Three-Dimensional Morphogenesis in Canine Gut-on-a-Chip Using Intestinal Organoids Derived from Inflammatory Bowel Disease Patients
Summary
Integration of canine intestinal organoids and a Gut-on-a-Chip microfluidic system offers relevant translational models for human intestinal diseases. Protocols presented allow for 3D morphogenesis and dynamic in vitro modeling of the gut, aiding in the development of effective treatments for intestinal diseases in dogs and humans with One Health.
Abstract
Canine intestines possess similarities in anatomy, microbiology, and physiology to those of humans, and dogs naturally develop spontaneous intestinal disorders similar to humans. Overcoming the inherent limitation of three-dimensional (3D) organoids in accessing the apical surface of the intestinal epithelium has led to the generation of two-dimensional (2D) monolayer cultures, which expose the accessible luminal surface using cells derived from the organoids. The integration of these organoids and organoid-derived monolayer cultures into a microfluidic Gut-on-a-Chip system has further evolved the technology, allowing for the development of more physiologically relevant dynamic in vitro intestinal models.
In this study, we present a protocol for generating 3D morphogenesis of canine intestinal epithelium using primary intestinal tissue samples obtained from dogs affected by inflammatory bowel disease (IBD). We also outline a protocol for generating and maintaining 2D monolayer cultures and intestine-on-a-chip systems using cells derived from the 3D intestinal organoids. The protocols presented in this study serve as a foundational framework for establishing a microfluidic Gut-on-a-Chip system specifically designed for canines. By laying the groundwork for this innovative approach, we aim to expand the application of these techniques in biomedical and translational research, aligning with the principles of the One Health Initiative. By utilizing this approach, we can develop more physiologically relevant dynamic in vitro models for studying intestinal physiology in both dogs and humans. This has significant implications for biomedical and pharmaceutical applications, as it can aid in the development of more effective treatments for intestinal diseases in both species.
Introduction
Intestinal epithelial morphogenesis has been largely studied through laboratory animal models, which are costly, time-consuming, and do not accurately represent human developmental processes1. Furthermore, conventional static 2D cell culture models lack the ability to mimic the complex spatial organization of a 3D epithelial architecture2. As a result, there is a need for a protocol to induce in vitro 3D morphogenesis using intestinal epithelial cells from human-relevant animal models to advance our understanding of gut epithelial architecture.
Companion dogs have developed intestinal anatomy and microbiome compositions that are remarkably similar to humans due to their shared environment and diet during domestication3. In addition to this similarity, both humans and dogs share various chronic morbidities that are thought to be attributed to intestinal health. Dogs, like humans, can spontaneously develop chronic conditions such as obesity, cognitive dysfunction, diabetes mellitus, inflammatory bowel disease (IBD), and colorectal adenocarcinoma4,5,6,7,8,9,10. Despite the development and use of human and murine epithelial cells in previous Gut-on-a-Chip studies2,11,12,13,14, canine intestinal epithelium has not been utilized until now. Our novel approach, utilizing canine intestinal organoid epithelium in a dynamic culture system with a 3D epithelial morphogenesis, has significant implications for both canine and human medicine.
Recent advancements in intestinal organoid culture have led to the establishment of canine intestinal organoid culture15. This culture system involves culturing intestinal stem cells under defined morphogen conditioning, resulting in a 3D model with self-renewing properties derived from adult stem cells16. However, conducting transport assays or host-microbiome cocultures poses difficulties with this 3D model because of the enclosed nature of the intestinal lumen17. To address this, researchers have generated a 2D monolayer derived from intestinal organoids, allowing for exposure of the luminal surface18,19. However, both 3D organoids and 2D monolayers are maintained under static conditions, which do not accurately reflect the in vivo biomechanics of the intestinal microenvironment. Combining patient-derived canine organoids technology with in vitro 3D morphogenesis presents an opportunity for translational research into chronic multifactorial diseases. This approach enables researchers to develop more effective treatments benefiting both humans and dogs and further advance translational research, aligning with the One Health Initiative, which is a collaborative approach that recognizes the interconnectedness of human, animal, and environmental health. It promotes interdisciplinary cooperation to address complex health challenges and achieve optimal health outcomes for all. By understanding the interdependencies between humans, animals, and ecosystems, the initiative aims to mitigate risks from emerging infectious diseases, environmental degradation, and other shared health concerns20,21,22.
This protocol outlines comprehensive methods for culturing canine intestinal epithelial cells obtained from patient organoids on a Gut-on-a-Chip microdevice with a polydimethylsiloxane (PDMS)-based porous membrane. Establishing the 3D epithelial morphogenesis by integrating canine intestinal organoids and this Gut-on-a-Chip technology enables us to study how the gut develops and maintains its cellular organization and stem cell niche. This platform offers a valuable opportunity to investigate the impact of microbiome communities on gut health and understand how these communities generate microbial metabolites that contribute to intestinal pathophysiology14,23. These advancements can now be extended to canine intestinal samples, providing researchers with opportunities to explore the intricate relationship between the gut microbiome and host physiology. This opens up avenues for gaining valuable insights into the underlying mechanisms of intestinal pathophysiology and understanding the potential role of microbial metabolites in both canine and human health, as well as various disease conditions. The protocol used for canine Gut-on-a-Chip is reproducible, making it a suitable experimental model for comparative medicine, as this approach enables investigation of host-microbiome interactions, pathogen infections, and probiotic-based therapeutic effects in both dogs and humans.
Protocol
The study was approved and conducted in accordance with the Institutional Animal Care and Use Committee of Washington State University (ASAF# 6993). In this protocol, we utilized a well-established Gut-on-a-Chip microfluidic device made of PDMS which was fabricated in-house2 (Figure 1D). Detailed methods of the fabrication of the Gut-on-a-Chip microdevice can be found in previous reports2,24. This protocol demonstrates a unique integration of intestinal organoids and a microfluidic system (Figure 2).
1. Surface activation of a Gut-on-a-Chip made of PDMS
- Prepare 1% polyethyleneimine (PEI) solution by adding 1 mL of 50% PEI solution into 49 mL of distilled (DI) water in a 50 mL conical tube. Invert the tube two to three times to mix the solution well, then filter the solution using a 0.2 µm syringe filter.
NOTE: Keep it stored at 4 °C. - Prepare 0.1% glutaraldehyde (GA) solution by adding 100 µL of 50% GA solution into 49.9 mL of DI in a 50 mL conical tube. Invert the tube two to three times to mix the solution well, then filter the solution using a 0.2 µm syringe filter.
NOTE: Store it at 4 °C and make sure to protect it from exposure to direct light. - Place the Gut-on-a-Chip device in a dry oven set at 60 °C and incubate it for a minimum of 30 min to eliminate any remaining moisture.
- Expose the Gut-on-a-Chip device to UV and Ozone treatment for 60 min using a UV/Ozone generator.
NOTE: To ensure optimal activation of the PDMS surfaces during treatment, maintain a distance of approximately 3 cm or less between the UV lamp and the device. Avoid overcrowding of devices in the generator to accomplish efficient surface activation. - Secure the upper microchannel's inlet, bypass, and outlet tubing by using binder clips to clamp them. Also clamp the inlet tubing for the lower microchannel.
- Disconnect the outlet tubing for the lower microchannel. Ensure that the bypass tubing of the lower microchannel remains open.
- Using a P100 micropipette, introduce 100 µL of a 1% PEI solution through the outlet hole of the lower microchannel.
NOTE: It is recommended to perform this process while the device is still warm from UV/Ozone exposure. Observe the PEI solution flowing through the microchannel and the PEI solution drops to come out from the bypass tubing for the lower microchannel. - Reconnect the outlet tubing for the lower microchannel.
- Secure the lower microchannel's inlet, bypass, and outlet tubing by using binder clips to clamp them. Also clamp the inlet tubing for the upper microchannel.
- Disconnect the outlet tubing for the upper microchannel. Ensure that the bypass tubing of the upper microchannel remains open.
- Using a P100 micropipette, introduce 100 µL of a 1% PEI solution through the outlet hole of the upper microchannel.
NOTE: Observe the PEI solution flowing through the microchannel and the PEI solution drops to come out from the bypass tubing for the upper microchannel. - Reattach the outlet tubing of the upper microchannel.
- Once both upper and lower microchannels are filled with 1% PEI solution, allow the device to incubate at room temperature (RT) for 10 min.
- Perform steps 1.5-1.12 with 0.1% GA solution.
- Once both upper and lower microchannels are filled with 0.1% GA solution, allow the device to incubate at RT for 20 min.
- Perform steps 1.5-1.12 with DI water to remove any excess surface activation solution.
- Place the chip in a dry oven at 60 °C and allow it to dry overnight.
NOTE: Make sure to remove the binder clips from all the tubing to avoid any impingement of the tubing. This drying process is critical to the even surface activation of the microchannel within the Gut-on-a-Chip12.
2. Extracellular matrix (ECM) coating and culture medium preparation for Gut-on-a-Chip culture
- Prepare an ECM mixture with organoid basal medium so that the final concentration of Collagen I and Matrigel are 60 µg/mL and 2 % (vol/vol), respectively.
NOTE: Prepare 50 µL of ECM mixture per gut chip (i.e., 20 µL of ECM mixture for each upper and lower microchannel and 10 µL extra). Prepare this on the day of use and store this solution at 4 °C or place it on ice until it is ready for use. - Take out the Gut-on-a-Chip that has been treated with UV/Ozone, PEI, and GA from the dry oven.
NOTE: Inspect under a phase-contrast microscope to see if there is any residual moisture. - Allow the Gut-on-a-Chip to cool down in a biosafety cabinet for 10 min.
- Secure the upper microchannel's inlet, bypass, and outlet tubing by using binder clips to clamp them. Also clamp the inlet tubing for the lower microchannel.
- Disconnect the outlet tubing for the lower microchannel.
- Using a P100 micropipette, introduce 20 µL of ECM mixture through the outlet hole of the lower microchannel.
NOTE: Observe the ECM mixture flowing through the microchannel without any air bubble entrapment. If there is any air bubble present, introduce additional ECM mixture to the lower microchannel until the bubble is gone. - Reattach the outlet tubing for the lower microchannel.
- Secure the lower microchannel's inlet, bypass, and outlet tubing by using binder clips to clamp them. Also clamp the inlet tubing for the upper microchannel.
- Disconnect the outlet tubing for the upper microchannel.
- Using a P100 micropipette, introduce 20 µL of ECM mixture through the outlet hole of the upper microchannel.
- Reattach the outlet tubing for the upper microchannel.
- Place the chip in a humidified CO2 incubator with 5% CO2 at 37 °C for 1 h to form the ECM layer onto the PDMS membrane treated with PEI and GA (Figure 3A).
NOTE: Make sure all the tubing is clamped during the incubation. - During the incubation of the ECM coating, prepare two 1 mL syringes containing cold seeding medium, which is the organoid culture medium lacking A8301 but containing 10 µM Y-27632 and 2.5 µM CHIR99021, as previously reported in Gut-on-a-Chip derived from human organoids2.
NOTE: A8301 was removed from the medium to enhance the attachment of cells to the coated ECM as previously reported2. Day 0 of Gut-on-a-Chip culture only requires upper microchannel flow. Therefore, prepare a full syringe for the upper channel while a minimum of 0.2 mL for the lower channel. - Once the ECM coating is completed, take the Chip out of the incubator and release the clamp to the bypass tubing connected to the lower microchannel.
- Connect a 1 mL syringe filled with seeding medium to the blunt-end needle that is linked to the lower microchannel of the Gut-on-a-Chip. Carefully introduce the seeding medium (~50 µL) to the bypass tubing. Once the tubing is filled with the introduced medium, secure the bypass tubing connected to the lower microchannel with a binder clip.
- Release the clamp of the outlet tubing connected to the lower microchannel. Carefully introduce the seeding medium into the lower microchannel allowing it to flow smoothly through the system. Secure the outlet tubing connected to the lower microchannel with a binder clip after perfusion.
- Next, open the bypass tubing connected to the upper microchannel.
- Connect a 1 mL syringe filled with seeding medium to the blunt-end needle that is linked to the upper microchannel of the Gut-on-a-Chip. Carefully introduce the seeding medium (~50 µL) to the bypass tubing. Once the tubing is filled with the introduced medium, secure the bypass tubing connected to the upper microchannel with a binder clip.
- Release the outlet tubing connected to the lower microchannel. Carefully introduce the seeding medium into the upper microchannel allowing it to flow smoothly through the system. Secure the outlet tubing connected to the upper microchannel with a binder clip after perfusion.
3. Canine intestinal organoid cell preparation for seeding
NOTE: To generate the Gut-on-a-Chip model, canine colon organoids (referred to as colonoids) derived from IBD patient dogs were utilized in this protocol. These colonoids were derived from three to five small fragments of biopsied colonic tissue, following a previously reported method15,18. For optimal results, it is crucial to use canine colonoids that have undergone a minimum of three culture passages to establish stable organoids suitable for in vitro applications. It is recommended to culture the canine colonoids for a duration of at least 3-4 days to facilitate adequate differentiation of multi-lineage cells within the organoids, ensuring their functional maturity and suitability for subsequent experiments in the Gut-on-a-Chip model. The maximum limit of the passage for this work is fewer than 20, as indicated by a previous study that demonstrated the unaltered phenotype and karyotype throughout 20 consecutive passages25. The signalment of these donors is presented in Supplementary Table S1.
- Culture the canine colonoids in 24-well plates embedded in 30 µL of Matrigel15,18 with organoid culture medium listed in the Table of Materials, which is modified from previously reported medium15,26,27. The conditioned medium was obtained by cultivating Rspo1 cells and HEK293 cells engineered to secrete Noggin28.
- Discard the organoid culture medium through vacuum suction and introduce 500 µL of Cell Recovery Solution at an ice-cold temperature into each well. Incubate for 30 minutes at 4 °C.
NOTE: Typically, three wells out of a 24 well-plate of mature canine intestinal organoids provide a sufficient quantity of dissociated cells to seed a single Gut-on-a-Chip device with 40-50 organoids/one field of view at x10 magnification. - Mechanically disrupt the Matrigel domes for 5 s using a P1000 micropipette. Subsequently, gather the organoid suspension in a 15 mL conical tube.
- Centrifuge the conical tube at 200 × g and 4 °C for 5 min, followed by the removal of the supernatant.
- Introduce 1 mL of trypsin-like protease at room temperature, supplemented with 10 µM Y-27632, and resuspend the cell pellet by pipetting using a P1000 micropipette.
- Place the cell suspension in a water bath set at 37 °C and incubate it for 10 min while periodically shaking the mixture.
- Introduce 5 mL of warm organoid basal medium and vigorously pipette the cell suspension using a P1000 micropipette until it becomes cloudy, without any visible cell clumps.
- Pass the cell suspension through a cell strainer with a 70 µm cutoff to eliminate any Matrigel debris and large cell clusters.
- Centrifuge the conical tube at 200 × g and 4 °C for 5 min, followed by resuspension of the pellet in seeding medium. For the seeding of one canine Gut-on-a-Chip device, use 20 µL of seeding medium to resuspend the cell pellet (i.e., use 20 µL of seeding medium when seeding one Gut-on-a-Chip device).
- Modify the concentration of viable cells to 1 × 107 cells/mL with seeding medium as previously reported2. Conduct viability assessment using a hemocytometer by combining 10 µL of the cell suspension with 10 µL of Trypan blue, and then observe the cells under a microscope.
NOTE: It is crucial to adjust the cell concentration appropriately to ensure optimal cell attachment and the formation of a uniform monolayer on the chip membrane. If the initial number of cells is insufficient, it may result in delayed or unsuccessful establishment of a confluent monolayer. Conversely, if the number of cells is excessive, non-adherent cells can aggregate into clumps within the channel, leading to undesirable concentration effects.
4. Seeding and formation of a 2D cell monolayer
- Disconnect the outlet tubing for the upper microchannel. Ensure that the bypass tubing of the upper microchannel remains open. Secure both the inlet and outlet of the lower microchannel by using binder clips to clamp them.
- Using a P100 or P20 micropipette, introduce 20 µL of the cell suspension from Protocol 3 to the outlet hole of the upper microchannel (Figure 3B Seeding).
- Secure the bypass and inlet tubing of the upper microchannel by using binder clips to clamp it. Then, reattach the outlet tubing to the outlet hole of the upper microchannel, ensuring that the tubing remains open throughout the process to avoid any pressure being applied to the upper microchannel. After this step, secure the outlet tubing of the upper channel slowly using a binder clip to clamp it.
- Verify using a microscope that the cells are evenly distributed throughout the upper microchannel.
NOTE: It is important to clamp the tubing to stop the movement of the medium within the channel to allow stable static conditions until the desirable cell attachment is accomplished. - Place the chip in a humidified CO2 incubator at 37 °C.
NOTE: It takes approximately 3 h for canine intestinal organoid cells to attach to the ECM coating (Figure 3B Attachment). - Connect the syringe attached to the upper microchannel of the Gut-on-a-Chip to a syringe pump positioned within a CO2 incubator.
NOTE: Carefully flush the medium into the microchannel using the knob of the syringe pump and remove unbound cells. Examine the chip under a microscope to ensure that any unattached cells have been effectively washed away. - Initiate the flow of culture medium at 30 µL/h with seeding medium. This continuous flow is only for the upper microchannel initially until the 2D monolayer establishment on a Gut-on-a-Chip. For the lower microchannel, leave the microchannels clamped and the medium unflushed.
- From the day after the cells are seeded, change the culture medium to organoid culture medium, which contains A8301 and lacks 10 µM Y-27632 and 2.5 µM CHIR99021, from the seeding medium.
- Once the monolayer is established, initiate the continuous medium flow to the lower microchannel as well. It usually takes 2 to 3 days for canine intestinal organoid epithelial cells to establish epithelial monolayers (Figure 3C).
5. Establishment of 3D morphogenesis in canine Gut-on-a-Chip
NOTE: After the confluent monolayers are formed in Gut-on-a-Chip, medium flow of both the upper channel and lower channel and cell strain were introduced to initiate 3D morphogenesis to 2D monolayer as presented in Figure 2.
- Introduce the organoid culture medium into both the upper and lower microchannels to initiate the development of 3D morphogenesis in a Gut-on-a-Chip system. To achieve shear stress of 0.02 dyne/cm2 in the existing Gut-on-a-Chip design (i.e., 500 µm height microchannel), increase the flow rate to 50 µL/h29.
- Initiate 10% of cell strain and 0.15 Hz of frequency, as recommended before2, using a computer-regulated bioreactor applying cyclic strain to cells cultured in vitro. This process would apply vacuum suction to the Gut-on-a-Chip device.
- Sustain these culture conditions for a minimum of 2 to 3 days. The 3D morphogenesis of the canine intestinal monolayer usually occurs 2 to 3 days after Dural channel flow and vacuum suction have been started (Figure 3C).
6. Characterization of canine Gut-on-a-Chip
- Live cell imaging
- Remove any air bubbles in the Gut-on-a-Chip by gently flowing the medium using the syringe pump.
- Disconnect the Gut-on-a-Chip device from the syringe pump.
NOTE: Avoid any maneuver that can apply pressure within the Gut-on-a-Chip. - Position the device onto a microscope to capture images of the established 3D epithelium. For visualizing the structure of the 3D epithelial layers using phase-contrast, use 10x and 20x objectives (Figure 3C).
- Immunofluorescence staining
- Prepare the blocking solution by dissolving 1 g of BSA in 50 mL of Phosphate buffer saline (PBS) to make 2% BSA. Pass the solution through a syringe filter of 0.2 µm for filtration. Keep this solution stored at 4 °C.
- Prepare the permeabilizing solution by combining 150 µL of Triton X-100 with 50 mL of blocking solution, resulting in a final concentration of 0.3% Triton X-100. Pass the solution through a syringe filter of 0.2 µm for filtration. Keep this solution stored at 4 °C.
- Perform fixation of the cells by injecting 100 µL of a 4% PFA into both the upper and lower microchannels as described in steps 2.4-2.11.
- Rinse the cells by injecting 100 µL of PBS into both upper and lower microchannels as described in steps 2.4-2.11.
- Perform permeabilization of the cells by injecting 100 µL of 0.3% Triton into both upper and lower microchannels as described in steps 2.4-2.11. Place the device at RT for 30 min.
- Rinse the cells by injecting 100 µL of PBS into both upper and lower microchannels as described in steps 2.4-2.11.
- Perform blocking of the cells to prevent nonspecific binding by injecting 100 µL of 2% BSA to both upper and lower microchannels as described in steps 2.4-2.11. Place the device at RT for 1 h.
- Inject 20 µL of the primary antibody solution diluted in 2% BSA and place it at RT for 3 h, followed by overnight incubation at 4 °C.
NOTE: Make sure all the tubing is clamped during the incubation at 4 °C for overnight. The concentration of a primary antibody should be 2-5 times higher than the recommended concentration for monolayer or 3D organoid staining (Supplementary Figure S1). - Rinse the cells by injecting 100 µL of PBS to both upper and lower microchannels as described in steps 2.4-2.11.
- Inject 20 µL of the secondary antibody solution diluted in 2% BSA and place it at RT for 1 h.
NOTE: Make sure all the tubing is clamped during the incubation. Starting from this step, it is necessary to shield the device setup with aluminum foil to prevent photobleaching. - Rinse the cells by injecting 100 µL of PBS to both upper and lower microchannels as described in steps 2.4-2.11.
- Prepare a combined solution for F-actin and DAPI (diamidino-2-phenylindole) counterstaining. Inject 20 µL of the combined solution into both upper and lower microchannels as described in steps 2.4-2.11.
- Rinse the cells by injecting 100 µL of PBS into both upper and lower microchannels as described in steps 2.4-2.11.
Perform fluorescence imaging of the architecture of the 3D epithelial cells using a fluorescence microscope or a confocal microscope.
7. Epithelial barrier function
- Remove any air bubbles in the Gut-on-a-Chip by gently flowing the medium using the syringe pump. Make sure all the tubing is open during the measurement.
- Remove the Gut-on-a-Chip from the syringe pump and place it at RT for at least 10 min.
- Remove Ag/AgCl electrodes from 70% EtOH solution.
- Place two Ag/AgCl electrodes into the upper inlet and lower outlet, respectively, to measure the resistance of the epithelial layer using a multimeter.
- Place two Ag/AgCl electrodes into the lower inlet and upper outlet, respectively. Report the mean of these two values as a resistance value for the Gut-on-a-Chip.
NOTE: The blank TEER should be measured on a Gut-on-a-Chip with only ECM coating without the epithelium. - Calculate transepithelial electrical resistance (TEER) value (kΩ × cm2) can be calculated using equation (1).
TEER = (Ωt - Ωblank) × A (1)
Where Ωt is a resistance value measured at the specific time point since the start of the experiment, Ωblank is a resistance value measured at the time without the epithelium, and A is the area of the surface covered by a cell layer (approximately 0.11 cm2 for this Gut-on-a-Chip design29).- Calculate normalized TEER using equation (2).
TEER =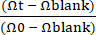 (2)
(2)
Where Ω0 is a resistance value at the initial reading time point of the experiment as reported previously30 (Figure 4C).
- Calculate normalized TEER using equation (2).
Representative Results
This protocol reliably facilitates the spontaneous development of 3D intestinal morphogenesis in a Gut-on-a-Chip system. This approach utilizes canine intestinal epithelial cells obtained from intestinal organoids derived from dogs affected by inflammatory bowel disease (IBD) (Figure 1B). Occasional clustering of 3D morphogenesis of canine intestinal epithelial cells can be observed throughout the microchannel following 6-9 days of medium flow (Figure 3C). These morphological changes can be monitored using phase-contrast techniques.In this study, we utilized organoids derived from two dogs diagnosed with IBD. Notably, successful 3D morphogenesis was observed in two biological replicates, each of which was performed with two technical replicates. The findings from this study provide a foundation for future investigations involving intestinal organoids derived from other canine donors. These results demonstrate the potential applicability and repeatability of our experimental approach, which has previously been reported in human samples. These findings provide further confirmation that Gut-on-a-Chip technology is applicable to canine intestinal epithelial cells as previously reported with the studies utilizing human intestinal epithelial cells2.
This protocol showed that immunofluorescence staining can be used to evaluate the 3D structure of organoid-derived monolayers that have formed villus-like structures in microfluidic chips using conventional fluorescence microscopy (Figure 4 A,B). This protocol can be adapted to validate the differentiated and spatially organized cellular phenotypes through immunofluorescence staining. The visualization of a 3D morphogenesis within a Gut-on-a-Chip provides an excellent opportunity to investigate the host response during various pathological interactions14,23,31. When combined with epithelial cells derived from patient donors, as previously described in humans, this technology can be utilized to construct personalized models of intestinal diseases13. Through the integration of immunofluorescence imaging with targeted RNA visualization techniques like fluorescence in situ hybridization, it can be feasible to visually analyze the transcriptomes and proteomes of the host within a Gut-on-a-Chip system.
Preserving the integrity of the intestinal membrane is vital for maintaining intestinal homeostasis, and the Gut-on-a-Chip platform provides a valuable advantage by allowing for precise monitoring and quantification of this crucial function. The measurement of TEER using the Gut-on-a-Chip technology offers several benefits. For instance, previous studies have successfully evaluated TEER while co-culturing intestinal cells with non-pathogenic and probiotic bacteria32, as well as under leaky-gut conditions23. This allows researchers to study the impact of different conditions on the intestinal barrier function and to identify potential interventions to promote intestinal health.
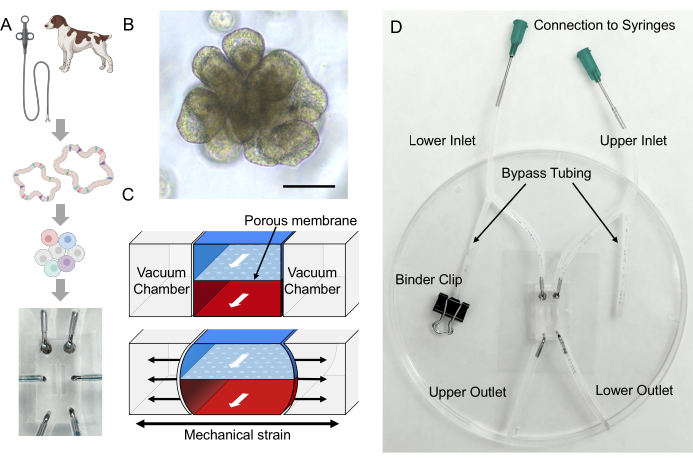
Figure 1: Establishment of patient-derived canine IBD Gut-on-a-Chip. (A) Integration of the patient-derived intestinal organoids and the Gut-on-a-Chip platform. Endoscopy biopsy can be performed to isolate intestinal crypt cells to develop donor-specific intestinal organoids. Epithelial cells can be dissociated into single cells from the organoids, then seeded into a PDMS-based Gut-on-a-Chip and cultured in a unique dynamic microenvironment. (B) Representative images of colonoids from an IBD dog. Scale bar = 100 µm. (C) This schematic illustrates a Gut-on-a-Chip device consisting of a porous membrane placed between upper and lower microchannels. The upper microchannel is indicated by the blue area, while the lower microchannel is indicated by the red area. Vacuum chambers are present on each side of the microchannel, which deform the porous membrane to mimic peristaltic motion24. (D) A setup of canine Gut-on-a-Chip includes a PDMS-based Gut-on-a-Chip assembled with tubing that is placed on a cover slip2,24. The bypass tubing is critical to avoid pressure built-up within the microchannel during the handling (i.e., connecting to syringes). Binder clips are used to clamp the tubing. Volume-sensitive materials can be infused via the open holes of the upper or lower outlet. Organoid culture medium can be infused by connecting the syringes to the blunt end needles and the flow through the upper and lower inlet. Abbreviations: IBD = inflammatory bowel disease; PDMS = polydimethylsiloxane. Please click here to view a larger version of this figure.
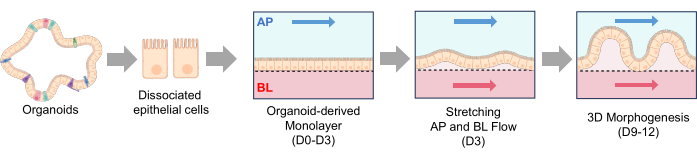
Figure 2: Formation of villus-like structures in canine IBD Gut-on-a-Chip. The dissociated epithelial cells were seeded in an ECM-coated Gut-on-a-Chip. Once the dissociated cells were attached to the PDMS membrane, apical flow was initiated for 3 days (D0-D3). When a confluent 2D monolayer is formed (D3), basolateral flow with frequent stretching is initiated (Stretching, AP, and BL Flow). After 2-3 days of dual flow and membrane stretching, the 2D monolayer begins to develop 3D morphogenesis, and villus-like structures are formed after 9 days of culture (3D morphogenesis, D9-D12). Abbreviations: ECM = extracellular matrix; PDMS = polydimethylsiloxane; AP = apical; BL = basolateral. Please click here to view a larger version of this figure.
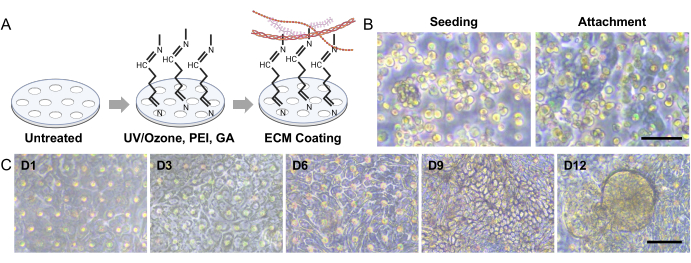
Figure 3: Canine intestinal organoid seeding and 3D morphogenesis in a Gut-on-a-Chip. (A) The experimental steps for the surface activation of a porous membrane in a PDMS-based Gut-on-a-Chip. The utilization of UV/Ozone treatment, PEI, and GA treatment in conjunction facilitates the cross-linking of amines present in ECM solutions. This process leads to the stable immobilization of ECM proteins onto the porous membrane. (B) The phase contrast images demonstrate the morphologies of cells immediately after seeding (left) and 3-5 h after seeding (right). The porous membrane after 3 h of seeding displays thinner and darker areas where single cells have attached, highlighting the attachment process. (C) The phase contrast images depict the 3D morphogenesis of intestinal monolayers within a Gut-on-a-Chip system. These monolayers were derived from dogs affected with IBD and these organoid cells were cultured for a period of 12 days under dynamic conditions, which involved fluid flow and stretching motions. Scale bars = 50 µm (B,C). Abbreviations: IBD = inflammatory bowel disease; ECM = extracellular matrix; PDMS = polydimethylsiloxane; PEI = polyethylenimine; GA = glutaraldehyde. Please click here to view a larger version of this figure.
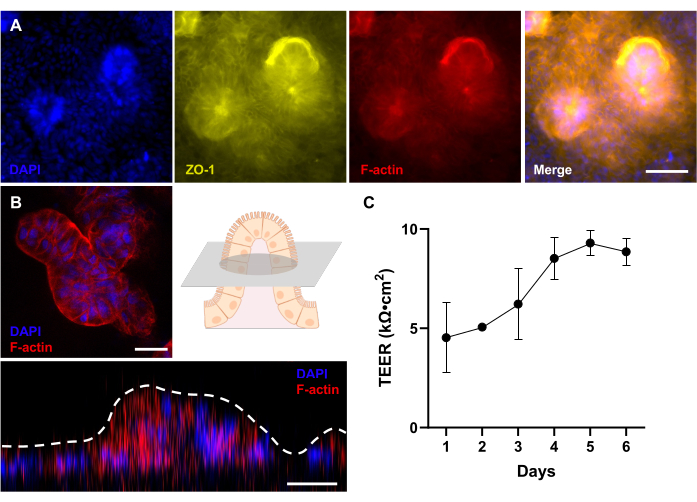
Figure 4: Evaluation of the 3D morphological development in patient-derived canine Gut-on-a-Chip. (A) Immunofluorescence imaging of a canine IBD Gut-on-a-Chip, showcasing a top-down view of a fully developed 3D epithelium after 12 days of culture, which is evaluated by a fluorescence microscope. The tight junction protein (ZO-1) is visualized in yellow; the brush-border membrane (F-actin) appears in red; and the nuclei are stained with DAPI and appear blue. (B) Immunofluorescence imaging of a canine IBD Gut-on-a-Chip using a confocal microscope with a long-distance lens. As shown in the schematic, a fluorescent image of a cross section of a villus-like structure of a fully developed 3D epithelium after 12 days of culture is shown. In addition, Z-stacking shows a side view of the 3D epithelium, which reveals the formation of villus-like structures. The brush-border membrane (F-actin) appears in red, and the nuclei are stained with DAPI and appear blue. (C) Intestinal barrier function was evaluated and measured by TEER in patient-derived canine Gut-on-a-Chips. Stable TEER values were reached on Day 5 of culture on Gut-on-a-Chip. Error bars express the SEM of the measurements. TEER value was measured among two biological replicates with one technical replicate. Scale bars = 50 µm (A), 25 µm (B). Abbreviations: IBD = inflammatory bowel disease; DAPI = 4',6-diamidino-2-phenylindole; TEER = transepithelial electrical resistance. Please click here to view a larger version of this figure.
Supplementary Figure S1: Characterization of anti-ZO-1 polyclonal antibody in canine colonoid-derived monolayers and on Gut-on-a-Chip devices. (A) Immunofluorescence staining of ZO-1 in yellow with F-actin in red and their overlay image. Scale bar = 25 µm. (B) Immunofluorescence visualization of ZO-1 in yellow in 'Leaky Gut Chips' was conducted under various conditions, including stimulation with probiotic bacteria (LGG + Cytokines or VSL#3 + Cytokines) and germ-free controls without probiotic stimulation (Cytokines). Scale bar = 50 µm. This figure is reproduced from Min et al.23. Please click here to download this File.
Supplementary Table S1: A summary of tissue donors' information. A summary table of the donors' age, sex, breed, histopathological evaluation, and canine IBD activity index (CIBDAI) score. The CIBDAI is a numeric scoring system used to infer clinical severity in canine IBD33. Please click here to download this File.
Discussion
This study marks the pioneering demonstration of the compatibility of canine intestinal organoids with the development of a canine IBD Gut-on-a-Chip model. The integration of intestinal organoids and organoid-derived monolayer cultures into a microfluidic system (i.e., Gut-on-a-Chip system) has further evolved the technology, enabling the creation of in vitro intestinal models that closely mimic physiological dynamics and are more representative of biological conditions. In particular, since there are very few reports of Gut-on-a-Chip culture using IBD-derived organoids in humans, the current study using canine IBD-derived Gut-on-a-Chip may provide leading insights into the study of IBD in humans.
The successful development of canine intestinal epithelial 3D morphogenesis on a Gut-on-a-Chip requires careful attention to several critical steps. First, the hydrophobic surface of PDMS microfluidic channels may impede ECM adhesion and subsequent cell attachment, necessitating surface activation of PDMS prior to ECM coating and cell seeding (see protocol section 1). To achieve a stable monolayer culture, the removal of excess unattached cells is crucial following cell attachment (protocol steps 4.6-4.7). Additionally, dynamic stimulation, such as constant medium flow and peristaltic-like vacuum motion, is necessary for 3D morphogenesis of the intestinal epithelium (protocol step 5.2). Careful handling is essential to avoid air bubbles in the microchannel during any steps of the Gut-on-a-Chip culture.
If encountering poor cell seeding into the Gut-on-a-Chip, it could be due to a low cell number or poor cell attachment. To troubleshoot low cell numbers, it is important to inspect the health of prepared intestinal organoids by observing their growth in Matrigel. Cell viability can be assessed by Trypan blue staining after cell dissociation to ensure no more than 20% of cells are dead. If viable cell numbers are insufficient, optimizing organoid medium conditions can be attempted. Another possibility is incomplete organoid dissociation, resulting in an excess of cell clumps larger than 70 µm that become trapped by the filter. To resolve this, one option is to extend the duration of pipetting during cell dissociation. Alternatively, the 15 mL conical tube can be gently agitated every minute while undergoing treatment with a trypsin-like protease. Poor cell attachment to the Gut-on-a-Chip may be due to improper ECM coating. During the coating process, it is advised to carefully check for the presence of air bubbles and prevent their formation by gently adding more coating solution as needed. Overcrowding of cells and a failure to wash away unattached cells can result in an insufficient initial monolayer. In such a case, a mild pulsing can be applied when pushing the syringe plunger. These troubleshooting steps can help identify and address issues during the Gut-on-a-Chip culture process.
While this Gut-on-a-Chip platform enables the creation of undulated 3D epithelial layers, we recognize the need for additional biological complexity to replicate the intestinal microenvironment fully. It is crucial to consider the interactions between epithelial and mesenchymal cells, the deposition of ECM for 3D regeneration, and the presence of crypt-villus characteristics that establish a suitable stem cell niche. Stromal cells, such as fibroblasts, play a vital role in the production of ECM proteins and the regulation of intestinal morphogenesis34,35,36. The inclusion of mesenchymal cells in this model has the potential to enhance both morphogenesis and the efficiency of cell attachment. Endothelial layers, which encompass capillary vasculature and lymphatic vessels, play a crucial role in governing molecular transport and the recruitment of immune cells37,38. The inclusion of patient-derived immune cells could be essential in modeling intestinal diseases as it allows for the demonstration of the interplay between innate and adaptive immunity as well as the establishment of tissue-specific immunity39. Following the completion of 3D morphogenesis on Gut-on-a-Chip, the organoid culture medium can be modified to an organoid differentiation medium. This can be a viable approach to induce additional cellular differentiation, depending on the experimental objectives.
Imaging the 3D microarchitecture in situ is challenging due to the long working distance required, which can be overcome with a long-distance objective. Additionally, the layer-by-layer microfabrication and bonding methods make it difficult to access upper layers for examination with SEM. For the current Gut-on-a-Chip design, one syringe pump per Gut-on-a-Chip microdevice is needed, occupying CO2 incubator space and preventing large-scale experiments. Innovations are needed to increase scalability for a user-friendly platform and high-throughput screening.
These current protocols allow for the spontaneous development of 3D epithelial layers in vitro, surpassing the limitations of traditional 3D organoids, 2D monolayers, and static microdevice culture systems. This dynamic in vitro intestinal microenvironment can be controlled by introducing co-culture of diverse cell types. Previous studies have explored methods for manipulating the Gut-on-a-Chip microenvironment, including co-culturing intestinal microbiome14,23 and peripheral mononuclear cells30. This reconstituted microenvironment has numerous potential applications, including drug testing, fundamental mechanistic studies, and disease modeling. The reconstructed microenvironment holds significant potential for a wide range of applications, such as drug testing23,40,41 and disease modeling12,13,14,30, as well as fundamental mechanistic investigations of intestinal morphogenesis42. A variety of assays can be performed by either collecting supernatants for assessment of metabolites43, by collecting cells for genomic examination2,32, or by visually examining the cells using live-cell dyes or fixation for subsequent immunofluorescence imaging23,44.
This study presents a reproducible protocol for developing 3D morphogenesis of canine intestinal epithelial layers in a Gut-on-a-Chip platform. The resulting 3D epithelial structure provides a more realistic representation of the intestinal microenvironment, which has immense potential for applications in various biomedical studies. By utilizing this intestinal architecture, we can conduct more translational research and potentially yield promising outcomes.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank WSU Small Animal Internal Medicine service (Dr. Jillian Haines, Dr. Sarah Guess, Shelley Ensign LVT) and WSU VTH Clinical Studies Coordinator Valorie Wiss for their support in case recruitment and sample collection from citizen scientists (patient donors). This work was supported in part by the Office of The Director, National Institutes Of Health (K01OD030515 and R21OD031903 to Y.M.A.) and Japan Society for the Promotion of Science Overseas Challenge Program for Young Researchers (202280196 to I.N.). Figure 1A and Figure 3A were created with BioRender.com.
Materials
| Organoid basal medium | |||
| Advanced DMEM/F12 | Gibco | 12634-010 | |
| GlutaMAX | Gibco | 35050-061 | 2 mM, glutamine substitute |
| 1 M HEPES | VWR Life Science | J848-500ML | 10 mM |
| 100x penicillin–streptomycin | Corning | MT30009CI | 1x |
| Organoids and organoid medium | |||
| A-83-01 | PeproTech | 9094360 | 500 nM |
| B27 supplement | Gibco | 17504-044 | 1x |
| CHIR99021 | Reprocell | 04-0004-base | 2.5 µM |
| HEK293 cells engineered to secrete Noggin | Baylor College of Medicine | ||
| Murine EGF | PeproTech | 315-09-1MG | 50 ng/mL |
| Murine Wnt-3a | PeproTech | 315-20-10UG | 100 ng/mL |
| N-Acetyl-L-cysteine | Sigma | A9165-25G | 1 mM |
| N2 MAX Media supplement | Gibco | 17502-048 | 1x |
| Nicotinamide | Sigma | N0636-100G | 10 mM |
| Noggin Conditioned Medium | NA | NA | 10% vol/vol |
| Primocin | InvivoGen | ant-pm-1 | 100 µg/ml |
| R-spondin1 (Rspo1) cells | Trevigen | 3710-001-01 | Rspo1 cells |
| R-Spondin-1 Conditioned Medium | NA | NA | 20% vol/vol |
| SB202190 | Sigma-Aldrich | S7067-25MG | 10 µM |
| Y-27632 | StemCellTechnologies | 72308 | 10 µM |
| [Leu15 ]-Gastrin I human | Sigma-Aldrich | G9145-.5MG | 10 nM |
| Reagents | |||
| 4% Paraformaldehyde solution | Fisher Scientific | AAJ19943K2 | |
| Alexa Fluor 647 Phalloidin | Thermo Fisher Scientific | A22287 | x250 dilution |
| Anti-Rabbit IgG H&L labeled with Alexa Fluor 555 | Abcam | ab150078 | x1,000 dilution |
| Anti-ZO-1 polyclonal antibody | Thermo Fisher Scientific | 61-7300 | x50 dilution |
| Cell Recovery Solution | Corning | 354253 | |
| Collagen I, Rat Tail 3 mg/mL | Gibco | A10483-01 | |
| Diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | 62248 | x1,000 dilution |
| EMS Glutaraldehyde Aqueous 50% | Electron Microscopy Sciences | 16320 | |
| Matrigel Matrix | Corning | 356255 | |
| Poly(ethyleneimine) solution | Sigma | 408700-250ML | |
| TrypLE Express | Gibco | 12604-021 | |
| Materials and Equipment | |||
| 24-well culture plates | Corning | 3524 | |
| 87V Industrial Multimeter | Fluke Corporation | ||
| Centrifuge | Eppendorf | 5910R | |
| CO2 incubator | Eppendorf | C170i | |
| DMi8 fluorescence microscope | Leica microsystems | DMi8 | |
| Dry oven | Fisher Scientific | 15-103-0519 | |
| FlexCell FX-5000 Tension system | Flexcell International Corporation | ||
| Inverted phase-contrast microscope | Leica microsystems | DMi1 | |
| SP8-X inverted confocal microscope | Leica microsystems | SP8-X | |
| Syringe pump | Braintree Scientific | model no. BS-8000 120V | |
| Syringe, 3 mL sterile | BD Biosciences | 14-823-435 | |
| Syringes, 1 mL sterile | BD Biosciences | 14-823-434 | |
| UV/ozone generator | Jelight Company | model no. 30 | |
| Software | |||
| LAS X imaging software | Leica microsystems |
References
- Shanks, N., Greek, R., Greek, J. Are animal models predictive for humans. Philosophy, ethics, and humanities in medicine: PEHM. 4, 2 (2009).
- Shin, W., Kim, H. J. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nature protocols. 17 (3), 910-939 (2022).
- Coelho, L. P., et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. BMC Biome. 6 (72), 1-11 (2018).
- German, A. J. The growing problem of obesity in dogs and cats. The Journal of Nutrition. 136, 1940-1946 (1940).
- Patronek, G. J., Waters, D. J., Glickman, L. T. Comparative longevity of pet dogs and humans: Implications for gerontology research. Journals of Gerontology – Series A Biological Sciences and Medical Sciences. 52 (3), B171-B178 (1997).
- Lutz, T. A. Mammalian models of diabetes mellitus, with a focus on type 2 diabetes mellitus. Nature reviews. Endocrinology. 19 (6), 350-360 (2023).
- Allenspach, K., Culverwell, C., Chan, D. Long-term outcome in dogs with chronic enteropathies: 203 cases. Veterinary Record. 178 (15), 368 (2016).
- Patnaik, A. K., Hurvitz, A. I., Johnson, G. F. Canine gastrointestinal neoplasms. Veterinary Pathology. 14 (6), 547-555 (1977).
- Saito, T., et al. Immunohistochemical analysis of beta-catenin, e-cadherin and p53 in canine gastrointestinal epithelial tumors. Journal of Veterinary Medical Science. 82 (9), 1277-1286 (2020).
- Kopper, J. J., et al. Harnessing the biology of canine intestinal organoids to heighten understanding of inflammatory bowel disease pathogenesis and accelerate drug discovery: A One Health approach. Frontiers in Toxicology. 3, 1-13 (2021).
- Jalili-firoozinezhad, S., et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature Biomedical Engineering. 3 (7), 520-531 (2019).
- Shin, W., et al. Robust formation of an epithelial layer of human intestinal organoids in a polydimethylsiloxane-based Gut-on-a-Chip microdevice. Frontiers in Medical Technology. 2, (2020).
- Shin, Y. C., et al. Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal Interface-on-a-Chip. Micromachines. 11 (7), 663 (2020).
- Tovaglieri, A., et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome. 7 (1), 43 (2019).
- Chandra, L., et al. Derivation of adult canine intestinal organoids for translational research in gastroenterology. BMC Biology. 17 (1), 1-21 (2019).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C., Smith, J. G. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunology. 8 (2), 352-361 (2015).
- Ambrosini, Y. M., et al. Recapitulation of the accessible interface of biopsy-derived canine intestinal organoids to study epithelial-luminal interactions. PLoS ONE. 15 (4), 1-17 (2020).
- Gabriel, V., et al. Canine intestinal organoids in a dual-chamber permeable support system. Journal of Visualized Experiments. 2 (181), 1-24 (2022).
- Yamada, A., et al. Confronting emerging zoonoses: the one health paradigm. Confronting Emerging Zoonoses: The One Health Paradigm. , 1 (2014).
- Lerner, H., Berg, C. The concept of health in One Health and some practical implications for research and education: what is One Health. Infection Ecology & Epidemiology. 5 (1), 25300 (2015).
- Garcia, S. N., Osburn, B. I., Jay-Russell, M. T. One Health for food safety, food security, and sustainable food production. Frontiers in Sustainable Food Systems. 4, 1-9 (2020).
- Min, S., et al. Live probiotic bacteria administered in a pathomimetic Leaky Gut Chip ameliorate impaired epithelial barrier and mucosal inflammation. Scientific Reports. 12 (1), 22641 (2022).
- Kim, H. J., Huh, D., Hamilton, G., Ingber, D. E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a Chip. 12 (12), 2165-2174 (2012).
- Qu, M., et al. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Research. 31 (3), 259-271 (2021).
- Sahoo, D. K., et al. Differential transcriptomic profiles following stimulation with lipopolysaccharide in intestinal organoids from dogs with inflammatory bowel disease and intestinal mast cell tumor. Cancers. 14 (14), 3525 (2022).
- Gabriel, V., et al. Standardization and maintenance of 3D canine hepatic and intestinal organoid cultures for use in biomedical research. Journal of Visualized Experiments. (179), 1-28 (2022).
- Heijmans, J., et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell reports. 3 (4), 1128-1139 (2013).
- Shin, W., et al. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Frontiers in Bioengineering and Biotechnology. 7, 1-13 (2019).
- Shin, W., Kim, H. J. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proceedings of the National Academy of Sciences of the United States of America. 115 (45), E10539-E10547 (2018).
- Park, G. S., et al. Emulating host-microbiome ecosystem of human gastrointestinal tract in vitro. Stem Cell Reviews and Reports. 13 (3), 321-334 (2017).
- Kim, H. J., Li, H., Collins, J. J., Ingber, D. E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proceedings of the National Academy of Sciences of the United States of America. 113 (1), E7-E15 (2016).
- Jergens, A. E., et al. A scoring index for disease activity in canine inflammatory bowel disease. Journal of Veterinary Internal Medicine. 17 (3), 291-297 (2003).
- Roulis, M., Flavell, R. A. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation; research in biological diversity. 92 (3), 116-131 (2016).
- Göke, M., Kanai, M., Podolsky, D. K. Intestinal fibroblasts regulate intestinal epithelial cell proliferation via hepatocyte growth factor. The American Journal of Physiology. 274 (5), G809-G818 (1998).
- Powell, D. W., Pinchuk, I. V., Saada, J. I., Chen, X., Mifflin, R. C. Mesenchymal cells of the intestinal lamina propria. Annual Review of Physiology. 73, 213-237 (2011).
- Pappenheimer, J. R., Michel, C. C. Role of villus microcirculation in intestinal absorption of glucose: coupling of epithelial with endothelial transport. The Journal of Physiology. 553, 561-574 (2003).
- Agace, W. W. T-cell recruitment to the intestinal mucosa. Trends in Immunology. 29 (11), 514-522 (2008).
- Ambrosini, Y. M., Shin, W., Min, S., Kim, H. J. Microphysiological engineering of immune responses in intestinal inflammation. Immune Network. 20 (2), 13 (2020).
- Kasendra, M., et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Scientific Reports. 8 (1), 1-14 (2018).
- Sontheimer-Phelps, A., et al. Human Colon-on-a-Chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cellular and Molecular Gastroenterology and Hepatology. 9 (3), 507-526 (2020).
- Shin, W., et al. Human intestinal morphogenesis controlled by transepithelial morphogen gradient and flow- dependent physical cues in a microengineered Gut-on-a-Chip. iScience. 15, 391-406 (2019).
- Gijzen, L., et al. An Intestine-on-a-Chip model of plug-and-play modularity to study inflammatory processes. SLAS Technology. 25 (6), 585-597 (2020).
- Kim, H. J., Lee, J., Choi, J. H., Bahinski, A., Ingber, D. E. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip microfluidic device. Journal of Visualized Experiments. (114), 3-9 (2016).

