used in step 7 is EBV-negative, so upon quantification of the EBV genome copy numbers in negative controls (that were not subjected to this protocol), no EBV DNA should be detected.
However, EBV DNA should be detected in both the cells and the culture medium of the experimental wells (that were subjected to this protocol), for the following reasons: EBV that infects BC-3 cells will replicate inside these cells, accounting for the fact that EBV DNA will be detected in the cell pellet and upon replication, BC-3 cells will shed viral particles to the outside; thus, EBV DNA will be detected in the culture medium supernatant (after DNA extraction).
Discussion
The production of EBV particles is necessary for understanding the biology of this virus as well as its associated diseases. Here we described the production of these particles from the P3HR-1 cell line. This cell line is not the only EBV-producer line; in fact, EBV particles have also been isolated from B95-8 cells21,22 as well as the Raji cell line18,19. The EBV lytic cycle has been induced in these cells with n-butyrate. Alternatively, phorbol esters such as 12-O-tetradecanoylphorbol-13-acetate (TPA), also known as phorbol 12-myristate-13-acetate (PMA), can be used. Overall, the protocol detailed here can be used employing these cells instead of the P3HR-1 line. However, the B95-8 line was derived from a cotton-top tamarin (Saguinus oedipus) that is currently classified as a critically endangered primate species23; hence, this line is rarely sold by vendors with restrictions on its sale and shipping. A license is also required by the convention on the international trade in endangered species of wild fauna and flora (CITES) for all orders outside the United Kingdom. While the Raji cell line is commercially available, the synthesis of replicative viral DNA and the production of viral capsid antigen (VCA) do not occur in these cells after induction by chemicals24,25. However, these can be readily induced by complementation after superinfection with EBV isolated from P3HR-1 cells20.
Due to the absence of a direct plaque assay for the enumeration of EBV particles26, we relied on a real-time polymerase-chain reaction. There are other methods of quantification that may work equally well for our purpose, such as immunofluorescence or PCR employing fluorescent probes that may offer enhanced specificity. The choice of real-time PCR using genome copy standards is based on the fact that this is readily accessible to most laboratories, less laborious, less technically challenging, and more cost-efficient27. Hence, other methods of quantification could work equally well and may be used based on availability and experience.
One of the most critical steps in the protocol detailed here is the plating of cells in step 2.1 and seeding with a cell number within the range recommended for the size of the plate. A low number of cells yields a low concentration of viral particles, while a large number of cells would crowd the plate, inhibit the growth and replication of cells, and hence slow the lytic cycle of the virus. This is why extreme care should be taken while counting the cells in step 1, calculating the required volume of suspension A, and finally plating the cells in step 2.
A limitation of the protocol is the assumption that one copy of the EBV genome corresponds to one viral particle. While this assumption is plausible, whether such particles are defective or incomplete cannot be detected using this assay. Since there are no plaque formation assays for EBV as there are for many other viruses, DNA extraction followed by real-time PCR remains an accepted method to quantify the viral preparation. Another limitation is that the virus is induced from a continuous mammalian cell line that may accumulate mutations upon many rounds of passaging. Such mutations may affect the virus itself, resulting possibly in altered in vivo or ex vivo properties of the virus. A potential solution would be to regularly sequence the cellular and viral genome. Alternatively, cells can be discarded after a few passages and the culture can be restarted anew from a liquid nitrogen-stored cell sample with a low number of previous passages. It is also worth noting that the EBV particles obtained from the P3HR-1 cell line lack the EBNA2 antigen, which is usually expressed in B lymphocytes that are latently infected with EBV, as part of six viral nuclear proteins in total28. The EBNA2 protein activates the promoters of genes that express proteins associated with the type III of EBV latency. However, recent studies have shown that EBNA2-deficient EBV is able to form tumors in CBH mice, and that the P3HR-1 cell line hence remains a useful model of EBV positive lymphomas29. On the other hand, EBV from P3HR-1 cells can be used to study the roles played by EBNA2 while using an EBNA2-positive EBV as control. In summary, EBV obtained from a continuous P3HR-1 cell line has a significant research value; it can be used to study the biology, infectivity, virulence, as well as many other properties of EBV.
Divulgations
The authors have nothing to disclose.
Acknowledgements
Funding for this work was supported by grants to ER from the Asmar Research Fund, the Lebanese National Council for Scientific Research (L-CNRS), and the Medical Practice Plan (MPP) at the American University of Beirut.
Materials
| 0.2 mL thin-walled PCR tubes | Thermo Scientific | AB0620 | Should be autoclaved before use |
| 0.2-10 µL Microvolume Filter Tips | Corning | 4807 | Should be autoclaved before use |
| 0.5-10 µL Pipette | BrandTech | 704770 | |
| 10 mL Disposable Serological Pipette | Corning | 4488 | |
| 1000 µL Filtered Pipette Tips | QSP | TF-112-1000-Q | |
| 100-1000 µL Pipette | Eppendorf | 3123000063 | |
| 100×20 mm Cuture Plates | Sarstedt | 83.1802 | |
| 10-100 µL Pipette | BrandTech | 704774 | |
| 15 mL Conical Tubes | Corning | 430791 | |
| 200 µL Filtered Pipette Tips | QSP | TF-108-200-Q | |
| 20-200 µL Pipette | Eppendorf | 3123000055 | |
| 50 mL Conical Tubes | Corning | 430828 | |
| CFX96 Real-Time C-1000 Thermal Cycler | Bio-Rad | 184-1000 | |
| DMSO | Amresco | 0231 | |
| DNase/RNase Free Water | Zymo Research | W1001-1 | |
| EBER Primers | Macrogen | N/A | Custom Made Primers |
| EBV DNA Control (Standards) | Vircell | MBC065 | |
| Ethanol (Laboratory Reagent Grade) | Fischer Chemical | E/0600DF/17 | |
| Fetal Bovine Serum | Sigma | F9665 | |
| Fresco 21 MicroCentrifuge | Thermo Scientific | 10651805 | |
| Glycogen Solution | Qiagen | 158930 | |
| Hemocytometer | BOECO | BOE 01 | |
| Inverted Light Microscope | Zeiss | Axiovert 25 | |
| iTaq Universal SYBR Green Supermix | Bio-Rad | 172-5121 | |
| Microcentrifuge Tube | Costar (Corning) | 3621 | Should be autoclaved before use |
| P3HR-1 Cell Line | ATCC | HTB-62 | |
| Penicillin-Streptomycin Solution | Biowest | L0022 | |
| Phenol | VWR | 20599.297 | |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | P8139 | |
| Pipette Filler | Thermo Scientific | 9501 | |
| Precision Wipes | Kimtech | 7552 | |
| RPMI-1640 Culture Medium | Sigma | R7388 | |
| SL 16R Centrifuge | Thermo Scientific | 75004030 | |
| Sodium Acetate | Riedel-de Haën (Honeywell) | 25022 | |
| Spectrophotomer | DeNovix | DS-11 | |
| Tris-HCl | Sigma | T-3253 | |
| Trypan Blue Solution | Sigma | T8154 | |
| Water Jacketed CO2 Incubator | Thermo Scientific | 4121 |
References
- Epstein, M. A., Achong, B. G., Barr, Y. M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1 (7335), 702-703 (1964).
- Sample, J., et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. Journal of Virology. 64 (9), 4084-4092 (1990).
- Chang, M. S., Kim, W. H. Epstein-Barr virus in human malignancy: a special reference to Epstein-Barr virus associated gastric carcinoma. Cancer Research and Treatment. 37 (5), 257-267 (2005).
- Manet, E., Schwab, M. . Encyclopedia of Cancer. , 1602-1607 (2017).
- Babcock, G. J., Decker, L. L., Volk, M., Thorley-Lawson, D. A. EBV persistence in memory B cells in vivo. Immunity. 9 (3), 395-404 (1998).
- Khan, G., Miyashita, E. M., Yang, B., Babcock, G. J., Thorley-Lawson, D. A. Is EBV persistence in vivo a model for B cell homeostasis. Immunity. 5 (2), 173-179 (1996).
- Jha, H. C., Banerjee, S., Robertson, E. S. The role of gammaherpesviruses in cancer pathogenesis. Pathogens. 5 (1), 18 (2016).
- El-Sharkawy, A., Al Zaidan, L., Malki, A. Epstein-Barr virus-associated malignancies: roles of viral oncoproteins in carcinogenesis. Frontiers in Oncology. 8, 265 (2018).
- Vereide, D., Sugden, B. Insights into the evolution of lymphomas induced by Epstein-Barr virus. Advances in Cancer Research. 108, 1-19 (2010).
- Vereide, D. T., Sugden, B. Lymphomas differ in their dependence on Epstein-Barr virus. Blood. 117 (6), 1977-1985 (2011).
- Houen, G., Trier, N. H. Epstein-Barr virus and systemic autoimmune diseases. Frontiers in Immunology. 11, 587380 (2020).
- Ortiz, A. N., et al. Impact of Epstein-Barr virus infection on inflammatory bowel disease (IBD) clinical outcomes. Revista Espanola de Enfermedades Digestivas. 114 (5), 259-265 (2021).
- Caplazi, P., et al. Mouse models of rheumatoid arthritis. Veterinary Pathology. 52 (5), 819-826 (2015).
- Kiesler, P., Fuss, I. J., Strober, W. Experimental models of inflammatory bowel diseases. Cellular and Molecular Gastroenterology and Hepatology. 1 (2), 154-170 (2015).
- Warde, N. Experimental arthritis: EBV induces arthritis in mice. Nature Reviews Rheumatology. 7 (12), 683 (2011).
- Jog, N. R., James, J. A. Epstein Barr virus and autoimmune responses in systemic lupus erythematosus. Frontiers in Immunology. 11, 623944 (2020).
- Shimizu, N., Yoshiyama, H., Takada, K. Clonal propagation of Epstein-Barr virus (EBV) recombinants in EBV-negative Akata cells. Journal of Virology. 70 (10), 7260-7263 (1996).
- Hsu, C. H., et al. Induction of Epstein-Barr virus (EBV) reactivation in Raji cells by doxorubicin and cisplatin. Anticancer Research. 22, 4065-4071 (2002).
- Nutter, L. M., Grill, S. P., Li, J. S., Tan, R. S., Cheng, Y. C. Induction of virus enzymes by phorbol esters and n-butyrate in Epstein-Barr virus genome-carrying Raji cells. Recherche en cancérologie. 47 (16), 4407-4412 (1987).
- Fresen, K. O., Cho, M. S., zur Hausen, H. Recovery of transforming EBV from non-producer cells after superinfection with non-transforming P3HR-1 EBV. International Journal of Cancer. 22 (4), 378-383 (1978).
- Glaser, R., Tarr, K. L., Dangel, A. W. The transforming prototype of Epstein-Barr virus (B95-8) is also a lytic virus. International Journal of Cancer. 44 (1), 95-100 (1989).
- Sairenji, T., et al. Inhibition of Epstein-Barr virus (EBV) release from P3HR-1 and B95-8 cell lines by monoclonal antibodies to EBV membrane antigen gp350/220. Journal of Virology. 62 (8), 2614-2621 (1988).
- Savage, A., et al. An assessment of the population of cotton-top tamarins (Saguinus oedipus) and their habitat in Colombia. PLoS one. 11 (12), 0168324 (2016).
- Kallin, B., Klein, G. Epstein-Barr virus carried by raji cells: a mutant in early functions. Intervirology. 19 (1), 47-51 (1983).
- Fresen, K. O., Cho, M. S., Hausen, H. Z. Recovery of transforming EBV from non-producer cells after superinfection with non-transforming P3HR-1 EBV. International Journal of Cancer. 22 (4), 378-383 (1978).
- Bounaadja, L., Piret, J., Goyette, N., Boivin, G. Evaluation of Epstein-Barr virus, human herpesvirus 6 (HHV-6), and HHV-8 antiviral drug susceptibilities by use of real-time-PCR-based assays. Journal of Clinical Microbiology. 51 (4), 1244-1246 (2013).
- Buelow, D., et al. Comparative evaluation of four real-time PCR methods for the quantitative detection of Epstein-Barr virus from whole blood specimens. Journal of Molecular Diagnostics. 18 (4), 527-534 (2016).
- Wu, D. Y., Kalpana, G. V., Goff, S. P., Schubach, W. H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. Journal of Virology. 70 (9), 6020-6028 (1996).
- Li, C., et al. EBNA2-deleted Epstein-Barr virus (EBV) isolate, P3HR1, causes Hodgkin-like lymphomas and diffuse large B cell lymphomas with type II and Wp-restricted latency types in humanized mice. PLoS Pathogens. 16 (6), 1008590 (2020).

