Cell Patterning Using Magnetic-Archimedes Strategy
Summary
This protocol describes an ink-free, label-free, substrate-independent, high-throughput cell patterning method based on the Magnetic Archimedes effect.
Abstract
Cell patterning, allowing precise control of cell positioning, presents a unique advantage in the study of cell behavior. In this protocol, a cell patterning strategy based on the Magnetic-Archimedes (Mag-Arch) effect is introduced. This approach enables precise control of cell distribution without the use of ink materials or labeling particles. By introducing a paramagnetic reagent to enhance the magnetic susceptibility of the cell culture medium, cells are repelled by magnets and arrange themselves into a pattern complementary to the magnet sets positioned beneath the microfluidic substrate.
In this article, detailed procedures for cell patterning using the Mag-Arch-based strategy are provided. Methods for patterning single-cell types as well as multiple cell types for co-culture experiments are offered. Additionally, comprehensive instructions for fabricating microfluidic devices containing channels for cell patterning are provided. Achieving this feature using parallel methods is challenging but can be done in a simplified and cost-effective manner. Employing Mag-Arch-based cell patterning equips researchers with a powerful tool for in vitro research.
Introduction
Cell patterning is evolving into an intuitive and powerful technology for in vitro studies1. By manipulating cell positions in culture plates, it provides solutions for a variety of experiments, including cell migration2, biomimetic multicellular co-culture3, organoid assembly4, biomaterial studies5, and more. In most situations, an ink-free, label-free method is preferred for cell patterning because it offers ease of operation and high cell viability for subsequent investigations.
The Mag-Arch effect is a physical phenomenon wherein diamagnetic objects in paramagnetic liquids tend to move toward regions with weak magnetic fields6. Living cells are naturally diamagnetic, while cell culture media can be made paramagnetic by adding soluble paramagnetic elements, such as gadopentetate dimeglumine (Gd-DTPA), commonly used intravenously in nuclear magnetic resonance imaging as a contrast agent7. Consequently, cells are expected to be repelled by the surrounding paramagnetic medium and move toward regions where magnetic fields are weaker8. A patterned magnetic field can be easily generated using a set of neodymium magnets. Ideally, cell patterns are assembled in opposition to the magnet patterns. Technically, this is defined as a label-free method because the only additional reagent, Gd-DTPA, remains in the extracellular environment and does not bind to cells. Thus, potential influences on subsequent cell culture can be easily avoided by replacing the culture medium. Compared to other methods1,3,9,10, the Mag-Arch-based strategy does not require bio-ink components or the application of specific particles to positively label the cells. Furthermore, it has been shown to work on multiple substrates for cell adhesion and is capable of high-throughput cell patterning4.
This article presents a detailed protocol for cell patterning using the Mag-Arch-based method, covering everything from device fabrication to adjusting the cell pattern. In addition to the patterns we have demonstrated, users can easily create various cell patterns using magnets and Gd-DTPA solution. Furthermore, protocols for assembling complex co-culture patterns and manipulating cells in enclosed microfluidic chips, are also provided.
Protocol
1. Assembling the magnet sets
- Assemble the magnet sets for strip patterns.
- Choose flat rectangular magnets, as depicted in Figure 1A. The dimensions of the rectangular magnets used for this demonstration are 1.5 mm × 10 mm × 35 mm (thickness × height × length) (see Table of Materials). The thickness of the magnets determines the gaps between the cell stripes.
- Cut 2 mm-thick silicone plates (see Table of Materials) into 2 mm × 8 mm × 30 mm rectangles. Ensure that the latter two dimensions of these silicone plates are slightly smaller than those of the rectangular magnets mentioned above to optimize the assembly. The thickness of the silicone plates determines the width of the cell stripes.
- Assemble the rectangular magnets and the silicone plates layer by layer, as illustrated in Figure 1A. Each magnet set consists of 4 magnets and 3 silicone plates. Ensure that the poles of each adjacent magnet are opposite so that the magnet sets can automatically align due to the intrinsic attractive force.
- Sterilize the magnets before use.
NOTE: It is recommended to wash the magnet sets with pure water and then immerse them in 75% ethanol for 10 min. The overall dimensions of these magnet sets are 12 mm × 10 mm × 35 mm, designed to fit standard 6-well cell culture plates.
- Assemble the magnet sets for dot-array patterns
- Choose miniature cylindrical magnets, as shown in Figure 1B. The dimensions of the cylindrical magnets used for this demonstration are Φ1.5 mm × 10 mm (diameter × height), magnetized in the axial direction.
- Assemble the cylindrical magnets as indicated in Figure 1B. Ensure that each magnet set consists of 36 cylindrical magnets arranged in a 6 × 6 grid. Also, ensure that the poles of each adjacent magnet are opposite to allow the magnet sets to align automatically due to the intrinsic attractive force.
- Sterilize the magnets before use, following the same procedure as in step 1.1.4. The overall dimensions of these magnet sets are 9 mm × 10 mm × 9 mm, designed to fit standard 6-well cell culture plates.
2. Cell patterning on glass slides
- Prepare the cell culture device.
- Use 2 mm-thick silicone plates to create silicone molds. Employ a puncher to create a 1 cm × 1 cm rectangular hole in the plate. Trim the silicone plate around the hole to craft a round mold (diameter = 25 mm) with the hole at the center, as depicted in Figure 1C.
- Thoroughly clean the silicone molds using an ultrasonic cleaner. Sterilize the molds by washing them with pure water for 10 min, followed by replacing the water with 75% ethanol for another 10 min. Finally, dry the molds in a sterilizer box at 65 °C for 60 min.
- Attach the sterilized silicone mold to a Φ25 mm round glass cell slide and gently press it so that the silicone adheres to the glass, as shown in Figure 1C. The resulting cell culture device will have a glass underside and a 200 µL culture cavity. The dimensions of the device are Φ25 mm × 2.15 mm, making it suitable for standard 6-well cell culture plates.
NOTE: The cell culture device described above can be replaced with other Petri dishes with undersides thinner than 0.5 mm, such as confocal dishes. - Place the magnet sets on a 6-well plate. Non-magnetic tweezers are recommended for ease of operation.
NOTE: All subsequent steps should be performed under sterile conditions from this point onward until the observation of cell patterns. - Position the cell culture device on top of the magnet set in the well of the 6-well plate. Ensure that the cell culture device is placed horizontally, and the underside is in close contact with the magnet set.
- Prepare the cell culture medium containing Gd-DTPA
- Prepare commercially available Gd-DTPA injection (see Table of Materials), usually with a concentration of 500 mM. Dilute the injection by adding 30 µL of Gd-DTPA injection to 470 µL of complete cell culture medium to achieve a target concentration of 30 mM.
NOTE: Depending on local regulations and availability, other gadolinium-based contrast agents (GBCAs) with the same target concentration may yield similar results11.
- Prepare commercially available Gd-DTPA injection (see Table of Materials), usually with a concentration of 500 mM. Dilute the injection by adding 30 µL of Gd-DTPA injection to 470 µL of complete cell culture medium to achieve a target concentration of 30 mM.
- Create the cell patterns.
- Harvest cells for patterning. In this demonstration, human umbilical vein endothelial cells (HUVECs, see Table of Materials) were used for patterning (Figure 2). Centrifuge the cells for 5 min at 200 x g (at room temperature) to collect them from suspension and resuspend them in the Gd-DTPA-containing medium. Count the cell density.
- Adjust the cell density to approximately ~2 × 105 cells/mL with Gd-DTPA-containing medium. Gently mix the cell suspension and add 200 µL to the cavity of each cell culture mold to create a brim-full liquid surface.
- Carefully transfer the plate into a cell culture incubator and incubate for 3-6 h until the cells adhere well to the substrate. Avoid slamming the incubator door to prevent interference with the assembly of cell patterns. For cells with poor adhesion rates, overnight treatment with GBCAs is generally safe12.
- Cell patterning is considered complete when cells adhere to the bottom of the cavity of the cell culture device. Replace the Gd-DTPA-containing medium with complete culture medium.
- Remove the cell culture device from the magnet set. One can either observe the cell pattern immediately or transfer the device into a new culture plate for further cultivation.
3. Co-culture patterning by magnet sideways: fabrication of the moving template
NOTE: The following procedure is presented to take advantage of Mag-Arch-based cell patterning and explore the possibility of more applications.
- Prepare the device for stripe cell patterning following step 1 and step 2. However, reduce the cell density to 1 × 105 cells/mL and replace the silicone plates that separate the magnets with thinner ones, making each stripe cell pattern thinner.
NOTE: This provides sufficient area for laying multiple cells in stripe patterns. As a reference, we suggest using 1 mm silicone plates instead of 2 mm to separate the magnets. - Pattern the first type of cells as illustrated in steps 2.2-2.3. This step also requires 3-6 h for cell attachment.
NOTE: To distinguish different cell types in the co-culture system, users may label cells with fluorescent dyes such as DiI, DiD, or cell trackers (CMTPX, CMFDA, CMAC, etc.) before patterning. For example, for DiI and DiD staining, add 1 µL of the stock solution (10 mM) (see Table of Materials) per 1 mL of FBS-free medium and mix well to obtain a working solution. Replace the culture medium with the working solution to cover adherent cells. After incubating for 30 min and washing with PBS three times, proceed with cell harvesting. - After patterning the first type of cells, move the cell culture device 1 mm from the magnets below to the right, as illustrated in Figure 3Aii.
- Gently wash the glass slides with 200 µL of PBS twice. Avoid letting the slides dry.
- Pattern the second type of cells in the same manner as illustrated in steps 2.2-2.3.
- Move the cell culture device 1 mm from the magnets below to the right, as illustrated in Figure 3Aiii, and wash the glass slides with 200 µL of PBS again. Pattern the third type of cells in the same manner as illustrated in steps 2.2-2.3.
- After patterning all types of cells, gently wash the glass slides with 200 µL of PBS twice and replace with complete cell culture medium.
- Remove the cell culture device from the magnet set. One can then observe the cell pattern or transfer the device to a new culture plate for further cultivation.
4. Co-culture patterning by adjusting the concentration of Gd-DTPA
NOTE: GBCAs do not significantly affect cell adhesion or subsequent growth at working concentrations (≤75 mM). Additionally, cell patterns are influenced by the concentration of Gd-DTPA: higher concentrations result in smaller/thinner cell patterns. Thus, it is possible to create co-culture systems by simply adjusting the concentration of Gd-DTPA. This example demonstrates patterning concentric circular arrays.
- Pattern the first type of cells as illustrated in steps 2.2-2.3 using miniature cylindrical magnets from step 1.2. Label the cells with fluorescent dyes such as DiI, DiD, or cell trackers (see step 3) before harvesting them to distinguish different cell types in the co-culture system.
NOTE: One will need a higher concentration of Gd-DTPA (50 mM instead of 30 mM) and a lower cell density, approximately 1 × 105 cells/mL. - After patterning the first cell array, gently wash the glass slides with 200 µL of PBS twice. Avoid allowing the slides to dry.
- Pattern the second type of cells following the same procedure as before. Keep the glass slides relatively motionless on the magnets to prevent dislocation. Attaching a silicone plate with a hollow that fits the magnet sets to the undersurface of the glass slide is recommended.
NOTE: Here, one will need a lower concentration of Gd-DTPA (20 mM instead of 30 mM) so that the second cell patterns are expected to be larger than the first, as illustrated in Figure 3B. - After patterning all types of cells, gently wash the glass slides with 200 µL of PBS twice and replace the medium with complete cell culture medium.
- Remove the cell culture device from the magnet set. One can then observe the cell pattern or transfer the device to a new culture plate for further cultivation.
5. Cell patterning in microfluidic chip
NOTE: The Mag-Arch-based method has been demonstrated to work in enclosed narrow chambers in our previous study8. Here is an example of patterning dot arrays in a microfluidic channel.
- Fabrication of the microfluid chip
- Customize 40 mm × 75 mm × 20 mm Polytetrafluoroethylene (PTFE) molds (see Table of Materials) containing a rectangular cavity of 24 mm × 50 mm × 8 mm, as illustrated in Figure 4A.
- Flatten a 0.5 mm thick silicone plate. Cut the silicone plates according to the shape of the fluid channel, which has a rectangular cavity of 15 mm × 20 mm × 0.5 mm with triangular buffer areas on both the inlet and outlet sides, as illustrated in Figure 4A,B.
- Customize 40 mm × 75 mm rectangular glass plates. Clean the glass plates with air plasma for 30 s.
- Immediately after air plasma cleaning, cover the glass plates with 0.5 mL of a commercially available anti-adhesion buffer (see Table of Materials) and allow them to air dry. Wash the glass plates once with ethanol and let them air dry.
- Attach a silicone plate prepared in step 5.1.2 securely to the middle of a glass plate, as illustrated in Figure 4A.
- Prepare polydimethylsiloxane (PDMS) prepolymer according to the manufacturer's instructions (see Table of Materials). For each PDMS chip, weigh 10 g of the base constituent and 1 g of the firming agent into a 50 mL centrifuge tube.
- Mix the agents thoroughly with a glass stirring rod until small bubbles are uniformly distributed in the colloid. The mixing takes 5-10 min, depending on the colloid volume. Centrifuge the colloid prepolymer for 1 min at 500 x g (at room temperature) to eliminate bubbles.
- Slowly pour ~10 mL of the prepolymer into the cavity to fill the PTFE mold.
- Carefully place the mold into a vacuum reservoir and keep it horizontal to prevent the prepolymer from flowing away.
- Remove residual air bubbles from the prepolymer with a vacuum pump. The vacuuming takes 120-180 min.
- If needed, pour more prepolymer to further fill the mold cavity. Vacuum for another 60 min.
- Carefully cover the PTFE mold with the glass plate, with the silica side facing the cavity, as illustrated in Figure 4Bi.
- Ensure that all bubbles are eliminated. Transfer the mold to a 60 °C drying oven for the prepolymer to solidify overnight.
- Remove the mold from the oven. After cooling, demold the solidified PDMS cover carefully. Create an inlet and an outlet with a Φ1 mm needle puncher.
- Wash the PDMS cover and 24 mm × 50 mm cover slides according to step 2.1.2.
NOTE: Proceed with the following steps under sterile conditions. - Attach a PDMS cover and a cover slide to create a microfluid chip containing a flow channel of approximately ~500 µm in height. The bottom side should consist of a ~0.15 mm glass cover slide to enable cell patterning with the Mag-Arch-based strategy.
- Assemble commercially available sterile adapters into both the inlet and outlet of the microfluid chip8.
- Prepare a 2% (w/v, dissolved in PBS) gelatin coating buffer. Sterilize the buffer by autoclaving.
- Inject the gelatin coating buffer into the chip via the inlet adapter using a 1 mL syringe. If bubbles emerge, flush them out with extra coating buffer.
- Place the chip into a 10 cm cell culture dish. Add 1 mL of PBS around the dish bottom to avoid drying. Transfer the dish to a cell culture incubator. The coating takes 30 min.
- Flush out the coating buffer with 2 mL of Gd-contained medium (30 mM) via the inlet.
- Gently inject 1 mL of cell suspension containing 30 mM Gd-DTPA with a 1 mL syringe.
- Pattern cells as illustrated in steps 2.2-2.3 using miniature cylindrical magnets from step 1.2.
NOTE: However, attaching a silicone plate with a hollow that fits the magnet sets to the undersurface of the glass slide is recommended, as shown in Figure 4Ci. - After patterning, gently refresh the Gd-DTPA-contained medium with 1 mL of complete medium by flushing with a 1 mL syringe.
NOTE: The process of cell patterning in microfluidics, from steps 5.7-5.9, takes about 180 min, which is similar to cell patterning on standard glass slides. - Quantify the patterns under a microscope. If needed, connect the microfluid chip to a circulating culture device for further cultivation.
NOTE: In our experience, cells are able to grow for at least 72 h when supported by a simple micropump-based device, which provides the microfluidics with a continuous flow of fresh culture medium in a cell incubator8.
Representative Results
Rectangular (1.5 mm × 10 mm × 35 mm) and cylindrical (Φ1.5 m × 10 mm) magnets were selected to create cell patterns as a demonstration. Users have the flexibility to modify the size and shape of magnets or assemble them differently to create diverse cell patterns. In Figure 1A,B, the magnets were assembled, with the magnetic poles depicted in blue (south) and red (north) for clarity. In this configuration, magnets attract each other laterally and align themselves, as illustrated in Figure 2. Figure 1C,D illustrates the structure of the cell culture device and the cell patterning procedure.
Figure 2 displays mono-type cell patterns. GFP-labeled HUVECs were utilized for observation under a fluorescent microscope. The cells were organized into stripe and dot array patterns using corresponding magnet sets. For HUVECs, which adhere rapidly to glass slides (within 120-180 min), the entire procedure was completed in 4 h. Following the protocol resulted in patterns with well-defined edges and high uniformity. To determine the viability, the cells were treated with Gd-DTPA for 12 h, which is much longer than 3-6 h in step 2. However, both Live/Dead staining and CCK8 assay8 showed no significant decrease in cell viability. A relatively high concentration of Gd-DTPA (50 mM) induced a statistical difference, but still preserved a living rate of 90.76% ± 1.78% (Supplementary Figure 1).
Building upon the mono-type cell patterning protocol, multi-type cell patterning examples were provided for potential co-culturing applications. In this scenario, HUVECs, A2780 ovarian cancer cells, and smooth muscle cells (SMCs) were employed. To distinguish between them, the cells were labeled with GFP, DiD, and DiI before patterning. By following step 3, a tripartite cell pattern of side-by-side stripes was generated (Figure 3A). Conversely, step 4 was used to create concentric dot arrays by adjusting the concentration of Gd-DTPA (Figure 3B). The first layer of cells was stained with DiI (red) and patterned with 50 mM Gd-DTPA, while the second layer of cells was labeled with GFP (green) and patterned with 25 mM Gd-DTPA. Consequently, the dot size of the first layer was smaller, surrounded concentrically by the second layer of dot-patterned cells. Different cell types exhibited varying attachment and spreading rates, with HUVECs attaching and spreading quickly, A2780s attaching rapidly but spreading more slowly, and SMCs attaching and spreading relatively slowly. These results demonstrated that various cell types could form cell patterns in 3 h and be utilized in co-culture experiments.
Furthermore, it was demonstrated that cell patterning using a magnetic field was compatible with enclosed narrow culture devices, such as microfluidic chips. By following step 5, microfluidic chips were fabricated, and dot arrays were generated within them (Figure 4).
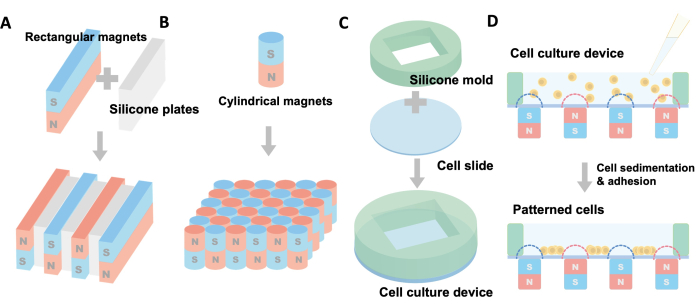
Figure 1: Setup and schematic diagram of mag-arch-based cell patterning. (A) Assembly of magnet sets for creating stripe cell patterns. (B) Assembly of magnet sets for generating dot array cell patterns. (C) Setup of the cell culture device. (D) Step-by-step procedure for cell patterning. Please click here to view a larger version of this figure.
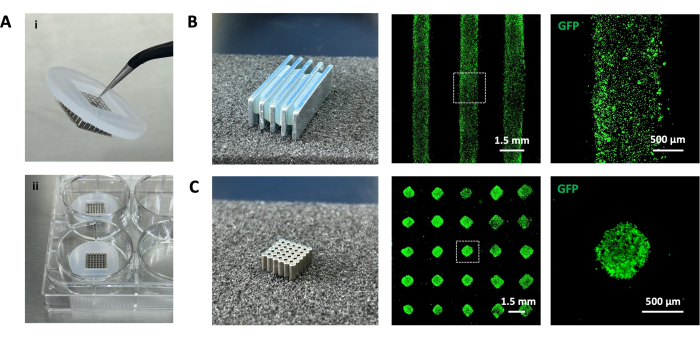
Figure 2: Assembly of devices and patterning HUVECs into stripe and dot array patterns. (A) Magnet sets confined within cell culture devices (i) and placed in a cell culture plate (ii). (B) and (C) Magnet sets and the corresponding cell patterns. Cells were labeled with green fluorescent protein (GFP) for visualization of the cell pattern. Scale bars = 1.5 mm; insets = 500 µm. Please click here to view a larger version of this figure.
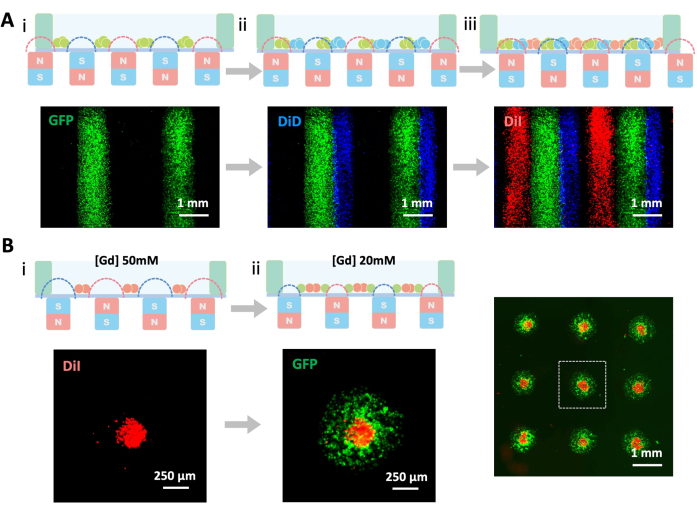
Figure 3: Patterning co-culture systems with step-by-step strategy. (A) Co-culture patterning using magnet sideways (i–iii). Cells were labeled with GFP (green), DiD (blue), and DiI (red) to distinguish different cell types. Scale bars = 1 mm. (B) Co-culture patterning achieved by adjusting Gd-DTPA concentration; (i) 50 mM, (ii) 20 mM. Scale bars = 1.5 mm; insets = 250 µm. Please click here to view a larger version of this figure.
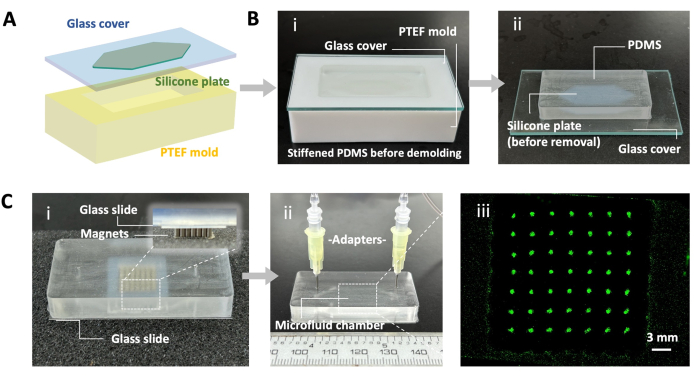
Figure 4: Cell patterning in a microfluidic chamber. (A) Schematic diagram of the microfluidic mold. (B) Fabrication of microfluidics using polydimethylsiloxane (PDMS) (i,ii). (C) Cell patterning within the microfluidic device (i,ii) and a representative result showing dot array cell patterns (iii). Cells were labeled with green fluorescent protein (GFP). Scale bar = 3 mm. Please click here to view a larger version of this figure.
Supplementary Figure 1: Influence of Gd-DTPA on cell viability. HUVECs were treated with varying concentrations of Gd-DTPA for 12 h and then subjected to Live/Dead staining or CCK-8 assay. (A) Live/Dead staining of HUVECs. Scale bars = 200 µm. (B) Histogram depicting Live/Dead staining results. (C) Histogram showing CCK-8 assay results. Please click here to download this File.
Discussion
The Mag-Arch-based cell patterning provides a user-friendly solution for most biomedical laboratories. This method advances parallel to characters of ink-free, label-free, substrate-independent, and the ability for high-throughput patterning8,13. For mono-type cell patterning, it patterns cells in a one-step way. The procedure finishes simply by refreshing culture mediums.
Previous studies have used magnetic particles to label cells and attract them with magnets to form precise patterns14,15. However, the presence of magnetic particles on cells raised concerns about potential effects on cell behaviors. Mag-Arch-based cell patterning takes the opposite strategy by turning extracellular liquids paramagnetic, rather than cells. This strategy makes it much easier to remove the extra paramagnetic reagents by refreshing the culture medium. Studies have generated cellular spheres and dot arrays with Mag-Arch-based cell patterning11,16. Compared to existing Mag-Arch-based studies, the methods presented by this protocol can customize the shape of patterns freely. Moreover, the protocol presents strategies for fabricating co-culture systems. It has also been proven to work inside enclosed narrow cell culture chambers, as we tested in microfluidics.
Rather than parallel methods, which require professional bioprinting equipment17, customized templates18, or surface modification of complex19, the Mag-Arch-based method requires only two necessities: magnets and GBCAs. The surface of the magnet pattern determines the cell pattern reversely. Several patterns of stripes and dot arrays as basic were demonstrated. Users may generate patterns at liberty with different shapes of magnet sets, which are abundantly commercially available. To attain an ideal result, it is recommended to adopt magnets that supply sufficient magnetic force. In our practice, we adopted N52 neodymium-iron-boron magnets, whose remanence was over 1430 mT and surface magnetism was over 100 mT on poles. For GBCAs, Gd-DTPA was adopted because it is stable in physiological conditions and cheaply available in most countries and areas. Other GBCAs could be adopted alternatively. Macrocyclic non-ionic GBCAs, such as gadobutrol and gadoteridol, could be a better choice for lower cytotoxicity when patterning vulnerable cells for long-term treatment11,12.
The limitation of Mag-Arch-based cell patterning mainly lies in the working area of the magnetic field generated by magnets. Following the inverse-square formula, the magnetic field decreases sharply with distance8. As a consequence, the Mag-Arch method fails to assemble ideal cell patterns on general polystyrene cell culture dishes or plates, whose bottoms are thicker than 1 mm. Thus, the protocol has to work on thinner cell culture surfaces, such as glass slides or confocal cell culture dishes. When patterning inside microfluidics, it is also required that the bottom slides of microfluidics should be thinner than 0.5 mm. For establishing co-culture systems, the method might be time-consuming, for each additional cell type increases the time by 3-6 h for cell attachment.
Overall, this protocol provides a simplified way for cell patterning, which could be replicated in most laboratories without any special equipment. Users may adopt it as a powerful tool for studying cell behaviors, mimicking multicellular microenvironments, or testing the cell affinity of biomaterials8.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This study is financially supported by the National Key R&D Program of China (2021YFA1101100), the National Natural Science Foundation of China (32000971), the Fundamental Research Funds for the Central Universities (No. 2021FZZX001-42), and the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (Grant No. SN-ZJU-SIAS-004).
Materials
| A2780 ovarian cancer cells | Procell | CL-0013 | |
| Cell culture medium (DMEM, high glucose) | Gibco | 11995040 | |
| Cover slides | Citotest Scientific | 80340-3610 | For fabricating microfluidics. Dimension: 24 mm × 50 mm |
| DiD | MedChemExpress (MCE) | HY-D1028 | For labeling cells with red fluorescence (Ex: 640 nm) |
| DiI | MedChemExpress (MCE) | HY-D0083 | For labeling cells with orange fluorescence (Ex: 550 nm) |
| Fetal Bovine Serum (FBS) | Biochannel | BC-SE-FBS07 | |
| Gadopentetate dimeglumine (Gd-DTPA) | Beijing Beilu Pharmaceutical | H10860002 | |
| Gelatin | Sigma Aldrich | V900863 | |
| Glass cell slides | Citotest Scientific | 80346-2510 | Diameter: 25 mm; thickness: 0.19-0.22 mm |
| Glass plates | PURESHI hardware store | For fabricating microfluidics. Dimension: 40 mm × 75 mm | |
| Human Umbilical Vein Endothelial Cells (HUVECs) | Servicebio | STCC12103G-1 | |
| Neodymium-iron-boron magnets (N52) | Lalaci | ||
| Non-toxic glass plate coating (Gel Slick Solution) | Lonza | 1049286 | For convenience of demolding when fabricating microfluidics |
| Phosphate Buffered Saline (PBS) | Servicebio | G4200 | |
| Plasma cleaner | SANHOPTT | PT-2S | |
| Polydimethylsiloxane (PDMS) kit | DOWSIL | SYLGARD 184 Silicone Elastomer Kit | For fabricating microfluidics |
| Polytetrafluoroethylene (PFTE) mold | PURESHI hardware store | Customized online, for fabricating microfluidics | |
| Silicon plate | PURESHI hardware store | ||
| Smooth Muscle Cells (SMC) | Procell | CL-0517 | |
| Ultrasonic cleaner | Sapeen | CSA-02 |
References
- Christian, J., et al. Control of cell adhesion using hydrogel patterning techniques for applications in traction force microscopy. J Vis Exp. 179, e63121 (2022).
- Abbas, Y., Turco, M. Y., Burton, G. J., Moffett, A. Investigation of human trophoblast invasion in vitro. Hum Reprod Update. 26 (4), 501-513 (2020).
- Park, M., et al. Modulation of heterotypic and homotypic cell-cell interactions via zwitterionic lipid masks. Adv Healthc Mater. 6 (15), 1700063 (2017).
- Ren, T., Chen, P., Gu, L., Ogut, M. G., Demirci, U. Soft ring-shaped cellu-robots with simultaneous locomotion in batches. Adv Mater. 32 (8), e1905713 (2020).
- Ren, T., Steiger, W., Chen, P., Ovsianikov, A., Demirci, U. Enhancing cell packing in buckyballs by acoustofluidic activation. Biofabrication. 12 (2), 025033 (2020).
- Ge, S., et al. Magnetic levitation in chemistry, materials science, and biochemistry. Angew Chem Int Ed Engl. 59 (41), 17810-17855 (2020).
- Puluca, N., et al. Levitating cells to sort the fit and the fat. Adv Biosyst. 4 (6), e1900300 (2020).
- Ren, T., et al. Programing cell assembly via ink-free, label-free magneto-archimedes based strategy. ACS Nano. 17 (13), 12072-12086 (2023).
- Li, Y. C., et al. Programmable laser-assisted surface microfabrication on a poly(vinyl alcohol)-coated glass chip with self-changing cell adhesivity for heterotypic cell patterning. ACS Appl Mater Interfaces. 7 (40), 22322-22332 (2015).
- Chliara, M. A., Elezoglou, S., Zergioti, I. Bioprinting on organ-on-chip: Development and applications. Biosensors (Basel). 12 (12), 1135 (2022).
- Moncal, K. K., Yaman, S., Durmus, N. G. Levitational 3D bioassembly and density-based spatial coding of levitoids. Adv Funct Mater. 32 (50), 2204092 (2022).
- Parfenov, V. A., et al. Magnetic levitational bioassembly of 3D tissue construct in space. Sci Adv. 6 (29), eaba4174 (2020).
- Dell, A. C., Wagner, G., Own, J., Geibel, J. P. 3D bioprinting using hydrogels: Cell inks and tissue engineering applications. Pharmaceutics. 14 (12), 2596 (2022).
- Ino, K., Ito, A., Honda, H. Cell patterning using magnetite nanoparticles and magnetic force. Biotechnol Bioeng. 97 (5), 1309-1317 (2007).
- Okochi, M., Matsumura, T., Honda, H. Magnetic force-based cell patterning for evaluation of the effect of stromal fibroblasts on invasive capacity in 3d cultures. Biosens Bioelectron. 42, 300-307 (2013).
- Mishriki, S., et al. Rapid magnetic 3D printing of cellular structures with mcf-7 cell inks. Research (Wash D C). 2019, 9854593 (2019).
- Ozturk-Oncel, M. O., Leal-Martinez, B. H., Monteiro, R. F., Gomes, M. E., Domingues, R. M. A. A dive into the bath: Embedded 3D bioprinting of freeform in vitro models. Biomater Sci. 11, 5462-5473 (2023).
- Sahni, G., Yuan, J., Toh, Y. C. Stencil micropatterning of human pluripotent stem cells for probing spatial organization of differentiation fates. J Vis Exp. 112, e54097 (2016).
- Joddar, B., et al. Engineering approaches for cardiac organoid formation and their characterization. Transl Res. 250, 46-67 (2022).

