Microsampling in Targeted Mass Spectrometry-Based Protein Analysis of Low-Abundance Proteins
Summary
A protocol is presented for the determination of low-abundance biomarkers from dried serum samples exemplified with the biomarker progastrin-releasing peptide (ProGRP). Antibody-coated magnetic beads are used for the selective cleanup and enrichment of a proteotypic ProGRP peptide. The captured peptide is subsequently analyzed by liquid chromatography-tandem mass spectrometry.
Abstract
This paper presents a protocol with detailed descriptions for efficient sample cleanup of low-abundance proteins from dried samples. This is performed using bead-based proteolysis prior to proteotypic peptide affinity-capture and liquid chromatography tandem mass spectrometry (LC-MS/MS) determination. The procedure can be applied to both conventional dried samples using paper cards (e.g., dried blood spots [DBSs] and dried serum spots [DSSs]), as well as samples collected with newer sampling methods such as volumetric absorptive microsampling (VAMS). In addition to describing this procedure, the preparation of both trypsin beads and antibody-coated beads is presented in a step-by-step manner in this work. The advantages of the presented procedure are time-efficient proteolysis using beads and selective robust cleanup using peptide affinity-capture. The current procedure describes the determination of the low-abundance small-cell lung cancer (SCLC) biomarker, progastrin-releasing peptide (ProGRP), in dried serum (both DSSs and VAMS). Detailed procedures for bead preparation make it easier to implement the workflow in new applications or other laboratories. It is demonstrated that the results may be dependent on the sampling material; for the present project, higher signal intensities were seen for samples collected using VAMS compared to DSSs.
Introduction
Microsampling has been around for more than 100 years since Ivar Bang described glucose monitoring from DBSs in 19131. After Guthrie and Susi introduced DBSs in 1963 for the determination of phenylalanine in newborns2, the technique has become increasingly widespread. The first reports of DBSs for the sampling and storage of proteins were made in the early 1970s3,4, and a decade later, in the 1980s, we found the first report of mass spectrometry (MS) for the determination of proteins from DBSs5. Despite this early introduction, it was not until after the turn of the century that MS determination of proteins from DBSs and other microsampling techniques became more widespread.
In a clinical context, it is of interest to determine proteins in the diagnosis and follow-up of diseases, as well as for treatment monitoring and doping purposes. This targeted determination of protein analytes by MS from small amounts of dried samples is still challenging, and often requires extensive sample preparation prior to analysis.
Targeted quantitative determination of proteins by MS is commonly performed by applying the bottom-up approach, digesting the proteins to peptides prior to analysis. This procedure produces a myriad of peptides, which makes direct analysis of the digested biological sample challenging. A way of circumventing this is to apply a selective affinity cleanup step upfront of MS analysis either before or after the digestion6,7,8. In this way, the protein of interest (or its proteotypic peptide, if the affinity capture step is performed after the digestion) is selectively isolated from the sample matrix prior to analysis, providing lower detection limits9.
Microsampling using DBS cards has certain advantages compared to conventional blood samples, including low sample volume, less invasive sampling, and increased storage stability. However, the sample matrix is different and can introduce other challenges in the analysis (e.g., dried vs. liquid sample matrix and capillary blood vs. serum or plasma)10,11. Another challenge that is observed with DBSs is the so-called hematocrit effect, where the blood hematocrit affects the sample volume further processed for analysis, and hence introduces interindividual variability in the analysis12. Newer microsampling units, such as VAMS introduced in 201413, address this issue by collecting a fixed volume of blood instead of a blood drop.
This protocol describes a setup for the analysis of low-abundance biomarkers from dried microsamples. After elution, the dried sample is digested and, subsequently, the proteotypic peptide is isolated by peptide affinity capture. The model analyte is the SCLC biomarker ProGRP. As ProGRP cannot be determined reliably from whole blood, serum was used as the sample matrix. Representative results from both DSSs and serum samples collected using VAMS are shown.
Protocol
Serum from healthy blood donors was used for the preparation of standard solutions. The use of serum from healthy blood donors was performed in strict accordance with Norwegian law. Informed consent was obtained from all subjects. Serum samples were analyzed using methods in accordance with relevant guidelines and regulations. The protocol described is a modified version of the method described in previous work14. An overview of the composition of buffers and solutions and how to prepare them can be found in Supplemental Table S1, while the Table of Materials contains materials, equipment, and reagents used in this protocol.
1. Preparation of antibody-coated magnetic beads
- Calculate the amount of antibody necessary to prepare the desired number of beads. Use 1 mg of antibody per 50 mg of magnetic beads.
NOTE: Typically, 20 µL of a 20 mg/mL bead suspension is used per sample in subsequent experiments (see step 5.1). - Calculate the antibody volume necessary to prepare the desired number of beads.
NOTE: The antibody volume is dependent on antibody concentration and needs to be calculated for each antibody. - Transfer the desired volume of antibody solution to a 5 mL low-protein-binding microcentrifuge tube. As an example, if the antibody solution contains 1 mg/mL antibody, use 0.4 mL to prepare 1.0 mL of bead suspension (20 mg beads/mL with 1 mg antibody/50 mg beads).
NOTE: The antibodies should be in an amine-free buffer. Use low-protein-binding microcentrifuge tubes for all solutions containing proteins to avoid analyte loss due to adsorption. - Add a small magnetic stirring bar to the antibody solution and adjust the pH to 2.5 under continuous stirring. Use a pH meter with a micro electrode to measure the pH and adjust the pH by stepwise addition of defined volumes of 1.0 M HCl (start by adding 10 µL portions of 1.0 M HCl and reduce the volume as the pH approaches 2.5). Record the total volume of 1.0 M HCl necessary to adjust the pH to 2.5.
- Remove the micro electrode and incubate the acidified antibody on ice on a magnetic stirrer for 1 h.
NOTE: Acid treatment of the antibody is performed to promote the correct orientation of the antibodies on the bead. The concentration of HCl may be reduced to 0.5 M or 0.2 M HCl, depending on the buffer concentration in the antibody solution. - Neutralize the antibody solution. Use a pH meter with a micro electrode to measure the pH and adjust the pH to 7 by stepwise addition of defined volumes of 1.0 M NaOH (start with a 10 µL portion and reduce the volume as the pH approaches 7). Record the total volume of 1.0 M NaOH added.
NOTE: The concentration of NaOH should be the same as the HCl concentration used in step 1.4. NaOH is highly corrosive; use protective equipment, including gloves and appropriate eye protection. - Calculate the desired volume of tosyl activated magnetic beads to be used.
NOTE: Typically, it is recommended to use 20 mg beads/mL when performing small-scale coupling. A minimum of 5 mg of beads should be used. - Mix the magnetic bead suspension thoroughly on a vortex mixer and withdraw a volume containing the desired amount of bead suspension. As an example, to prepare 1 mL of bead suspension, withdraw a volume containing 20 mg of beads (667 µL if the concentration of beads is 30 mg/mL).
- Place the bead suspension on a magnetic rack for 1 min and remove the supernatant. Wash the beads 2x with equal volume of Type 1 H2O (667 µL if a 30 mg/mL bead solution is used to prepare 1 mL of 20 mg/mL antibody-coated beads). Mix using a vortex mixer after each addition of wash solution and place on the magnetic rack for 1 min prior to removing the supernatant.
- Add the following to the magnetic beads: acid-treated antibody, 0.5 M borate buffer (pH 9.5) (1/5 of total volume as the final concentration should be 0.1 M; this equals 0.2 mL to prepare 1 mL of bead suspension), and coupling buffer for antibody coupling (to prepare 1 mL of antibody-coated magnetic beads, the volume of coupling buffer should equal 1 mL – volume of acid treated antibody – 0.2 mL borate buffer). Mix using a vortex mixer.
NOTE: The volume of acid-treated antibody equals the volume of antibody solution (0.4 mL in preparation of 1 mL of bead suspension using a 1 mg/mL antibody solution) + the volume of 1.0 M HCl used to adjust the pH to 2.5 + the volume of 1.0 M NaOH used to adjust the pH to 7).
NOTE: The composition of 0.5 M borate buffer (pH 9.5) and coupling buffer for antibody coupling can be found in Supplemental Table S1. - Rotate at ambient temperature overnight using an end-over-end sample mixer, preferably with reciprocal rotation and vibration.
- Centrifuge for 10 min at 239 × g. Place the tube on the magnet for 2 min and remove the supernatant.
- Wash the beads 2 x 2 h and once overnight by rotating in storage buffer for antibody beads at ambient temperature using an end-over-end sample mixer. Use the same volume as the total volume in step 1.10. Place the tube on the magnet for 1 min and remove the supernatant between each wash.
NOTE: The composition of storage buffer for antibody beads can be found in Supplemental Table S1. - Store in the desired buffer (e.g., the same buffer as in step 1.13) using the desired stock concentration of beads (typically 20 mg/mL). Store in the fridge.
2. Preparation of 2 mL of trypsin immobilized beads (20 mg/mL beads)
- Add 20 mL of 1 mM HCl to 2 mL of NHS-activated agarose beads to wash the beads.
- Mix and centrifuge at 2,655 × g for 5 min. Remove the supernatant.
- Add 20 mL of 0.1 M phosphate buffer (pH 7.8). Mix using a vortex mixer and centrifuge at 2,655 × g for 5 min. Remove the supernatant.
NOTE: The composition of 0.1 M phosphate buffer (pH 7.8) can be found in Supplemental Table S1. - Add 2 mL of 20 mg/mL trypsin (dissolved in cold coupling buffer for trypsin coupling).
NOTE: The composition of coupling buffer for trypsin coupling can be found in Supplemental Table S1. Store the buffer in the fridge. - Incubate for 25 min at 22 °C and 1,100 rpm using a temperature-controlled mixer. Centrifuge at 2,655 × g for 5 min. Remove the supernatant.
- Add 2 mL of modification buffer for the preparation of trypsin beads and incubate for 20 min at 22 °C and 1,100 rpm using a temperature-controlled mixer. Centrifuge at 2,655 × g for 5 min. Remove the supernatant.
NOTE: The composition of the modification buffer for the preparation of trypsin beads can be found in Supplemental Table S1. Store the buffer in the fridge. - Add 10 mL of blocking buffer for the preparation of trypsin beads and incubate for 10 min at 22 °C and 1,100 rpm using a temperature-controlled mixer. Centrifuge at 2,655 × g for 5 min. Remove the supernatant.
NOTE: The composition of blocking buffer for the preparation of trypsin beads can be found in Supplemental Table S1. Store the buffer in the fridge. - Add 2 mL of storage buffer, mix using a vortex mixer, and store in the fridge until use.
NOTE: The composition of storage buffer for trypsin beads can be found in Supplemental Table S1. Store the buffer in the fridge.
3. DSS/VAMS sampling and subsequent extraction of dried serum
- Prepare standards by spiking the serum with ProGRP (or the protein of interest) at the appropriate level. Keep the spiking volume ≤ 1% of the total volume.
NOTE: Here, a stock solution of 295 µg/mL ProGRP in water was used for the preparation of spiked serum standards. - Mix the standards on a vortex mixer.
- Apply 10 µL of serum (spiked standards) onto DBS cards or allow 10 µL of serum to be collected by VAMS (10 µL) using the manufacturer's guidelines. For DSS, make sure that the spot is within the dotted circle.
- Allow to air dry at ambient temperature for at least 2 h.
- Cut out the whole spot and transfer it to a 2 mL low-protein-binding microcentrifuge tube or remove the VAMS from the holder and place it in a 2 mL low-protein-binding microcentrifuge tube.
- Add 1,000 µL of 100 mM ammonium bicarbonate (ABC) solution. Extract the dried serum from the spot/VAMS for 1 h at 22 °C using a temperature-controlled mixer at 1,000 rpm.
- Transfer the extract to a new 1.5 mL low-protein-binding microcentrifuge tube for tryptic proteolysis.
4. Digestion of DSS/VAMS extracts
- Use 30 µL of trypsin beads (as prepared in section 2) per sample. Transfer a volume containing trypsin beads for all samples to a new 1.5 mL low-protein-binding microcentrifuge tube, centrifuge at 2,655 × g for 5 min, and remove the supernatant.
- Wash the trypsin beads 2x with 1 mL of cold 50 mM ABC, before resuspending in the same volume that was pipetted out. Centrifuge at 2,655 × g for 5 min and remove the supernatant between steps.
- Add 30 µL of washed trypsin beads to each DSS/VAMS extract to initiate digestion.
- Incubate for 2 h at 37 °C and 1,000 rpm using a temperature-controlled mixer or similar. Centrifuge at 2,655 × g for 5 min and transfer the supernatant (not beads) to a new 2.0 mL low-protein-binding microcentrifuge tube.
- Add 25 µL of 14 ng/mL internal standard (IS) solution containing the stable isotope-labeled (SIL) peptide ALGNQQPSWDSEDSSNF[K_13C6_15N2] in 100 mM ABC solution.
5. Capture of proteotypic ProGRP peptide using antibody-coated magnetic beads
- Use 20 µL of antibody coated beads (20 mg/mL as prepared in section 1) per extraction. Transfer all antibody-coated beads to a new 1.5 mL low-protein-binding microcentrifuge tube, place on the magnet for 1 min, and remove the storage buffer.
- Wash the magnetic antibody-coated beads with 1 mL of phosphate-buffered saline (PBS), pH 7.4, containing 0.05% polysorbate 20, and 2 x 1 mL of PBS before resuspending in the same volume that was pipetted out. Mix on a vortex mixer and place on the magnet for 1 min between buffer exchanges.
NOTE: PBS contains 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 1.8 mM KH2PO4. It is recommended to prepare the solution at a 10x concentration for increased stability. For the composition of 100 mL of 10x PBS, see Supplemental Table S1. Dilution of the 10x PBS 10x will achieve a pH of ~7.4. - Add 20 µL of washed magnetic bead suspension to each low-protein-binding microcentrifuge tube containing a digested DSS/VAMS extract and IS. Perform immunoextraction for 1 h using an end-over-end sample mixer at ambient temperature.
- Wash the magnetic beads using the following solutions: 500 µL of PBS with 0.05% (v/v) polysorbate 20, 400 µL of PBS, 300 µL of 10 mM Tris HCl (pH 7.4), and 300 µL of 100 mM ABC.
- After the addition of each wash solution, remove the low-protein-binding microcentrifuge tube with beads and wash solution from the magnet rack and invert carefully until homogeneous. Then, place the low-protein-binding microcentrifuge tube on the magnet for 30 s, invert for 30 s, and place on the magnet for 1 min. Remove the wash solution.
- Spin down the remaining suspension using a microcentrifuge. Place on the magnet for 1 min and remove the remaining wash solution.
- Add 15 µL of 2% (v/v) formic acid in Type 1 H2O to each sample and incubate for 5 min at 22 °C and 1,000 rpm using a temperature-controlled mixer, or similar, to elute the captured peptides. Place on the magnet for 1 min and transfer the eluate to a new 1.5 mL low-protein-binding microcentrifuge tube. Repeat once and transfer the second eluate to the same low-protein-binding microcentrifuge tube as the first eluate.
- Add 20 µL of 100 mM ABC to each eluate (total volume of 50 µL).
- Centrifuge (microcentrifuge) and transfer 40 µL of the eluate to micro inserts for high-performance liquid chromatography (HPLC) vials.
6. Analysis by LC-MS/MS
- Use a micro LC triple quadrupole MS with electrospray ionization for high robustness.
- Prepare the LC-MS/MS system for the analysis by purging the pumps with mobile phase A (20 mM FA in H2O and MeCN, 95:5 v/v) and B (20 mM FA in H2O and MeCN, 5:95 v/v).
- Insert the analytical column (C18, 50 mm x 1 mm inner diameter [ID], 3 µm particles).
- Continue to prepare the instrument for analysis by following the start-up procedures for the specific instrument.
- In the instrument software, set the column oven temperature to 25 °C and the flow rate to 50 µL/min (100% mobile phase A).
- Equilibrate the column with mobile phase A for at least 15 min.
- Place the samples in the autosampler.
- Prepare the instrument method for analysis of the ProGRP-specific, tryptic peptide ALGNQQPSWDSEDSSNFK, and its IS.
NOTE: Several method parameters are instrument-specific and must be optimized on the specific instrument used. Details of the method parameters used here are described in steps 6.9 to 6.11. - For the gradient program in LC, use the following settings: flow rate: 50 µL/min; from time 0.0 to 3.0 min: 100% mobile phase A; from time 3.0 to 18.0 min: run a linear gradient from 0% to 50% mobile phase B; from time 18.0 to 18.1 min: increase mobile phase B from 50% to 100%; from time 18.1 to 20.0 min: 100% mobile phase B; from time 20.0 to 20.1 min: decrease mobile phase B from 100% to 0%; from time 20.1 to 30 min: 100% mobile phase A to re-equilibrate the column (use at least 10 column volumes).
- For MS/MS settings, run MS/MS in positive mode using instrument settings that ensure efficient ionization. To follow this protocol, use a spray voltage of +4,000 V, a heated capillary temperature of 270 °C, nitrogen as sheet gas (5 arbitrary units), and argon (2 mTorr) for collision-induced dissociation (CID) fragmentation. If the instrument allows, direct the LC flow to waste the first 2 min (0-2 min) and the last 1 min (29-30 min) of the run, instead of into the MS.
- Use selected reaction monitoring (SRM) transitions for (A) the signature peptide (ALGNQQPSWDSEDSSNFK): 1,005.45→913.3, 1,005.45→1,028.3, and 1,005.45→1,398.5, and (B) the IS SIL peptide (ALGNQQPSWDSEDSSNF[K_13C6_15N2]): 1,009.45→921.3, 1,009.45→1,036.3, and 1,009.45→1,406.5. Use optimized collision energy (35 V in this protocol) for all monitored transitions.
- Prepare a sequence containing the samples to be run using the software available for your LC-MS/MS system. Set the injection volume to 10 µL.
NOTE: Make sure to add blank samples and quality control samples as needed. - Press "Run sequence" in the instrument software to start the sequence.
- Monitor the peak area of the affinity-captured, ProGRP-specific, tryptic peptide ALGNQQPSWDSEDSSNFK, and its IS SIL peptide.
NOTE: Selection and confirmation of the signature peptide, ALGNQQPSWDSEDSSNFK, is described elsewhere14.
Representative Results
An overview of the analytical workflow using both DSS sampling and VAMS is shown in Figure 1. Except for the differences in sampling method, the procedures are identical. Images of the serum sampled using the two sampling methods can be seen in Figure 2.
Both sampling forms (VAMS and DSS) are suitable for the sampling of ProGRP-containing serum. This can be seen from Figure 3 where MS chromatograms of the proteotypic peptide and the IS SIL peptide from DSS and VAMS sampling are shown. In addition, the MS chromatogram after the analysis of a control sample consisting of 10 µL of spiked liquid serum sample processed in the same manner as the dried samples is included. The latter was diluted in the same volume as the volume of the DSS/VAMS extraction solution, subjected to digestion using trypsin beads and cleaned up using peptide affinity capture.
Comparing VAMS and DSS sampling (Figure 4), VAMS provides a higher proteotypic peptide/IS area ratio than DSS. This indicates that there might be a loss of the target protein ProGRP to the paper used for DSS (pure cellulose). When comparing to a control sample, where the serum is not dried prior to further processing and analysis (Figure 4), it is shown that VAMS provides similar area ratios as the control sample (two-tailed t-test, p≤ 0.65), indicating no loss to the sampling material, while DSS provides a significantly lower area ratio (two-tailed t-test, p≤ 0.005), indicating loss to the sampling material.
A brief evaluation was performed using VAMS. Linearity was shown from 10 to 1,000 ng/mL (R2 = 0.9996), with a limit of detection (LOD, S/N = 3) of 6.7 ng/mL. The LOD is considered satisfactory as the analysis was performed on a rather old triple quadrupole (2008) with a 1 mm ID column. The repeatability of all levels with an S/N > 10 was also considered satisfactory with an RSD between 7% and 17% (n = 3), using IS correction.
Affinity capture can be performed both before and after the digestion step by either capturing the protein of interest or its proteotypic peptide. The current procedure describes peptide affinity capture. An advantage of this approach compared to protein capture is that only the peptide of interest is captured, and an even more efficient sample cleanup is achieved. This is illustrated in Figure 5, showing a more complex full scan chromatogram with more noise after protein capture compared after peptide capture. The samples analyzed in Figure 5 are not sampled using DSS sampling or VAMS; however, serum is also the sample matrix, and affinity capture is performed using the same antibody used for peptide capture in the described procedure.
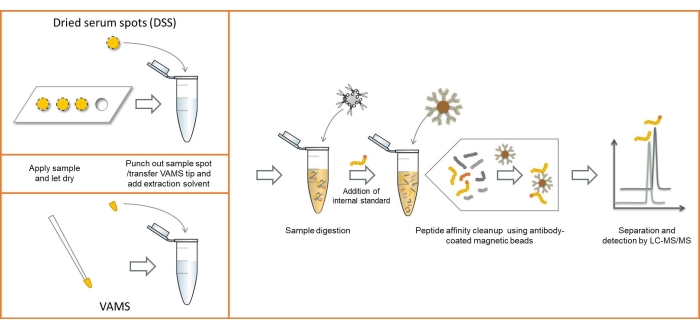
Figure 1: Overview of the analytical workflow using both DSS and VAMS sampling. Abbreviations: DSS = dried serum spot; VAMS = volumetric absorptive microsampling; LC-MS/MS = liquid chromatography-tandem mass spectrometry. Please click here to view a larger version of this figure.
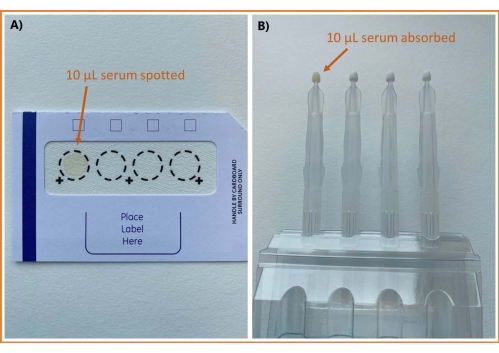
Figure 2: Images of serum sampled using different methods. (A) DBS cellulose card and (B) VAMS. Abbreviations: DBS = dried blood spot; VAMS = volumetric absorptive microsampling. Please click here to view a larger version of this figure.
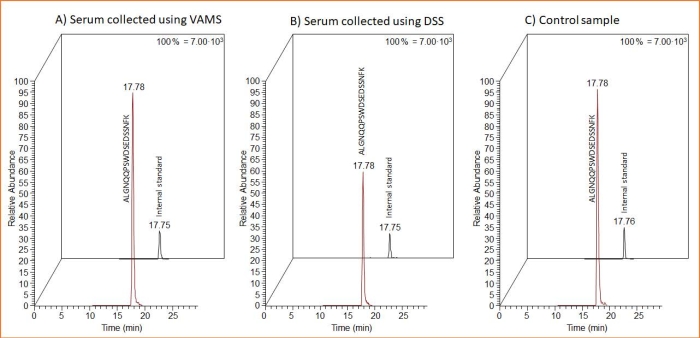
Figure 3: Representative MS chromatograms of the proteotypic peptide and the IS SIL peptide after DSS and VAMS sampling, as well as for a spiked serum sample added directly into the extraction solution. The MS chromatograms shows 10 µL serum samples spiked with 1.5 µg/mL of ProGRP and applied to (A) VAMS, (B) the cellulose sampling card (for DSS), or (C) directly to the extraction buffer (control sample). Twenty-five microliters of 14 ng/mL IS SIL peptide added to all samples prior to peptide affinity capture. Abbreviations: DSS = dried serum spot; VAMS = volumetric absorptive microsampling; MS = mass spectrometry; ProGRP = progastrin-releasing peptide; IS = internal standard; SIL = stable isotope-labeled. Please click here to view a larger version of this figure.
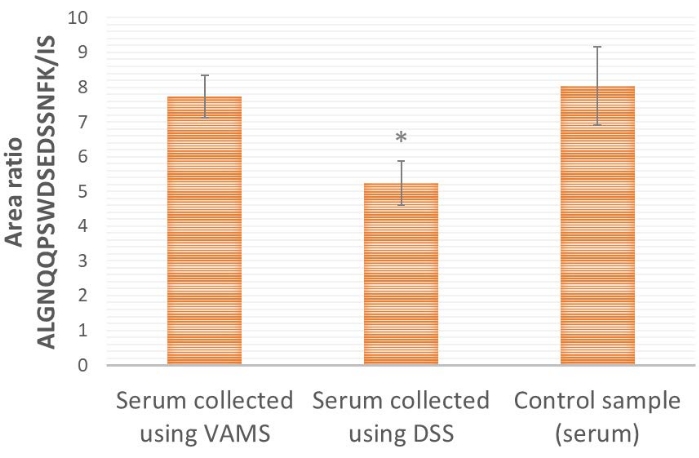
Figure 4: Representative results of the ALGNQQPSWDSEDSSNFK/IS area ratio for serum samples spiked with ProGRP and applied (10 µL) to VAMS, DSS, or directly to extraction solution (control sample). Concentration of ProGRP is 1.5 µg/mL, n = 4 for each condition; 25 µL of 14 ng/mL IS SIL peptide is added to all samples prior to peptide affinity capture. *indicates that the area ratio is significantly different from samples applied to VAMS (two-tailed t-test, p≤ 0.005). Error bars are ± standard deviation. Abbreviations: DSS = dried serum spot; VAMS = volumetric absorptive microsampling; ProGRP = progastrin-releasing peptide; IS = internal standard; SIL = stable isotope-labeled. Please click here to view a larger version of this figure.
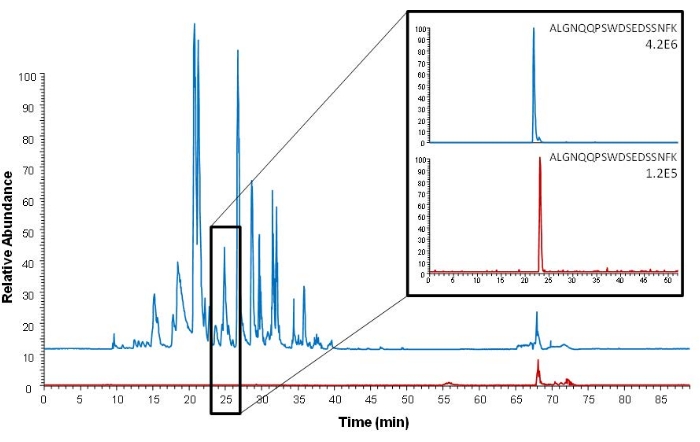
Figure 5: Comparison of base peak chromatograms (full scan Orbitrap analysis) after intact protein extraction (blue) and proteotypic epitope peptide extraction (red). Extracted ion chromatograms of the proteotypic epitope peptide (ALGNQQPSWDSEDSSNFK, m/z 1005.45) are shown on the right. Serum spiked with 150 ng mL−1 ProGRP was used as the sample. This figure is reprinted from Levernæs et al.14. Please click here to view a larger version of this figure.
Supplemental Table S1: An overview of the composition of buffers and solutions and how to prepare them. Please click here to download this File.
Discussion
The described protocol contains information about how to conduct several important steps in the analysis of low-abundance biomarkers from dried microsamples (DSS and VAMS), including the preparation of trypsin beads and antibody-coated magnetic beads. Based on previous experience, we always treat the antibody with acid prior to bead immobilization to improve the orientation of the antibodies15.
One of the critical steps in this procedure is the selection of the most suitable microsampling format. First, one must consider if the analyte in question can be determined from whole blood, or if the concentration is influenced by the blood cells and has to be determined in serum or plasma (as for the model analyte, ProGRP).
Both paper- and polymer-based approaches have advantages and limitations; for ProGRP, VAMS provides a clear advantage with respect to analyte recovery after extraction from the sampler. This can, however, probably be optimized using a different extraction solution for the DSS samples. Nevertheless, this potential interaction between the analyte and the sampling material is important to take into account as it might result in increased analytical variation and higher detection limits. As the IS used is an SIL peptide and first added after the digestion, IS corrects for the steps following digestion (e.g., affinity extraction and LC-MS/MS analysis). IS correction is not possible for extraction from DSS/VAMS and the digestion step.
Two types of beads are used in the procedure: trypsin beads for digestion after extraction of the serum sample from the sampler, and antibody-coated magnetic beads for capture of the proteotypic peptide after digestion. A major reason for using trypsin beads, in addition to speeding up the digestion, is to minimize residual trypsin activity in the sample during affinity capture. This is important to avoid tryptic proteolysis of the mAb during affinity capture.
Agarose beads were used for preparation of the trypsin beads, while magnetic beads were used for preparation of the antibody-coated beads. Agarose beads are less expensive than magnetic beads but have a limitation that separation of the beads from the solution requires centrifugation. This makes the separation of beads and supernatant less efficient than when using magnetic beads. In addition, automation of the workflow is difficult using agarose beads. However, magnetic NHS-activated beads are available and can be used for a more streamlined and automatable sample preparation workflow.
Microsampling is an important trend in the bioanalysis of both drugs and biomarkers. One challenge with the current approach is the limited amount of sample volume (10 µL), which may be of particular importance in the determination of very-low-abundance analytes such as ProGRP (pg/mL-low ng/mL level). However, this challenge may be circumvented using state-of-the-art analytical equipment. For these low-abundance analytes, the choice of sample preparation is crucial, and selective sample cleanup through antibody-based affinity capture is most often needed. As peptide capture has been shown to provide cleaner extracts and lower detection limits than protein capture (using the same antibody)14, the present method focuses on this approach in combination with microsampling. Another advantage of the peptide capture approach is that the IS SIL peptide also corrects for the affinity capture step.
In this work, an antibody targeting a protein was used for peptide capture. This is an advantage as the availability of off-the-shelf antibodies targeting proteins are higher than off-the-shelf antibodies targeting proteotypic peptides. However, for an anti-protein antibody to efficiently capture a proteotypic peptide, the epitope needs to be intact after protein digestion. In addition, for many antibodies, the exact epitope is not known, making the search for an anti-protein antibody tedious. This limits the number of available anti-protein antibodies applicable for peptide capture. The described procedure is demonstrated using serum as the matrix and ProGRP as the target analyte. The procedure is intended to be applicable to other matrixes and other target analytes. Instead of using a commercially available anti-protein antibody for the affinity capture of the proteotypic peptide, it is possible to also use custom-made, anti-peptide antibodies. The cleanup efficiency of peptide capture compared to protein capture is illustrated in Figure 5. By exchanging the agarose beads used for the preparation of trypsin beads with magnetic beads, the procedure should also be compatible with robotic sample preparation work stations on the market.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We greatly acknowledge Prof. Elisabeth Paus at the Norwegian Radium Hospital (Oslo University Hospital, Oslo, Norway) for providing the ProGRP standard and the anti-ProGRP monoclonal antibody, M18. The publication fee was covered by a grant from Apoteker Harald Conrad Thaulows legat. Trine Grønhaug Halvorsen and Léon Reubsaet are partners in the National Network of Advanced Proteomics Infrastructure (NAPI) consortium, which is funded by the Research Council of Norway INFRASTRUKTUR-program (project number: 295910).
Materials
| Acetic acid N-hydroxysuccinimide ester | Carbosynt (Staad, Switzerland) | FA33719 | Store in freezer below -20 °C |
| ALGNQQPSWDSEDSSNF[K_13C6 _15N2] (≥ 95%) |
Innovagen (Lund, Sweden) | Not applicable | Store in freezer below -20 °C |
| Ammonium bicarbonate BioUltra (≥ 99.5% ) | Sigma Aldrich (St. Louis, MO, USA) | 09830-500G | |
| Aquasil C18 column, 3 µm, 50 mm x 1 mm | Thermo scientific (Waltham, MA, USA) | 77503-051030 | Analytical column compatible with 100% aqueous mobile phase |
| Benzamidine (≥ 95.0% ) | Sigma Aldrich (St. Louis, MO, USA) | 12072 | Store in fridge at 2-6 °C |
| Calsium chloride dihydrate (≥ 99% ) | Sigma Aldrich (St. Louis, MO, USA) | 223506-500G | |
| Centrifuge 5804 | Eppendorf (Hamburg, Tyskland) | 5804000010 | |
| Cloned ProGRP isoform 1 | Radium hospital, Oslo University Hospital (Oslo, Norway) | Not applicable | Store in freezer below -20 °C |
| Disodium hydrogenphosphate dihydrate (pro analysis) | Sigma Aldrich (St. Louis, MO, USA) | 30435-500G | |
| Disodium hydrogenphosphate dodecahydrate (pro analysis) | Merck (Darmstadt, Tyskland) | 1.06579.0500 | |
| Dynabeads M-280 tosylactivated 10 mL | Invitrogen (Carlsbad, USA) | 14204 | Store in fridge at 2-6 °C |
| DynaMag-2 | Invitrogen (Carlsbad, USA) | 123-21D | |
| Ethanolamine (pro analysis, ≥ 99%) | Sigma Aldrich (St. Louis, MO, USA) | #02400 | |
| Formic acid (≥ 99% ) for LC-MS | VWR International (Radnor, PA, USA) | 84865.260 | |
| FTA DMPK-C cellulose card | Whatman (Kent, UK) | WB129243 | DBS card |
| HPLC vials, clear glass, 1.5 mL, 32 x 11.6 mm, Clean Pack | Nerliens Meszansky (Oslo, Norge) | LPP 11 09 0519 | |
| Hulamixer sample mixer | Invitrogen (Carlsbad, USA) | 101561503016 | Sample mixer with end-over-end mixing and reciprocal rotation and vibration |
| Human serum from healty blood donors | Bloodbank, Ullevål, Oslo University Hospital (Oslo, Norway) | Not applicable | Store in freezer below -20 °C |
| Hydrochloric acid fuming 37% (Emsure for analysis) | Merck (Darmstadt, Tyskland) | 1.00317.1000 | |
| LC-MS/MS system: Ultimate 3000 system (Autosampler, WPS-3000TRS; Micropump, LPG-3400M; Flow manager, FLM-3300, MIC, 1X2P-10P) and TSQ Quantum access. Controlled by Xcalibur 2.2 SP1.48 | Thermo scientific (Waltham, MA, USA) | Not applicable | |
| LiChrosolv Acetonitrile hypergrade for LC-MS | Merck (Darmstadt, Tyskland) | 1.00029.2500 | |
| LL Biotrode, Combined glass electrode | Metrohm (Herisau, Sveits) | 6.0224.100 | |
| Magnetic stirrer, Type M10 | Franz Morat KG (Eisenbach, Germany) | 10236 | |
| Micro inserts, glass (31 x 6 mm, 0.1 mL) | VWR International (Radnor, PA, USA) | 548-0006 | |
| MilliQ integral 3 with Q-POD | Merck Millipore (Molsheim, France) | ZRXQ003T0 | For production of Type 1 water |
| monoclonal antibody M18 | Radium hospital, Oslo University Hospital (Oslo, Norway) | Not applicable | Store in fridge at 2-6 °C |
| Neoteryx Mitra microsampler (10 μL) 4 sampler Clamshell | Fisher Scientific (Waltham, MA, USA) | NC1382947 | |
| NHS-activated sepharose beads 4 fast flow | Sigma Aldrich (St. Louis, MO, USA) | GE17-0906-01 | Agarose beads, store in fridge at 2-6 °C |
| Optifit, Refill pipet tips, 10 mL | Sartorius Biohit (Helsinki, Finland) | 613-2911 | |
| Optifit, Refill pipet tips, 10 μL | Sartorius Biohit (Helsinki, Finland) | 790012 | |
| Optifit, Refill pipet tips, 1,000 μL | Sartorius Biohit (Helsinki, Finland) | 791002 | |
| Optifit, Refill pipet tips, 200 μL | Sartorius Biohit (Helsinki, Finland) | 790202 | |
| pH glass electrode | Metrohm (Herisau, Sveits) | 6.0233.100 | |
| pH meter 744 | Metrohm (Herisau, Sveits) | 8.744.1003 | |
| Pipet 10 mL | Sartorius Biohit (Helsinki, Finland) | 725090 | |
| Pipet m10 µL | Sartorius Biohit (Helsinki, Finland) | 725020 | |
| Pipet m100 µL | Sartorius Biohit (Helsinki, Finland) | 725050 | |
| Pipet m1,000 µL | Sartorius Biohit (Helsinki, Finland) | 725070 | |
| Pipet m20 µL | Sartorius Biohit (Helsinki, Finland) | 725030 | |
| Potassium chloride (KCl ≥ 99.9%) | Sigma Aldrich (St. Louis, MO, USA) | P-3911 | |
| Potassium dihydrogenphosphate (pro analysis) | Merck (Darmstadt, Tyskland) | 1.04873.0250 | |
| Protein LoBind Eppendorf tube 0.5 mL | Eppendorf (Hamburg, Tyskland) | 525-0133 (0030 108.094) | |
| Protein LoBind Eppendorf tube 1.5 mL | Eppendorf (Hamburg, Tyskland) | 525-0132 (0030 108.116) | |
| Protein LoBind Eppendorf tube 2.0 mL | Eppendorf (Hamburg, Tyskland) | 525-0134 (0030 108.450) | |
| Protein LoBind Eppendorf tube 5.0 mL | Eppendorf (Hamburg, Tyskland) | 525-0792 (0030108.302) | |
| Scissors | Sigma Aldrich (St. Louis, MO, USA) | Z186716-1EA | |
| Sodium azide (BioUltra; ≥ 99.5% ) | Sigma Aldrich (St. Louis, MO, USA) | 71289-5G | |
| Sodium chloride (for analysis) | Merck (Darmstadt, Tyskland) | 1.06404.1000 | |
| Sodium dihydrogenphosphate monohydrate (pro analysis) | Merck (Darmstadt, Tyskland) | 1.06346.0500 | |
| Sodium hydroxide (AnalaR NORMAPUR) | VWR International (Radnor, PA, USA) | 28244.295 | |
| Sodium tetraborate decahydrate (≥ 99. %) | Sigma Aldrich (St. Louis, MO, USA) | S9640-500G | |
| Spectrafuge Mini Centrifuge | LABNET International (Edison, NJ, USA) | C1301 | |
| Stirring magnet, 25 mm x 6 mm Ø, circular | Leybold (Cologne, Germany) | 666 851 | |
| Stuart Scientific SA8 vortex mixer | Stuart (Staffordshire, UK) | Z648531-1EA | |
| SuperClear centrifuge tubes (15 mL) | VWR International (Radnor, PA, USA) | 525-0150 | |
| SuperClear centrifuge tubes (50 mL) | VWR International (Radnor, PA, USA) | 525-0155 | |
| Thermomixer comfort 1.5 mL | Eppendorf (Hamburg, Tyskland) | 53,55,27,831 | Temperature controlled mixer |
| Trizma base (reagent grade, ≥ 99.0 %) | Sigma Aldrich (St. Louis, MO, USA) | T6066 | Tris(hydroxymethyl)aminometan (tris) |
| Trizma HCl (reagent grade, ≥ 99.0%) | Sigma Aldrich (St. Louis, MO, USA) | T3253-100G | Tris(hydroxymethyl)aminometan HCl (tris HCl) |
| Trypsin (TCPK-treated from bovine pancreas, 10,000-15,000 BAEE units/mg Protein) | Sigma Aldrich (St. Louis, MO, USA) | T8802 | Store in freezer below -20 °C |
| Tween 20 | Sigma Aldrich (St. Louis, MO, USA) | P7949-500ML | polysorbate 20 |
| Tweezers | Sigma Aldrich (St. Louis, MO, USA) | TEM-78511-27 | |
| Vial caps, white, 9 mm | Nerliens Meszansky (Oslo, Norge) | LPP 09 15 0981 | |
| Mitra microsampler with VAMS (Volumetric adsorptive microsampling) technology, 10 µL, 4-sampler clamshell | Neoteryx (Torrance, CA, USA) |
References
- Bang, I. B. Fresenius. Zeitschrift für analytische Chemie. 52 (7), 521-523 (1913).
- Guthrie, R., Susi, A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 32 (3), 338-343 (1963).
- Laurell, C. B. A screening test for α1-antitrypsin deficiency. Scandinavian Journal of Clinical and Laboratory Investigation. 29 (3), 247-248 (1972).
- Thielmann, K., Aquino, A. M. Whole blood samples dried and stored on filter paper as substrate for the electrophoretic separation on hemoglobin S from hemoglobin A. A screening procedure. Clinica Chimica Acta. 35 (1), 237-238 (1971).
- Wada, Y., Fujita, T., Kidoguchi, K., Hayashi, A. Fetal hemoglobin variants in 80,000 Japanese neonates: High prevalence of Hb F Yamaguchi (AγT 80 Asp→Asn). Human Genetics. 72 (3), 196-202 (1986).
- Halvorsen, T. G., McKitterick, N., Kish, M., Reubsaet, L. Affinity capture in bottom-up protein analysis-overview of current status of proteolytic peptide capture using antibodies and molecularly imprinted polymers. Analytica Chimica Acta. 1182, 338714 (2021).
- Halvorsen, T. G., Reubsaet, L. Antibody based affinity capture LC-MS/MS in quantitative determination of proteins in biological matrices. TrAC Trends in Analytical Chemistry. 95, 132-139 (2017).
- Neubert, H., et al. Protein biomarker quantification by immunoaffinity liquid chromatography-tandem mass spectrometry: Current state and future vision. Clinical Chemistry. 66 (2), 282-301 (2020).
- Pan, S., et al. Mass spectrometry based targeted protein quantification: methods and applications. Journal of Proteome Research. 8 (2), 787-797 (2008).
- Enderle, Y., Foerster, K., Burhenne, J. Clinical feasibility of dried blood spots: Analytics, validation, and applications. Journal of Pharmaceutical and Biomedical Analysis. 130, 231-243 (2016).
- Londhe, V., Rajadhyaksha, M. Opportunities and obstacles for microsampling techniques in bioanalysis: Special focus on DBS and VAMS. Journal of Pharmaceutical and Biomedical Analysis. 182, 113102 (2020).
- Denniff, P., Spooner, N. The effect of hematocrit on assay bias when using dbs samples for the quantitative bioanalysis of drugs. Bioanalysis. 2 (8), 1385-1395 (2010).
- Denniff, P., Spooner, N. Volumetric absorptive microsampling: A dried sample collection technique for quantitative bioanalysis. Analytical Chemistry. 86 (16), 8489-8495 (2014).
- Levernæs, M. C. S., et al. Immunocapture sample clean-up in determination of low abundant protein biomarkers-a feasibility study of peptide capture by anti-protein antibodies. RSC Advances. 9 (60), 34902-34911 (2019).
- Conradie, J. D., Govender, M., Visser, L. Elisa solid phase: Partial denaturation of coating antibody yields a more efficient solid phase. Journal of Immunological Methods. 59 (3), 289-299 (1983).

