Quantitative Polymerase Chain Reaction (qPCR)-Based Rapid Diagnosis of Helicobacter pylori Infection and Antibiotic Resistance
Summary
The protocol presents a noninvasive method for the rapid diagnosis of Helicobacter pylori stomach infections through the string test and determines its antibiotic resistance to clarithromycin and levofloxacin using quantitative polymerase chain reaction (qPCR).
Abstract
Helicobacter pylori is a major human pathogen that infects approximately half of the global population and is becoming a serious health threat due to its increasing antibiotic resistance. It is the causative agent of chronic active gastritis, peptic ulcer disease, and gastric cancer and has been classified as a Group I Carcinogen by the International Agency for Research on Cancer. Therefore, the rapid and accurate diagnosis of H. pylori and the determination of its antibiotic resistance are important for the efficient eradication of this bacterial pathogen. Currently, H. pylori diagnosis methods mainly include the urea breath test (UBT), the antigen test, the serum antibody test, gastroscopy, the rapid urease test (RUT), and bacterial culture. Among them, the first three detection methods are noninvasive, meaning they are easy tests to conduct. However, bacteria cannot be retrieved through these techniques; thus, drug resistance testing cannot be performed. The last three are invasive examinations, but they are costly, require high skills, and have the potential to cause damage to patients. Therefore, a noninvasive, rapid, and simultaneous method for H. pylori detection and drug resistance testing is very important for efficiently eradicating H. pylori in clinical practice. This protocol aims to present a specific procedure involving the string test in combination with quantitative polymerase chain reaction (qPCR) for the rapid detection of H. pylori infection and antibiotic resistance. Unlike bacterial cultures, this method allows for easy, rapid, noninvasive diagnosis of H. pylori infection status and drug resistance. Specifically, we used qPCR to detect rea for H. pylori infection and mutations in the 23S rRNA and gyrA genes, which encode resistance against clarithromycin and levofloxacin, respectively. Compared to routinely used culturing techniques, this protocol provides a noninvasive, low-cost, and time-saving technique to detect H. pylori infection and determine its antibiotic resistance using qPCR.
Introduction
H. pylori is a spiral-shaped, highly motile, gram-negative bacterium that mainly lives in the pylorus region of the stomach1. It is a common pathogen that infects nearly 50% of the global population2. Most people with H. pylori infection have no clinical manifestations, and most develop different diseases after several years of infection, including chronic gastritis, peptic ulcers, gastric ulcers, and gastric cancer3. In several studies based on different populations, the efficacy of eliminating H. pylori for preventing stomach cancer and precancerous lesions has been demonstrated4,5. Therefore, the World Health Organization (WHO) International Agency for Research on Cancer has advised H. pylori eradication as a preventative measure6.
The use of noninvasive methods to identify H. pylori infection is a key component of treatment for most individuals with asymptomatic dyspepsia. The urea breath test (UBT), H. pylori fecal antigen test (SAT), and serological testing are popular noninvasive techniques. Among these, the UBT is the least intrusive and most accurate procedure available. UBT uses urease, abundantly present in H. pylori, to hydrolyze isotopically labeled urea into ammonia and carbon dioxide (13C or 14C). In contrast, the immunochromatographic assay (ICA)7 is convenient, simple, and noninvasive for sampling. However, the accuracy of the test is affected by several factors, such as the quality of the stool sample, the temperature, and the interval between the sample collection and testing. Another test based on the immune response is the serum H. pylori antibody test, which detects antibodies in a patient's serum. However, this test is not suitable for post-treatment analysis since the antibodies remain long after the bacteria have been cleared8. Another major drawback is that these methods only diagnose H. pylori infection and do not allow for drug resistance testing to guide sensitivity-based treatment.
For invasive testing methods, gastric biopsy tissue needs to be taken by endoscopy and then subjected to histology, the urease rapid test, and bacterial culture. These testing methods are also very limited due to several factors. Currently, these techniques are limited to elderly patients, patients at high risk for precancerous or malignant disease, and patients who have failed first-line therapy for gastroesophageal reflux disease or H. pylori infection9. Secondly, due to the unique growth characteristics of H. pylori, the success rate of bacterial culture only reaches 50%10. Thus, molecular detection methods offer new hope to overcome the high demands of invasive detection methods and guide sensitivity-based treatment. Among molecular detection methods, quantitative PCR has evolved tremendously in recent years. qPCR, unlike traditional PCR, does not require gel electrophoresis and accurately quantifies DNA/RNA in samples by adding primers and probes at the annealing stage. qPCR kits for the detection of H. pylori infection and drug resistance are now commercially available. Nevertheless, each method has its limitations; therefore, a patient's clinical diagnosis and treatment should be considered in conjunction with their symptoms, signs, history, other laboratory tests, and response to treatment.
Currently, the primary method of treating H.pylori infections is taking antibiotics, but lately, it is becoming increasingly difficult to treat these infections due to the rise in antibiotic resistance. Subsequently, a significant decline in H. pylori treatment efficacy has been observed globally, making H. pylori eradication a major public health issue11.
Clarithromycin and levofloxacin are the two broad-spectrum antibiotics used to treat infections caused by H.pylori, but several studies have reported widespread resistance against these two drugs in H.pylori isolates. A2143G, A2142G, and A2142C are three of the numerous point mutations found in the 2.9 kb 23S rRNA gene that result in clarithromycin resistance by preventing the macrolide from binding. At the same time, the mutation loci of the levofloxacin resistance gene are mainly located in the six mutation sites (A260T, C261A, T261G, G271A, G271T, A272G) of the gyrA gene12. The discovery of these resistance mechanisms based on genetic mutations has led to a gradual shift in the detection of H. pylori through cultural-based studies to molecular testing.
Overall, there is an urgent clinical need for a noninvasive, effective, and simultaneous diagnostic method for the detection of H. pylori infections and drug resistance. We adopted a combined string test and qPCR method to overcome the difficulties of sampling and achieve the goal of the simultaneous detection of H. pylori infection and drug resistance using different primer probes.
Protocol
The present study was conducted in conformity with ethical considerations established by the ethical committee of Guangdong Provincial People's Hospital, Southern Medical University, Guangzhou, China (Approval Number: KY-Q-2022-384-02). Patients in the age range of 18-60 years old were included in this study. Patients taking antibiotics, antibacterial Chinese medicines, drugs such as proton pump inhibitors (PPI), or H2 receptor antagonists, etc., within 2 weeks prior to testing were not included in this study. Those patients who had received treatment against H.pylori in the past 3 months were also excluded from this study. Those with serious heart, liver, kidney problems, severe neuropathy, or mental illness were also not allowed to participate in this study. There were no pregnant women and lactating mothers included in this study. Details of the supplies (reagents, chemicals, equipment, and software) used in this study are given in the Table of Materials.
1. String test for gastric fluid sampling
- Ask the patient to fast overnight before the sample collection.
- The next day, open the string test kit, take the capsule, and hold the loop at the end of the string.
- Tape the string onto the patient's cheek, and ask them to place the capsule on their tongue near the back of the throat and swallow it with a sip of water.
- Allow the capsule to be dissolved in the patient's stomach, thus exposing the string within the capsule to absorb gastric mucus.
- Ask a trained assistant to pull out the string carefully 1 h after the patient swallowed the capsule.
- Cut off the lower end of the string (40 cm) soaked in the gastric fluid, place it in the TSE (Tris/saline/EDTA) sample preservation solution, and then send it to the clinical laboratory at room temperature (RT) for the subsequent detection of H. pylori and the determination of the antibiotic resistance profiles using qPCR.
2. DNA extraction
- Place the specimen-carrying collection tubes on a test tube rack, properly labeled, and vortex them for 10 s.
- Turn the 32-well plate (nucleic acid extraction or purification kit) upside down several times to resuspend the magnetic beads. After a short vortex for 10 s, carefully remove the aluminum foil sealing from the plate.
- Transfer 200 µL of gastric fluid from each sample and H. pylori DNA as a positive control into the separate wells.
- Place the 32-well plate in the corresponding sample slot of the nucleic acid extractor machine for automatic DNA extraction.
- After DNA extraction and confirmation, initiate the antibiotic resistance testing for clarithromycin and levofloxacin through qPCR on the positive samples.
- Place the excessive gastric fluid and extracted DNA samples at −20 °C for long-term storage and future use.
- Perform all these steps in a biosafety cabinet to avoid any contamination.
3. qPCR for the detection of H. pylori and resistance to antibiotics (clarithromycin and levofloxacin)
- Perform qPCR for the detection of H. pylori and the suspected antibiotic resistance mutations in its genome.
- Thaw the qPCR reaction mixture (H. pylori nucleic acid detection kit) on ice, and mix it by flicking and spinning to prevent any loss of reagent.
- For each sample on a 32-well plate, mix 20 µL of PCR reaction mixture (genotyping reaction premix [containing the Taq enzyme, deoxyribonucleoside triphosphate, reaction buffer, and magnesium chloride], with ureA forward and reverse primers, and a ureA probe), and 5 µL of the extracted DNA.
- Run the 32-well qPCR plate on the qPCR machine. Program the thermal cycler: the reaction mixture will be first reacted sequentially at 42 °C and 95 °C (both for one cycle) for 2 min each; then, 40 cycles at 95 °C, denaturation for 10 s, and annealing and extension at 58 °C for 45 s.
- Set the fluorescence signal acquisition to FAM (H. pylori) and the data acquisition to the amplification extension period. After the reaction is completed, save the data for future results analysis.
- Analyze the data using specific software for qPCR, as the instrument will automatically select baseline thresholds.
- If the test result for H. pylori is positive, the machine will automatically start drug resistance testing on the sample. Replace the reagent kit with detection reagents for 23S rRNA gene and gyrA gene mutations in the H. Pylori (qPCR) kit.
NOTE: The reagent kit (detection reagents for 23S rRNA gene and gyrA gene mutations in Helicobacter pylori [qPCR]) includes the PCR reaction mixture (genotyping reaction premix [containing the Taq enzyme, deoxyribonucleoside triphosphate, reaction buffer, magnesium chloride], primers, and probes). All the negative quality control products are sterile, purified water. The positive quality control product in the H. pylori nucleic acid detection kit is inactivated H. pylori standard beads (ATCC 43504). The strong H. pylori-positive and weak positive quality control products in the kit (detection reagents for 23S rRNA gene and gyrA gene mutations in H. pylori) are the DNA of the inactivated H. pylori strain containing the target gene and the mutant gene. Similarly, based on the actual situation, all diagnostic standards, including the presence of H. pylori infection or drug resistance, are set to CT ≤ 35 and have a typical S-shaped curve. The concentration of the weak quality control is 1.0 x 103 copies/mL, while the concentration of the strong quality control is 1.0 x 108 copies/mL.
Representative Results
Detection of H. pylori infection and antibiotic resistance in stomach fluid by qPCR
We performed qPCR for the detection of H. pylori infection by amplifying the ureA gene and determined its antibiotic resistance profile by targeting point mutations in the 23S rRNA gene and gyrA gene (Table 1). The quality control CT values in all three groups of the qPCR experiments were within the recommended range, indicating that the samples were all in a normal state at the time of the experiment and that the test results were reliable. In this study, five samples with different test results (S1-S5) were selected to characterize the reliability of the experimental protocol. S1 represents a representative strain of H. pylori without infection, while the S2-S5 samples are those infected with H. pylori with different resistance results (Figure 1). We set the system not to perform further resistance testing with the samples not infected with H. pylori, so the S1 sample did not enter the resistance test after the system test showed a negative result for H. pylori. In terms of the samples positive for H. pylori infection, the S2 CT values were all within the detection range, indicating that the sample was H. pylori-positive and showed dual resistance to clarithromycin and levofloxacin, and clinicians were recommended to choose other methods for treatment at their discretion. The S3 CT values were within the detection range for H. pylori infection and the levofloxacin resistance test, while no CT values were detected in the clarithromycin resistance test, indicating that the S3 sample was from a levofloxacin-resistant patient. Similarly, the CT value of the S4 sample was within the detection range for H. pylori infection and clarithromycin resistance, while no CT value was detected for levofloxacin resistance, indicating that this patient was resistant to clarithromycin, and it was recommended that they take levofloxacin for treatment. Finally, the S5 sample test showed CT values within the detection range only for the detection of H. pylori infection, indicating that this patient was sensitive to both antibiotics and could be treated using either of the two drugs. Compared to the bacterial culture method, which also detects H. pylori infection and drug resistance, this method is safe and effective in detecting H. pylori infection and drug resistance without causing damage to the patient and can be used to guide the doctor in formulating an appropriate treatment plan.
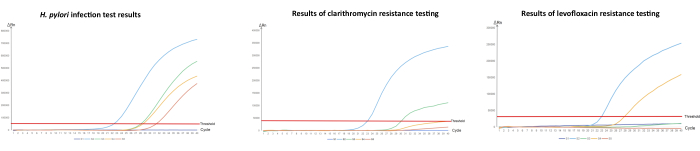
Figure 1: Detection of H. pylori and its antibiotic resistance in stomach fluid by qPCR.
(A) Quantitative PCR amplification of H. pylori infection, (B) detection of clarithromycin resistance, and (C) detection of levofloxacin resistance. "S" stands for sample. "S1" is a sample that came back negative for H. pylori infection and was also tested for antibiotic resistance; "S2" is an H. pylori-infected sample with resistance to both clarithromycin and levofloxacin; "S3" is also an H. pylori-positive sample but is clarithromycin-susceptible and levofloxacin-resistant; "S4" is also an H. pylori-positive sample but is clarithromycin-resistant and levofloxacin-susceptible; "S5" is an H. pylori-positive sample but is susceptible to both clarithromycin and levofloxacin. The concentration of the weak quality control is 1.0 x 103 copies/mL, while the concentration of the strong quality control is 1.0 x 108 copies/mL. Please click here to view a larger version of this figure.
| Sample | H. pylori | Clarithromycin | Levofloxacin | |||
| +/- | CT | +/- | CT | +/- | CT | |
| S1 | – | U | – | U | – | U |
| S2 | + | 22.61 | + | 22.77 | + | 23 |
| S3 | + | 28.32 | – | U | + | 30.18 |
| S4 | + | 28.76 | + | 27.67 | – | U |
| S5 | + | 31.59 | – | U | – | U |
Table 1: Table showing the qPCR results of the detection of H. pylori infection and resistance to clarithromycin and levofloxacin. This table presents the qualitative results for H. pylori infection, the detection of 23S rRNA gene mutations showing that the isolate is resistant to clarithromycin, and the detection of gyrA gene mutations showing that the isolate is resistant to levofloxacin. +/−, qualitative result; +, positive result; −, negative result.
Discussion
H. pylori detection can be performed using both invasive and noninvasive methods13. Commonly used invasive techniques such as histopathology, the rapid urease test, polymerase chain reaction (PCR), and bacterial culturing require endoscopy and biopsy.Serological tests, urea breath tests, and enzyme-linked immunosorbent assays (ELISA) are recommended among the noninvasive procedures14. While noninvasive methods are easy to perform, economical, and more comfortable for patients, the isolation and culturing of H. pylori to perform additional assays, such as strain identification and antibiotic susceptibility testing, require invasive tests. With the current rise in antibiotic resistance, previously used antibiotics are becoming ineffective and, thus, failing to treat infection caused by drug-resistant H. pylori isolates. In this case, retrieving H. pylori to perform antibiotic susceptibility testing may be required to select the antibiotics that will most likely successfully eliminate the resistant bacteria.
Routinely conducted culture-based susceptibility assays such as the agar dilution method and Epsilometer test (E-test) have several limitations: they are time-consuming, since H. pylori is a slow-growing bacteria, costly, skill-based, and require invasive techniques. Alternately different molecular-based techniques, such as fluorescence in situ hybridization (FISH) techniques and qPCR, can be used to identify several mutations, such as in the 23S rRNA gene, which encodes resistance against clarithromycin15,16. Three point mutation sites (A2142G, A2143G, A2142C) of the 23S rRNA resistance gene of H. pylori and six point mutation sites (A260T, C261A, T261G, G271A, G271T, A272G) of the gyrA resistance gene of quinolone antibiotics were selected for designing primers and probes to determine resistance against clarithromycin and levofloxacin, respectively.
The string test, which originated from the ENTEO-TEST, uses a capsule attached to a highly absorbent nylon string, which is swallowed to collect gastric secretions17. Currently, string tests have been used to diagnose tuberculosis18, to differentiate highly virulent Klebsiella pneumoniae (hvKp) from traditional Klebsiella pneumoniae19, to identify Gram-positive and Gram-negative bacteria and yeast, and to diagnose H. pylori from gastric fluid20. In this study, we combined the string test, a minimally invasive technique, with qPCR for the identification of H. Pylori infections in a clinical setup and performed susceptibility testing by targeting previously reported resistance-encoding mutations in the H. pylori genome. We chose the ureA gene because it is a housekeeping gene unique to H. pylori with no potential risk of cross-reaction with other organisms. All H. pylori isolates must have the ureA gene in order to survive in the human stomach, and knock-out experiments have shown that H. pylori without the ureA gene does not have the ability to colonize in the stomach.
For qPCR, quality control standards are extremely important. In the qPCR detection of H. pylori, the requirements of the quality control standards are as follows: a negative quality control product (no increase in fluorescence signal in the FAM detection pathway, no typical S-type amplification curve, CT value > 35.00 or no obvious signal); a positive quality control product (fluorescence signal growth curve of the FAM detection pathway showing an S-shaped curve, CT value ≤ 35.00), and the above requirements must be met simultaneously; otherwise, the test will be considered invalid and needs to be conducted again. Moreover, for the qPCR detection of H. pylori resistance to antibiotics (clarithromycin and levofloxacin), except for the CT value that distinguishes between positive and negative results, which changes to 30.00 for resistance detection, the other requirements are the same as above. Notably, the sequence information about the primers and probes is unavailable in this paper because it involves commercial property rights.
In order to select more efficient antibiotics for each patient, it is crucial to survey the local prevalence of H. pylori and its antibiotic resistance profile due to the increased antimicrobial resistance globally and, additionally, due to the variation in resistance patterns between various geographic areas. The major limitation of our study is that the sample size was very small and did not cover different geographical regions in China to show the prevalence of H. pylori infection and its complete antibiotic resistance profile, but the string test technique is well developed, and by combining it with qPCR, we achieved a simultaneous diagnosis of H. pylori infection and drug resistance detection. These results suggest that this approach could be used on a larger scale in different regions of the world.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Sanming Project of Medicine in Shenzhen (Grant No. SZSM201510050) and the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2022A1515220023). Research Foundation for Advanced Talents of Guandong Provincial People's Hospital (No. KJ012021097), and the National Natural Science Foundation of China (81871734, 82072380, 82272423). The funders had no role in the study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Materials
| 23S rRNA and gyrA gene point mutations detection kit (PCR-Fluorescence Probing) | Hongmed Infagen | Detection of Helicobacter pylori resistance to clarithromycin and levofloxacin | |
| ABI 7500 fluorescence quantitative PCR machine | Thermo Fisher Scientific | SEDA 20163220767 | Fluorescent quantitative PCR amplification |
| ABI 7500 software | Thermo Fisher Scientific | Data Analysis | |
| BSC-1500IIA2-X | BIOBASE | SEDA 20143222263 | Biosafety cabinet |
| DNA extraction kit | Daan Gene | ||
| E-Centrifuge | WEALTEC | Centrifuge the residual liquid off the wall of the tube. | |
| H. Pylori DNA detection kit (PCR-Fluorescence Probing) | Hongmed Infagen | Testing for H. pylori infection | |
| Stream SP96 automated nucleic acid extractor | Daan Gene | SEDA 20140104 | For DNA extraction |
| String test kit | Hongmed Infagen | It contains a capsule attached to a string, scissors, cotton swab, and sample preservation tube | |
| Ultra-low temperature freezers (DW-YL450) | MELING | SEDA 20172220091 | -20 °C for storing reagents |
| Vortex-5 | Kylin-bell | For mixing reagent |
References
- Proença-Modena, J. L., Acrani, G. O., Brocchi, M. Helicobacter pylori: Phenotypes, genotypes and virulence genes. Future Microbiology. 4 (2), 223-240 (2009).
- Hussein, R. A., Al-Ouqaili, M. T. S., Majeed, Y. H. Association between alcohol consumption, cigarette smoking, and Helicobacter pylori infection in Iraqi patients submitted to gastrointestinal endoscopy. Journal of Emergency Medicine, Trauma and Acute Care. 2022 (6), 12 (2022).
- Reshetnyak, V. I., Burmistrov, A. I., Maev, I. V. Helicobacter pylori: Commensal, symbiont or pathogen. World Journal of Gastroenterology. 27 (7), 545-560 (2021).
- Thrift, A. P., Wenker, T. N., El-Serag, H. B. Global burden of gastric cancer: Epidemiological trends, risk factors, screening and prevention. Nature Reviews Clinical Oncology. 20 (5), 338-349 (2023).
- Liou, J. M., et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: The Taipei global consensus. Gut. 69 (12), 2093-2112 (2020).
- IARC Helicobacter pylori Working Group. . Helicobacter Pylori Eradication as A Strategy for Preventing Gastric Cancer. , (2014).
- Vaira, D., et al. The stool antigen test for detection of Helicobacter pylori after eradication therapy. Annals of Internal Medicine. 136 (4), 280-287 (2002).
- Laheij, R. J. F., Straatman, H., Jansen, J. B. M. J., Verbeek, A. L. M. Evaluation of commercially available Helicobacter pylori serology kits: A review. Journal of Clinical Microbiology. 36 (10), 2803-2809 (1998).
- Malfertheiner, P., et al. Management of Helicobacter pylori infection – The Maastricht IV/ Florence consensus report. Gut. 61 (5), 646-664 (2012).
- Peng, X., et al. Gastric juice-based real-time PCR for tailored Helicobacter Pylori treatment: A practical approach. International Journal of Medical Sciences. 14 (6), 595-601 (2017).
- Thung, I., et al. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Alimentary Pharmacology and Therapeutics. 43 (4), 514-533 (2016).
- Zhang, Y., et al. Mutations in the antibiotic target genes related to clarithromycin, metronidazole and levofloxacin resistance in Helicobacter pylori strains from children in China. Infection and Drug Resistance. 13, 311-322 (2020).
- Hussein, R. A., Al-Ouqaili, M. T. S., Majeed, Y. H. Detection of Helicobacter pylori infection by invasive and noninvasive techniques in patients with gastrointestinal diseases from Iraq: A validation study. PLoS One. 16 (8), e0256393 (2021).
- Chen, Q., et al. Advanced sensing strategies based on different types of biomarkers toward early diagnosis of H. pylori. Critical Reviews in Analytical Chemistry. , (2023).
- Hussein, R. A., Al-Ouqaili, M. T. S., Majeed, Y. H. Detection of clarithromycin resistance and 23SrRNA point mutations in clinical isolates of Helicobacter pylori isolates: Phenotypic and molecular methods. Saudi Journal of Biological Sciences. 29 (1), 513-520 (2022).
- Xuan, S. H., Wu, L. P., Zhou, Y. G., Xiao, M. B. Detection of clarithromycin-resistant Helicobacter pylori in clinical specimens by molecular methods: A review. Journal of Global Antimicrobial Resistance. 4, 35-41 (2016).
- Perez-Trallero, E., Montes, M., Alcorta, M., Zubillaga, P., Telleria, E. Non-endoscopic method to obtain Helicobacter pylori for culture. Lancet. 345 (8950), 622-623 (1995).
- DiNardo, A. R., et al. Use of string test and stool specimens to diagnose pulmonary tuberculosis. International Journal of Infectious Diseases. 41, 50-52 (2015).
- Li, G., et al. Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. European Journal of Clinical Microbiology and Infectious Diseases. 39 (9), 1673-1679 (2020).
- Agbonlahor, D. E., Odugbemi, T. O., Udofia, P. O. Differentiation of gram-positive and gram-negative bacteria and yeasts using a modification of the "string" test. The American Journal of Medical Technology. 49 (3), 177-178 (1983).

