Development of Multiplex Real-Time RT-qPCR Assays for the Detection of SARS-CoV-2, Influenza A/B, and MERS-CoV
Summary
We present two probe-based in-house one-step RT-qPCR kits for common respiratory viruses. The first assay is for SARS-CoV-2 (N), Influenza A (H1N1 and H3N2), and Influenza B. The second is for SARS-Cov-2 (N) and MERS (UpE and ORF1a). These assays can be successfully implemented in any specialized laboratory.
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes Coronavirus disease 2019 (COVID-19) is a serious threat to the general public's health. During influenza seasons, the spread of SARS-CoV-2 and other respiratory viruses may cause a population-wide burden of respiratory disease that is difficult to manage. For that, the respiratory viruses SARS-CoV-2, Influenza A, Influenza B, and Middle East respiratory syndrome (MERS-CoV) will need to be carefully watched over in the upcoming fall and winter seasons, particularly in the case of SARS-CoV-2, Influenza A, and Influenza B, which share similar epidemiological factors like susceptible populations, mode of transmission, and clinical syndromes. Without target-specific assays, it can be challenging to differentiate among cases of these viruses owing to their similarities. Accordingly, a sensitive and targeted multiplex assay that can easily differentiate between these viral targets will be useful for healthcare practitioners. In this study, we developed a real-time reverse transcriptase-PCR-based assay utilizing an in-house developed R3T one-step RT-qPCR kit for simultaneous detection of SARS-CoV-2, Influenza A, Influenza B, and SARS-CoV-2, MERS-CoV. With as few as 10 copies of their synthetic RNAs, we can successfully identify SARS-CoV-2, Influenza A, Influenza B, and MERS-CoV targets simultaneously with 100% specificity. This assay is found to be accurate, reliable, simple, sensitive, and specific. The developed method can be used as an optimized SARS-CoV-2, Influenza A, Influenza B, and SARS-CoV-2, MERS-CoV diagnostic assay in hospitals, medical centers, and diagnostic laboratories as well as for research purposes.
Introduction
The pandemic of the ongoing coronavirus disease 2019 (COVID-19) is caused by the novel coronavirus known as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)1. Due to SAR-CoV-2's strong contagiousness and capacity for rapid transmission, the COVID-19 pandemic emerged in Wuhan City, China, and spread quickly throughout the world. This eventually led to the start of respiratory distress signs and even death2,3,4. COVID-19 has been declared a pandemic in more than 213 countries, expecting a steep increase in the number of confirmed cases, as evidenced by the papers published by different research studies3,5. COVID-19 is transmitted primarily by small respiratory droplets that infected individuals release into the environment and then get exposed to vulnerable individuals through inhalation or close contact with contaminated surfaces. When these droplets come into contact with the mucosa of the eyes, mouth, or nose, a person may become infected6. Statistics released by the World Health Organization (WHO) show that there have been more than 76 million confirmed cases of the pandemic worldwide, with a staggering 7 million deaths7. Thus, the United Nations classified the pandemic caused by COVID-19 disease as a disaster because of its direct impact on the lives of billions of people around the globe and had far-reaching economic, environmental, and social effects.
Public health initiatives including thorough testing, early detection, contact tracing, and case isolation have all been shown to be crucial in keeping this pandemic under control8,9,10,11. The winter months will increase the circulation of other respiratory viruses like Influenza A and B with COVID-19-like symptoms making it difficult to identify, track down, and isolate COVID-19 instances early on. Every year, Influenza A and B outbreak starts in the late fall or early January with a predictable seasonality12. Numerous epidemiological traits are shared by SARS-CoV-2 and Influenza viruses. Besides, sharing similarities in the susceptible populations which include children, the elderly, immunocompromised, and individuals with chronic comorbidities such as asthma, chronic obstructive pulmonary disease, cardiac and renal failure, or diabetes12,13. These viruses not only share vulnerable populations but also transmission routes of contact and respiratory droplets14. It is anticipated that patients may likely contract more than one of these respiratory viruses as flu season approaches14. For that, the screening of SARS-CoV-2 and the Influenza viruses needs to be done on symptomatic patients before they are isolated. Running separate tests for the three viruses (SARS-CoV-2, Influenza A, and Influenza B) is not possible due to the global lack of resources for nucleic acid extraction and diagnostics. In order to screen them all in one reaction, a method or test needs to be developed.
Middle East respiratory syndrome (MERS)-CoV is a human coronavirus (CoV) family member. The first MERS-CoV virus isolates came from a hospitalized patient in Saudi Arabia who had died in September 2012 due to acute respiratory issues15. There is evidence that suggests that a prominent reservoir host for MERS-CoV is dromedary camels. It has been proven that viruses from infected dromedary camels are zoonotic and thus can infect humans16,17. Humans infected with this virus can spread it to others through close contact18. As of January 26th, 2018, there had been 2143 laboratory-confirmed cases of MERS-CoV infection including 750 deaths globally19. The most typical MERS-CoV symptoms are coughing, fever, and shortness of breath. MERS-CoV infections have also been reported to exhibit pneumonia, diarrhea and gastrointestinal sickness symptoms20. Currently, no commercial vaccine or specific treatment for MERS-CoV is available. Therefore, prompt and precise diagnosis is essential for preventing the widespread MERS-CoV outbreaks and differentiating MERS-CoV from SARS-CoV-2 disease.
To date, many approaches have been proposed to detect these viruses such as multiplex RT-PCR21,22,23,24,25, CRISPR/Cas1226,27, CRISPR/Cas928, and CRISPR/Cas329, lateral flow immunoassay30, paper-based biomolecular sensors31, SHERLOCK testing in one pot32, DNA aptamer33, loop-mediated isothermal amplification (LAMP)19,34, etc. Each of the aforementioned methods has unique benefits and drawbacks in terms of sensitivity and specificity. Among these methods, the nucleic acid amplification-based test: multiplex qRT-PCR, is the most common and is considered to be the gold standard for the diagnosis of SARS-CoV-2, Influenza A, Influenza B, and MERS-CoV.
In this study, we designed and assessed various primer combinations and probes for the effective, accurate, and simultaneous detection of SARS-CoV-2, Influenza A, Influenza B, and SARS-CoV-2, MERS-CoV utilizing standard twist synthetic viral RNAs. The multiplexed assays developed for either MERS-CoV or SARS-CoV-2 target genes are recommended by the World Health Organization (WHO). These genes generally encode proteins and complexes that contribute to the formation of a replication/transcription complex (RTC)35 such as the region within the open reading frame 1a (ORF1a) that is used for MERS-CoV assay. In addition, structural proteins are encoded by the genes utilized in diagnostic assays such as the upstream region of envelope gene (upE) and nucleocapsid gene (N) which are used for MERS-CoV and SARS-Cov-2 assays, respectively35,36. We used in-house R3T one-step RT-qPCR kit to establish the RT-qPCR for the detection of viruses37. Virus detection, sensitivity, specificity, and dynamic range of our R3T one-step RT-qPCR kit and primer sets were tested and evaluated using 10-fold serial dilutions of the standard twist synthetic RNAs. The lowest practical detection limit was approximately 10 transcripts copies per reaction. As a result, the in-house R3T one-step RT-qPCR kit and primer/probe sets can be successfully used and implemented for routine simultaneous diagnosis of SARS-CoV-2, Influenza A, Influenza B, and SARS-CoV-2, MERS-CoV.
Protocol
1. Taq polymerase expression and purification
- Construct a plasmid with a cleavable hexa-histidine tag at the C-terminus of the enzyme.
- Transform 50 ng of the expression vector into E. coli BL21-(DE3) strain following the standard protocol38.
- Inoculate the transformed cells in four 6 L flasks each containing 2 L of 2YT media broth at 37 °C with shaking at 170 rpm until the OD 600 of 0.8 or cell number 6.4 x 108 is reached.
- Induce Taq polymerase expression with 0.5 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) and further incubate at 16 °C for 18 h with shaking.
- Harvest the cells by spinning them down at 4 °C at 7808 x g for 10 min. Resuspend the pellets in 200 mL of an ice-cold Taq polymerase lysis buffer (Table 1) in 5 mL/g of cell pellet.
- Incubate the cells with lysozyme (2 mg/mL of lysis buffer) and protease inhibitors for 45 min and then pass the lysate through a cell disruptor at 30 kPsi and centrifuge at 22,040 x g for 30 min at 4 °C to separate the cell debris.
- Heat the supernatant for 15 min at 85 °C. Spin down at 95,834 x g for 1 h at 4 °C. Collect the supernatant and filter it on ice using 0.45 µm filter.
- Perform protein purification using Fast Protein Liquid Chromatography (FPLC) by first passing the sample through a Ni-NTA HP 5 mL column at 4 mL/min flow rate using Taq polymerase buffer A (Table 2; Figure 1A).
- Wash with 10 column volumes (CV) of Taq polymerase buffer A and 4% of Taq polymerase buffer B (Table 2). Elute the bound protein with a linear gradient using Taq polymerase Buffer B over 20 CV in a 100 mL bottle.
- Pass the eluent in the 100 mL bottle through a cation exchange 5 mL column at 4 mL/min using Taq polymerase buffer C (Table 2; Figure 1A).
- Wash with 20 CV of 100% Taq polymerase buffer C and 5% Taq polymerase buffer D (Table 2).
- Elute with a linear gradient using Taq polymerase buffer D (Table 2) starting from 5% to 100%.
- Check the eluted fractions by performing SDS-PAGE gel electrophoresis 39 followed by gel staining 40 as previously described. Briefly, take 10 µL of each fraction and add an equal volume of 2x SDS loading dye. Denature the sample at 90oC for 10 min. Load the samples onto 10% SDS-PAGE gel and run the gel for 25 min at 200 V. Stain the gel using Coomassie Brilliant Blue and then de-stain.
- Collect all the fractions that contain purified Taq polymerase and dialyze against taq polymerase storage buffer (Table 3) at 4 °C. Briefly, prepare 2 L of the storage buffer and immerse the dialysis cassette in the buffer for 2 min to hydrate. Inject the collected fractions using a needle into the dialysis cassette and leave it overnight in the dialysis buffer at 200 rpm on the stirrer.
- After dialysis, measure the protein concentration using a spectrophotometer, make 10 µL aliquots, snap freeze in liquid nitrogen and store at -80 °C.
NOTE: Filter all buffers for purification using a 0.45 µm filter before loading them into the FPLC system.
2. MMLV-RT expression in insect cell expression system and purification
- Bacmid DNA generation and isolation
- Clone the sequence of MMLV-RT with a C-terminus cleavable TEV-8xHis-Strep tag as previously described41.
- Mix 100 ng of the expression vector into 50 μL of DH10Bac cells and mix gently by tapping the tube.
- Incubate the cells on ice for 15 min. Heat-shock the cells for 1 min at 42 °C.
- Immediately transfer on ice and add 400 μL of S.O.C medium. Incubate at 37 °C with shaking for 4 h.
- Plate 10 μL and 15 μL of the mixture on LB Agar plates containing three antibiotics; 50 μg/mL Kanamycin, 10 μg/mL Tetracycline and 7 μg/mL Gentamicin along with 40 μg/mL IPTG and 100 μg/mL X-Gal for blue-white selection.
- Incubate the plates at 37 °C for 48 h. Pick several white colonies and one blue colony as a control, and re-streak them on fresh LB agar plates containing the above antibiotics. Incubate the plates overnight at 37 °C.
- After confirming the white phenotype, pick a couple of colonies and inoculate them into 10 mL of LB media containing 50 μg/mL Kanamycin, 10 μg/mL Tetracycline, and 7 μg/mL Gentamicin in 50 mL tubes.
- Incubate the culture at 37 °C with shaking at 170 rpm overnight. Pellet down the cells by spinning them down at 22,040 x g for 10 min.
- Decant the supernatant and resuspend the pellet in 250 µL of resuspension solution from miniprep kit (this solution needs to be handled in accordance with the manufacturer's instructions), then transfer the solution to a 1.5 mL microcentrifuge tube.
- Add 250 µL of lysis solution, gently mix and incubate for 3 min. Add 350 µL of neutralizing solution, invert 2x-3x and centrifuge at 22,040 x g for 10 min.
- Transfer the supernatant to a fresh tube and add an equal volume of ice-cold isopropanol and incubate for 30 min at -20 °C.
- Pellet down the precipitated DNA by spinning at 22,040 x g for 10 min. Discard the supernatant and wash the pellet with 700 µL of 70% ice-cold ethanol, 2x.
- Remove the supernatant with a pipette without touching the pellet. Let the pellet air dry in the laminar flow hood for 5-10 min or until no liquid is seen in the tube. Dissolve the pellet in 100 µL of EB buffer.
- Bacmid transfection and virus amplification
- P1 virus preparation
- In a 6-well tissue culture plate, seed ~ 9 x 105 cells/well in 2 mL of insect cell culture medium.
- Incubate the plate for ~30 min at room temperature to allow the cells to attach.
- Prepare Bacmid/Fugene mix as follows: mix 1 µg of Bacmid DNA and 300 µL of insect cells media in one tube. In another tube, mix 8 µL of transfection reagent and 300 µL of insect cells media. Mix both mixtures by gentle pipetting and leave for ~30 min at room temperature for the bacmid/transfection reagent complex to form.
- Add bacmid complex mix (~210 µL) to each well dropwise and seal the plate with a transparent film.
- Incubate the plate at 27 °C for 3-4 days.
- Take the medium which that contains the virus, add FBS (final concentration 2%), filter using 0.45 µm filter and store at 4°C in the dark as P1 virus stock.
- P2 virus preparation
- Transfect 50 mL of insect cells at 2 x 106 cells/mL with 3 mL of P1 virus in a culturing flask.
- Incubate at 27 °C with shaking at 100 rpm for 4-6 days until the percentage of dead cells reaches 25%-30%.
- Centrifuge at 300 x g for 10 min and collect the supernatant that contains the virus.
- Add FBS to a final concentration of 2%, filter through 0.45 µm filter and aliquot 1 mL each and store at -80 °C as P2 virus stock.
- P3 virus preparation
- Transfect 100 mL of insect cells at 2 x 106 cells/mL with the 2 mL of P2 virus in a culturing flask.
- Incubate at 27 °C with shaking at 100 rpm for 3-4 days until the percentage of dead cells reaches 15%-20%.
- Centrifuge the cells at 300 x g for 10 min and collect the supernatant that contains the virus.
- Add FBS to a final concentration of 2%. Filter through a 0.45 µm filter. It can be stored in the dark at 4 °C for 1 month.
- P1 virus preparation
- Expression and purification of MMLV-RT in insect cells
- Add 5 mL of P3 virus to fresh insect cells at a density of 2 x 106 cells/mL (700 mL/2L flask) in total 6 flasks.
- Harvest the cells after 55-60 h of post-transfection by spinning at 7808 x g for 10 min.
- Resuspend the cell pellets in 200 mL of MMLV-RT lysis buffer (Table 4). Pass the lysate through a cell disruptor at 15 kPsi and centrifuge at 22,040 x g for 30 min at 4 °C.
- Transfer the supernatant into new tubes and spin down again at 95,834 x g at 4 °C for 1 h. Collect the supernatant and filter it on ice using a 0.45 µm filter.
- Start protein purification using FPLC by first passing the sample through Ni-NTA Excel 5 mL column (Figure 1B) at 4 mL/min flow rate by using MMLV-RT buffer A (Table 5).
- Wash the column with 10 CV of MMLV-RT buffer A and 4% of MMLV-RT buffer B (Table 5) and elute the protein with the linear gradient of MMLV-RT buffer B over 20 CV in a 100 mL bottle.
- Pass the eluted sample through the Strep 5 mL column (Figure 1B) equilibrated with MMLV-RT buffer C (Table 5).
- Wash the column with 10 CV of 100% MMLV-RT buffer C and elute the protein with a linear gradient with 20 CV of buffer D (Table 5).
- Check the eluted fractions by performing SDS-PAGE as done is steps 1.13.-1.14. and collect the fractions that contain purified MMLV-RT to be dialyzed against MMLV-RT storage buffer (Table 6) at 4 °C.
- Measure the protein concentration using Nanodrop, aliquot, snap freeze in liquid nitrogen and store at -80 °C.
NOTE: Filter all buffers for purification using a 0.45 µm filter before loading them into the FPLC system.
3. Preparation of in-house multiplex R3T one-step RT-qPCR kit components
- Buffer mix preparation
- Prepare the 2x buffer for the RT-qPCR reaction that contains the reagents previously shown in37. Prepare all the reagents in DNase/RNase-Free water and the buffer mix stored in a -20 °C freezer.
- Primer and Probe Sets and primers mixture preparation
NOTE: The probes and primers used are listed in Table 7. The Influenza and SARS-CoV-2 multiplexed kit contains 3 probes and 4 forward and reverse primer sets. The probes are labeled at the 5′ ends using reporters 6-carboxyfluorescein (FAM) for InfA, Texas Red-XN for SARS-CoV-2 (N gene) and Yakima Yellow for InfB. The MERS-CoV and SARS-CoV-2 multiplexed kit contains 3 probes and 3 forward and reverse primer sets. The probes are also labeled at the 5′ ends using reporters Texas Red-XN for MERS-CoV (ORF1a gene), VIC for MERS-CoV (UpE gene) and 6-carboxyfluorescein (FAM) SARS-CoV-2 (N gene).- Prepare a primer mix containing all the primers and probes listed in Table 7 with a final concentration of 6.7 µM for each forward and reverse primer and 1.25 µM for each probe in a 1.5 mL tube. Store the primer mix in the -20 °C freezer. Use elution buffer to make up the required volume.
- Enzyme mix preparation
- Prepare Taq polymerase (30 U/µL) and MMLV-RT (60 ng/µL) enzyme mix in the Taq polymerase storage buffer as shown in Table 3 to make up the required volume. Perform this step on ice and store the enzyme mix at -20 °C.
- RNA template
- Resuspend synthetic RNAs of SARS-CoV-2, Influenza A H1N1, Influenza A H3N2, Influenza B and MERS-CoV separately in 100 µL of 1x Tris-EDTA buffer (10 mM Tri-Cl and 1 mM EDTA (pH 8.0) to make a stock of 1 x 106 RNA copies/µL.
- Mix synthetic RNAs for RT-qPCR in the following configurations: 1) SARS-CoV-2, Influenza A and Influenza B by adding 5 µL of each virus stock to 50 µL volume to make 1 x 105 RNA copies/µL; 2) SARS-CoV-2, MERS-CoV by adding 5 µL of each virus stock to 50 µL volume to make 1 x 105 RNA copies/µL. Make 10-fold serial dilutions of each synthetic RNA mixture ranging from 1 x 105 RNA copies/µL to 10 RNA copies/µL.
NOTE: Make sure to use ice-cold DNase/RNase-Free water to prepare the serial dilution of the templates.
4. In-house multiplexed SARS-CoV-2, Influenza A, Influenza B and SARS-CoV-2, MERS-CoV one-step RT-qPCR test
NOTE: Disinfect workstation surfaces and use a 96-well plate template to plan the PCR plate layout.
- Preparation of 96-well plate
- Thaw 2x buffer and primer mix. Keep enzyme mix on ice.
- To each well, add 10 µL of 2x buffer mix, 1.5 µL of the primer mix, 1 µL of enzyme mix, 6.5 µL of DNase/RNase-Free Water, and 1 µL of corresponding RNA mixture that needs to be tested. The final volume in each well is 20 µL. Please refer to the prepared layout for help with this step.
NOTE: It is recommended to make a master mix based on the number of reactions that contain all the reagents except the RNA sample to avoid pipetting bias. In addition, perform the reactions in duplicate or triplicate to avoid technical errors. - Seal the plate with an adhesive PCR plate seal and centrifuge briefly for 1 min to collect all liquid at the bottom of the plate. Ensure that each well has the same volume of liquid and is free of bubbles before transferring the samples to the qPCR machine.
NOTE: All the PCR reactions need to be set up on ice.
- PCR program
- Open the real-time qPCR machine program and choose the following experimental properties:
The block type: Fast 96-well (0.1 mL)
Experiment setup: Standard curve
Reagents: TaqMan reagents
Run properties: Standard. - Define all the gene targets and their reporter dye as presented in Table 7. In addition, define sample names for each reaction to be tested for easier analysis of the amplification curves.
- Assign targets and samples to the program's plate layout based on the 96-well plate.
- Set the following cycle program in the PCR instrument as follows:
55 °C for 10 min
40 cycles: 94 °C for 1 min
94 °C for 10 s
68 °C for 10 s
68 °C for 20 s; here acquire fluorescence. - Transfer the plate to the real-time qPCR machine and place it in the holder ensuring the correct orientation of the plate and start the run.
- Choose the file location to save the experimental data.
- Open the real-time qPCR machine program and choose the following experimental properties:
- Data analysis
- It is essential to first examine the amplification plots produced by the qPCR program. Analyze the limit of detection to assess the minimum RNA concentration that can be detected.
- Plot the log of RNA copy number on the x-axis and the corresponding average Ct value on the y-axis to assess the sensitivity of the assay. The slope and the R2 indicate the reliability and efficiency of the reaction.
Representative Results
In recent years, there have been significant advances in the diagnostic approach for detecting common respiratory viruses using PCR approaches21,22,23,24,25. However, despite these advancements, the multiplexed approach, which allows for detecting multiple viruses in a single test, has not been widely implemented, particularly in the RT-qPCR platform. This method has been successfully implemented using synthetic RNA viruses and in-house optimization of reaction components37. The specificity, sensitivity, and reliability of the diagnostic assay were assessed to be high, through the use of a limit of detection analysis. The presented approach could greatly improve the efficiency and accuracy of respiratory virus diagnosis, reducing the need for multiple tests and minimizing the risk of misdiagnosis as in43. Further research is needed to explore this approach's benefits using patient samples and encourage its adoption in clinical practice.
Expression and purification of Histidine-tagged Taq DNA polymerase
Based on the established protocol of Taq DNA polymerase expression and purification44, N-terminal histidine-tagged Taq DNA polymerase plasmid was constructed under the control of a cold-shock promoter to ease its expression and purification process as explained above (Figures 1A and Figure 2A). Thus, by just altering IPTG concentration and temperature, the level of expression of this novel construct can be controlled easily. Incubation of cells containing the Taq DNA polymerase plasmid at 16°C with 1 mM IPTG showed enhanced expression of His-Taq DNA polymerase. In addition, the earlier established protocol44 required time-consuming polyethyleneimine (PEI) precipitation steps, which can be skipped with the construct used here as the final product purity was unaffected and comparable to commercially available Taq polymerase (Figure 2B). The purified Taq polymerase migrated slower than the commercial one due to the presence of the linker peptides between the enzyme and histidine tag at the N-terminus. Taq DNA polymerase was produced successfully with 0.98 mg/L of E. coli culture of pure protein.
Expression and purification of double His- and Strep-Tagged MMLV reverse transcriptase
The baculovirus expression system was used to express MMLV-RT with a C-terminal double His and Strep-tag in insect cells (Sf9) as described in Figure 2A. A previous study showed that the C-terminally tagged MMLV-RT demonstrates higher activity where both the N-terminally tagged protein and the C-terminally tagged protein were synthesized in silkworms utilizing the silkworm-baculovirus expression vector system (silkworm-BEVS)41. To produce a homogeneous MMLV-RT protein, the same strategies and protocol were utilized as presented in the above-mentioned study. As a result, enhanced expression and more than 95% pure protein were achieved with a yield of 7.5 mg/L of insect cell culture as shown by the SDS-PAGE gel result (Figure 2C).
Optimization and standardization of the multiplexed qRT-PCR
An optimized and developed in-house multiplexed RT-qPCR test was assembled and produced successfully after rounds of buffer and primer mix optimization to detect three different common respiratory viruses simultaneously (Figure 3)37. The first kit was designed to target the SARS-CoV-2 N gene, Influenza A H1N1 and H3N2, and Influenza B gene; and the second kit is for SARS-CoV-2 N gene, MERS ORF1a, and upE genes. Thus, both kits can successfully and simultaneously detect all three targets as the workflow presented here (Figure 3). Synthetic RNA samples of each of the viruses used in this protocol were amplified indicating the possibility of using such as a multiplexed kit for common respiratory viruses' detection. The sensitivity of this diagnostic test is similar to previously reported results in patient samples. In cases where patients display influenza-like symptoms, the use of a multiplexed real-time RT-qPCR has shown comparable results with the SARS-CoV-2, influenza A and influenza B multiplexed assay45.
Sensitivity and limit of detection of in-house multiplexed RT-qPCR kit toward common respiratory viruses
The effectiveness of the in-house multiplexed kit was assessed using two sets of primer mix and the corresponding synthetic RNA for each kit as illustrated in the multiplexed RT-qPCR workflow (Figure 3). Using each primer mix with 10-fold serial dilution of each synthetic RNA mix ranging from 10 to 105 copies/µL as a template, the sensitivity, specificity, and the limit of detection of both developed multiplexed RT-qPCR can be determined using standard curve analysis. This is done in three independent replicates of each test, to confirm the efficacy and consistency of the multiplexed RT-qPCR kit. As a result, the two kits developed here can successfully amplify all three target genes simultaneously with as low as 10 RNA copies/reaction as seen by the amplification curve and the limit of detection (LoD) (Figure 4 and Figure 5). The slope and the R2 values of each curve were used to evaluate the efficiency of individual assays (Figure 4 and Figure 5). The R2 values provided an estimate of the goodness of the linear fit to the data points. In an efficient qPCR assay, R2 should be very close to or greater than 0.90. The amplification efficiencies of Influenza A, Influenza B and SARS-CoV-2 or MERS-CoV and SARS-CoV-2 titrations were above 99% for all primer sets (Figure 4 and Figure 5). In addition, the small error bars prove the reproducibility of each multiplexed RT-qPCR kit. It is important to note both multiplexed kits did not show any primer dimer formation since there is no amplification with the negative control (data not shown). Thus, the design of both kits amplified all target genes successfully with reliability, specificity, and sensitivity.
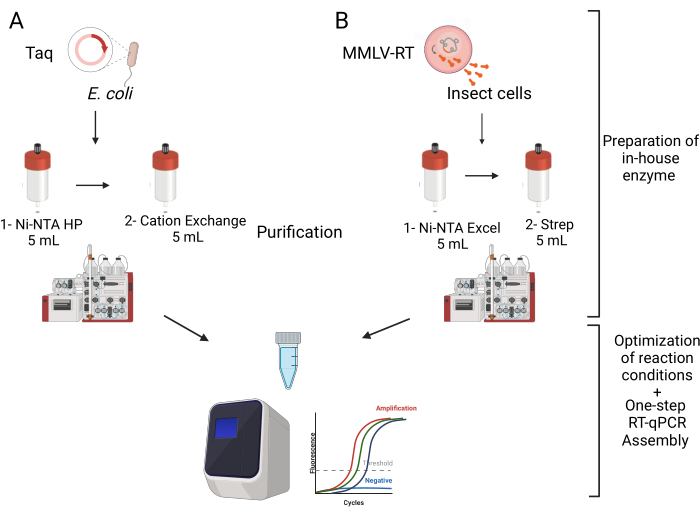
Figure 1: Schematic overview of the multiplexed one-step RT-qPCR kit assembly. The assembly of the kit starts with the expression and purification of the enzymes needed, Taq DNA polymerase and MMLV reverse transcriptase. Both enzymes needed to be passed through two columns as illustrated. Second, following the purification was the optimization of the reaction conditions and the thermal cycling program which resulted in optimal conditions for the multiplexed testing kit. Please click here to view a larger version of this figure.
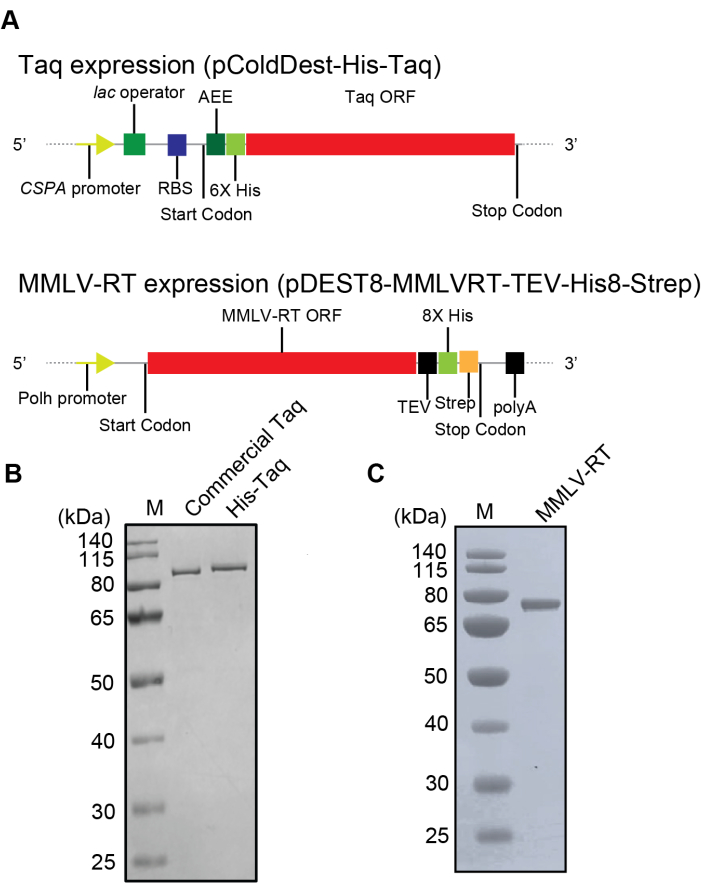
Figure 2: The plasmid constructs and the purification results of His-Taq Pol and C-His/Strep MMLV-RT. (A) Schematic representation of the recombinant His-Taq Pol and C-His/Strep MMLV-RT expression plasmids. Abbreviations: CSPA promoter – cold-shock protein A promoter; RBS – ribosome binding site; 6x His – His-tag with six histidines; TEE – translational-enhancing element; Taq ORF – Taq Pol open reading frame; Polh – polyhedrin promoter; MMLV-RT ORF – MMLV-RT open reading frame; TEV – etch virus protease targeting site; 8x His – His-tag with eight histidines; Strep – Strep-tag; polyA – SV40 late polyadenylation signal. (B) SDS-PAGE analysis of His-Taq Pol expressed in BL21(DE3) E. coli cells and Taq polymerase. (C) SDS-PAGE analysis of purified C-His/Strep MMLV-RT expressed in the Sf9 cells. Please click here to view a larger version of this figure.
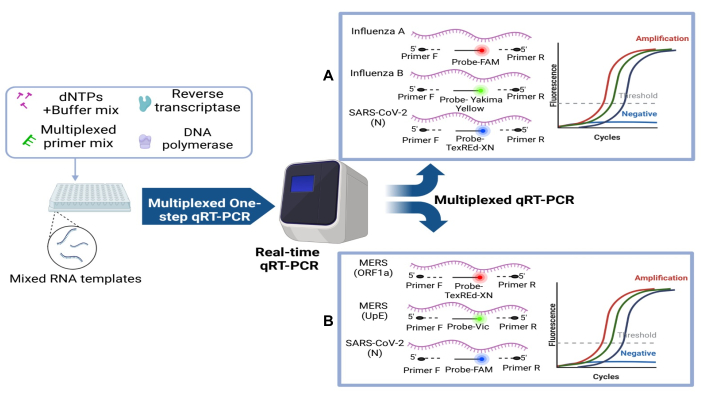
Figure 3: Schematic diagram of the two optimized, standardized, and developed multiplexed one-step RT-qPCR kits along with the reaction components. Each single multiplexed reaction is composed of dNTPs, buffer mix, enzyme mix, multiplexed primer mix, and the synthetic RNA template to be tested. (A) Influenza A/B, SARS-CoV-2 kit and (B) MERS ORF1a/upE, SARS-CoV-2. Please click here to view a larger version of this figure.
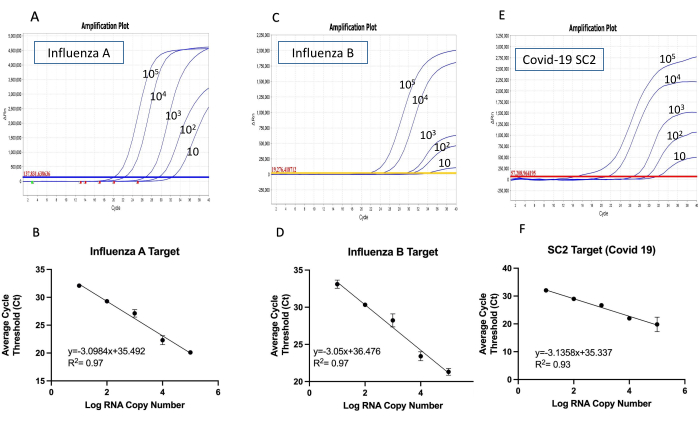
Figure 4: The amplification curves and the detection limit for Influenza A/B and SARS-CoV2 multiplex one-step RT-qPCR kit. The amplification curves of the multiplexed RT-qPCR were conducted using synthetic RNA mixture with 10-fold serial dilutions (10 to 105 copies/µL) with all targets (A) Influenza A, (C) Influenza B and (E) SARS-CoV-2. The reactions were carried out in triplicate, showing one replicate as an example. The limit of detection for (B) Influenza A, (D) Influenza B and (F) SARS-CoV-2 was determined by plotting the mean of three replicated Ct values against the log copy number. The coefficient of determination (R2) and the equation of the linear regression curve were calculated and shown in each panel. The error bars represent the standard deviation between three replicates. Please click here to view a larger version of this figure.
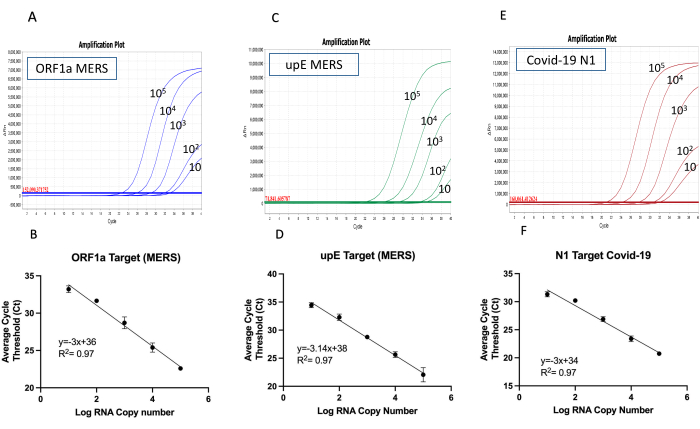
Figure 5: The amplification curves and the limit of the detection for MERS ORF1a/upE and SARS-CoV2 multiplex one-step RT-qPCR kit. The amplification curves of the multiplexed RT-qPCR were conducted using a synthetic RNA mixture with 10-fold serial dilutions (10 to 105 copies/µL) with all targets (A) MERS ORF1a, (C) MERS upE, and (E) SARS-Cov-2. The reactions were carried out in triplicate, showing one replicate as an example. The limit of detection for (B) MERS ORF1a, (D) MERS upE, and (F) SARS-CoV-2 was determined by plotting the mean of three replicated Ct values against the log copy number. The coefficient of determination (R2) and the equation of the linear regression curve were calculated and shown in each panel. The error bars represent the standard deviation between three replicates. Please click here to view a larger version of this figure.
Table 1: The list of the components for Taq polymerase lysis buffer. Please click here to download this Table.
Table 2: Taq polymerase purification buffers. The list of the buffer components needed for the two-column purification of Taq DNA polymerase. Please click here to download this Table.
Table 3: Taq polymerase storage buffer. The list of the buffer components required to dialyze Taq DNA polymerase after purification. Please click here to download this Table.
Table 4: The list of the components for MMLV-RT lysis buffer. Please click here to download this Table.
Table 5: MMLV-RT purification buffers. The list of the buffer components needed for the two-column purification of MMLV-RT. Please click here to download this Table.
Table 6: MMLV-RT storage buffer. The list of the buffer components needed to dialyze MMLV-RT after purification. Please click here to download this Table.
Table 7: Primers and probes information. The sequence information of the primers and probes needed for each multiplexed primer mix. Please click here to download this Table.
Discussion
There is a heavy economic burden on the healthcare system worldwide resulting from the high infection and mortality rates due to the spread of common respiratory viruses such as SARS-CoV-2, Influenza A/B and MERS-CoV variants12,19,20. Motivated by the sense of responsibility towards alleviating this burden, we realized the need for a quick, precise and accessible diagnostic assay such as RT-qPCR to distinguish between these common viruses in one test. By virtue of the multiplexed nature of qRT-PCR, it is feasible to diagnose and distinguish SARS-CoV-2 from other respiratory viruses, such as Influenza A/B and MERS-CoV viruses. Eventually, better and more precise treatment of patients can be achieved using the present multiplexed assay46. To recapitulate, this study describes the realization of an in-house one-step multiplex RT-qPCR test that can target two different combinations. Each combination is composed of three different respiratory viruses to limit the spread of such infectious viruses and reduce the economic burden on the healthcare system.
The present multiplexed RT-qPCR kit can target two combinations of common respiratory viruses (SARS-CoV-2, Influenza A/B) and (SARS-CoV-2, MERS UpE/ORF1a). The described protocol is easy, straightforward to follow and much cheaper than the commercially available one-step RT-qPCR kits. Another advantage was the multiplexed properties of the kit that can target multiple viral genetic targets by utilizing different probe and primer combinations. Thus, the versatility and simplicity of the kit enable straightforward implementation in resource-limited settings. Also, the kit is suitable for extensive testing resulting in accurate and precise diagnosis that can help limit the spread and co-infection of multiple respiratory viruses.
Preliminary assays were performed to ensure that all targets were amplified successfully and with high fidelity to limit the formation of non-specific products. First, we went through several rounds of optimization for the components of the buffer mix and their respective concentrations in the buffer mix. Second, we optimized the primers and their corresponding concentrations in both the primer mix and the PCR reaction. Third, we optimized the composition of the enzyme mix to maximize its activity and minimize the inhibitory effect of MMLV-RT on the activity of Taq polymerase47. Fourth, we optimized the thermal cycling conditions taking into account the limitations for the thermal stability of both enzymes. Incorporating the aforementioned improvements, the optimized version of our multiplexed RT-qPCR kit presented here was able to detect down to 10 RNA copies/reactions with high specificity, sensitivity and reproducibility.
Some precautions must be considered to ensure the high efficiency and successful implementation of the multiplex RT-qPCR assay, to prevent inhibitors and avoid pipetting errors. Since each facility is unique in its setup and instrumentation, here are some steps that might help while building such a kit: first, gloves need to be worn at all times to avoid the presence of any contamination and PCR inhibitor. Second, make sure to use aerosol-resistant filter tips and use a calibrated pipette. In addition, make sure to use PCR-grade water. The use of no-template control will help in verifying the presence of any contamination. It is important to note that the master mix needs to be prepared for all replicates to avoid possible contamination and pipetting variation. Other troubleshooting tips need to be considered to maintain probe quality such as aliquoting the stock and avoiding the use of water to dilute it. It is important to follow the manufacturer's recommendations on storing and diluting the primer and probe used. Thus, these simple tips will help in maintaining the success of the multiplex RT-qPCR assay and ensure its high efficiency.
Although the multiplexed RT-qPCR assays developed in this research have shown good results when tested with the synthetic viral RNAs, its effectiveness in a real clinical setting still needs to be evaluated and validated. Unfortunately, the scarcity of clinical specimens makes it difficult to determine the assay's specificities and sensitivity for the viruses included in this investigation. However, it is worth mentioning that the LOD (limit of detection), which indicates the minimum number of copies that can be detected in 100% of samples, was established through a statistically proven linear regression. This provides potential for clinical diagnosis and is an important finding of the research.
An in-house multiplex one-step RT-qPCR assay that is probe-based was successfully developed and validated with high efficacy in this study. SARS-CoV-2 can be easily detected and distinguished from other common respiratory viruses as explained above with high efficiency and reliability in just one test tube. Thus, the dependence on this multiplexed feature will help in boosting the throughput of the diagnosis and save time, cost, and sample compared to other testing approaches and singleplex PCR. All in all, the possibility of respiratory viruses circulating together is high and this multiplex one-step RT-qPCR will be very advantageous.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by King Abdullah University of Science and Technology through core funding and the National Term Grand Challenge (NTGC) to S.M.H.
Materials
| 0.45 μm filter cups | Thermo Scientific | 291-4545 | |
| 10X Tris-Glycine SDS running buffer | Novex | LC2675 | |
| 6-well tissue culturing plates | Corning | 353046 | |
| Ammonium sulfate | Fisher Scientific | A701-3 | |
| Ampicillin | Corning | 61-238-RH | |
| Cation exchange (HiTrap SP HP) 5 mL | Cytiva | 17-1152-01 | |
| D-(+)-Biotin, 98+% | Thermo Scientific | A14207.60 | |
| DH10Bac competent cells | Fisher Scientific | 10361012 | |
| Dialysis bag (Snakeskin 10,000 MWC) | Thermo Scientific | 68100 | |
| Dithiothreitol (DTT) | Thermo Scientific | R0862 | |
| Dnase/Rnase Free Distilled Water | Ambion | AM9930 | |
| dNTPs | Thermo Scientific | R0192 | |
| E. coli BL21(DE3) competent cells | Invitrogen | C600003 | |
| EDTA | Fisher Scientific | BP120-1 | |
| Elution Buffer | Qiagen | 19086 | |
| ESF 921 insect cell culture medium (Insect cells media) | Expression Systems | 96-001-01 | |
| FBS Solution | Gibco | A38400-01 | |
| Fugene (transfection reagent) | Promega | E2311 | |
| Gentamicin | Fisher Scientific | 15750060 | |
| Glycerol | Sigma Aldrich | G5516-500 | |
| IGEPAL CA-630 | Sigma Aldrich | I8896-100ml | |
| Imidazole | Sigma Aldrich | 56750-1Kg | |
| Influenza A (H1N1) synthetic RNA | Twist Bioscience | 103001 | |
| Influenza A (H3N2) synthetic RNA | Twist Bioscience | 103002 | |
| Influenza B synthetic RNA | Twist Bioscience | 103003 | |
| IPTG | Gold Biotechnology | I3481C100 | |
| Kanamycin | Gibco | 11815-032 | |
| LB Agar | Fisher Scientific | BP1425-500 | |
| LB Broth media | Fisher Scientific | BP1426-500 | |
| Lysozyme | Sigma Aldrich | L6876-10G | |
| Magnesium Chloride | Sigma Aldrich | 13152-1Kg | |
| MERS-CoV synthetic RNA | Twist Bioscience | 103015 | |
| MicroAmp Fast Optical 96-well Reaction plates with Barcode (0.1 mL) | Applied Biosystems | 10310855 | |
| Mini- PROTEAN TGX Precast Gel | Bio-Rad | 456-1093 | |
| Miniprep kit | Qiagen | 27106 | |
| Ni-NTA Excel (HisTrap Excel) 5 mL | Cytiva | 17-3712-06 | |
| Ni-NTA HP (HisTrap HP) 5 mL | Cytiva | 17-5248-02 | |
| Optical Adhesice Covers (PCR Compatible,DNA/Rnase/PCR Inhibitors Free | Applied Biosystems | 4311971 | |
| Potassium Chloride | Fisher Bioreagents | BP366-1 | |
| Primers and Probes | Integrated DNA Technologies, Inc. | ||
| Protease Inhibitor Mini tablets EDTA-Free | Thermo Scientific | A32955 | |
| Protein marker | Fermentas | 26616 | |
| RT-qPCR machine (QuantStudio 7 Flex) | Applied Biosystems | ||
| S.O.C medium | Fisher Scientific | 15544034 | |
| SARS-CoV-+A2:C442 synthetic RNA | Twist Bioscience | 102024 | |
| Sf9 insect cells | Gibco | A35243 | |
| Sodium Chloride | Sigma Aldrich | S3014-1Kg | |
| StrepTrap XT 5 mL | Cytiva | 29401323 | |
| Tetracycline | IBI Scientific | IB02200 | |
| Tris Base Molecular Biology Grade | Promega | H5135 | |
| Tris-HCl | Affymetrix | 22676 | |
| Tween 20 | Sigma Aldrich | P1379-100ml | |
| X-Gal | Invitrogen | B1690 |
References
- Hu, B., Guo, H., Zhou, P., Shi, Z. L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 19 (3), 141-154 (2021).
- Zhu, N., et al. A novel Coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 382 (8), 727-733 (2019).
- Huang, C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395 (10223), 497-506 (2020).
- Wu, F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 579 (7798), 265-269 (2020).
- Yang, S., et al. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann Transl Med. 8 (7), 450 (2020).
- El Hassan, M., et al. A review on the transmission of COVID-19 based on cough/sneeze/breath flows. Eur Phys J Plus. 137 (1), 1 (2022).
- . WHO Coronavirus (COVID-19) Dashboard Available from: https://covid19.who.int (2023)
- Kucharski, A. J., et al. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect Dis. 20 (10), 1151-1160 (2020).
- Reddy, K. P., et al. Cost-effectiveness of public health strategies for COVID-19 epidemic control in South Africa: a microsimulation modelling study. Lancet Glob Health. 9 (2), e120-e129 (2021).
- Cheng, H. Y., et al. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med. 180 (9), 1156-1163 (2020).
- Kretzschmar, M. E., et al. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health. 5 (8), e452-e459 (2020).
- Krammer, F., et al. Influenza. Nat Rev Dis Primers. 4 (1), 3 (2018).
- Yang, J., et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 94, 91-95 (2020).
- Lansbury, L., Lim, B., Baskaran, V., Lim, W. S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 81 (2), 266-275 (2020).
- Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 367 (19), 1814-1820 (2012).
- Azhar, E. I., et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 370 (26), 2499-2505 (2014).
- Ling, Y., Qu, R., Luo, Y. Clinical analysis of the first patient with imported Middle East respiratory syndrome in China. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 27 (8), 630-634 (2015).
- Nazer, R. I. Outbreak of Middle East Respiratory Syndrome-Coronavirus causes high fatality after cardiac operations. Ann Thorac Surg. 104 (2), e127-e129 (2017).
- Huang, P., et al. A rapid and specific assay for the detection of MERS-CoV. Front Microbiol. 9, 1101 (2018).
- Ezhilan, M., Suresh, I., Nesakumar, N. SARS-CoV, MERS-CoV and SARS-CoV-2: A diagnostic challenge. Measurement (Lond). 168, 108335 (2021).
- Ulloa, S., et al. A simple method for SARS-CoV-2 detection by rRT-PCR without the use of a commercial RNA extraction kit. J Virol Methods. 285, 113960 (2020).
- Kudo, E., et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biol. 18 (10), e3000867 (2020).
- Norz, D., Hoffmann, A., Aepfelbacher, M., Pfefferle, S., Lutgehetmann, M. Clinical evaluation of a fully automated, laboratory-developed multiplex RT-PCR assay integrating dual-target SARS-CoV-2 and influenza A/B detection on a high-throughput platform. J Med Microbiol. 70 (2), 001295 (2021).
- Yun, J., et al. Evaluation of three multiplex real-time reverse transcription PCR assays for simultaneous detection of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial virus in nasopharyngeal swabs. J Korean Med Sci. 36 (48), e328 (2021).
- Lu, X., et al. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 52 (1), 67-75 (2014).
- Broughton, J. P., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 38 (7), 870-874 (2020).
- Ali, Z., et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 288, 198129 (2020).
- Ali, Z., et al. Bio-SCAN: A CRISPR/dCas9-based lateral flow assay for rapid, specific, and sensitive detection of SARS-CoV-2. ACS Synth Biol. 11 (1), 406-419 (2022).
- Yoshimi, K., et al. CRISPR-Cas3-based diagnostics for SARS-CoV-2 and Influenza virus. iScience. 25 (2), 103830 (2022).
- Chen, Z., et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using Lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal Chem. 92 (10), 7226-7231 (2020).
- Kasetsirikul, S., et al. Detection of the SARS-CoV-2 humanized antibody with paper-based ELISA. Analyst. 145 (23), 7680-7686 (2020).
- Joung, J., et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot testing. N Engl J Med. 383 (15), 1492-1494 (2020).
- Chen, Z., Wu, Q., Chen, J., Ni, X., Dai, J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol Sin. 35 (3), 351-354 (2020).
- Jang, W. S., et al. Development of a multiplex Loop-Mediated Isothermal Amplification (LAMP) assay for on-site diagnosis of SARS CoV-2. PLoS One. 16 (3), e0248042 (2021).
- McBride, R., Fielding, B. C. The role of Severe Acute Respiratory Syndrome (SARS)-Coronavirus accessory proteins in virus pathogenesis. Viruses-Basel. 4 (11), 2902-2923 (2012).
- AlBalwi, M. A., et al. Evolving sequence mutations in the Middle East Respiratory Syndrome Coronavirus (MERS-CoV). J Infection Public Health. 13 (10), 1544-1550 (2020).
- Takahashi, M., et al. Quick and easy assembly of a One-Step qRT-PCR Kit for COVID-19 diagnostics using In-House enzymes. ACS Omega. 6 (11), 7374-7386 (2021).
- Sambrook, J., Fritsch, E. R., Maniatis, T. . Molecular cloning: A laboratory manual (2nd ed.). , (1989).
- Simpson, R. J. SDS-PAGE of Proteins. CSH Protoc. 2006 (1), (2006).
- Simpson, R. J. Staining proteins in gels with Coomassie blue. CSH Protoc. 2007, (2007).
- Takumi Yano, J. M. L., et al. Expression of the thermostable Moloney murine leukemia virus reverse transcriptase by silkworm-baculovirus expression system. J Asia-Pac Entomol. 22 (2), 453-457 (2019).
- van Kasteren, P. B., et al. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. 128, 104412 (2020).
- Shu, B., et al. Multiplex Real-Time reverse transcription PCR for Influenza A virus, Influenza B virus, and Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 27 (7), 1821-1830 (2021).
- Engelke, D. R., Krikos, A., Bruck, M. E., Ginsburg, D. Purification of Thermus aquaticus DNA polymerase expressed in Escherichia coli. Anal Biochem. 191 (2), 396-400 (1990).
- Pabbaraju, K., Wong, A. A., Ma, R., Zelyas, N., Tipples, G. A. Development and validation of a multiplex reverse transcriptase-PCR assay for simultaneous testing of Influenza A, Influenza B and SARS-CoV-2. J Virol Methods. 293, 114151 (2020).
- Hirotsu, Y., et al. Analysis of COVID-19 and non-COVID-19 viruses, including Influenza viruses, to determine the influence of intensive preventive measures in Japan. J Clin Virol. 132, 104634 (2020).
- Sellner, L. N., Coelen, R. J., Mackenzie, J. S. Reverse-Transcriptase inhibits Taq Polymerase-Activity. Nucleic Acids Res. 20 (7), 1487-1490 (1992).

