Msp1 Extraction Assay to Study the Removal of Mislocalized TA Proteins
Abstract
Source: Fresenius, H. L., Wohlever, M. L. Reconstitution of Msp1 Extraction Activity with Fully Purified Components. J. Vis. Exp. (2021).
This video demonstrates an in vitro protocol to study the Msp1-mediated removal of mislocalized TA proteins integrated into the lipid bilayer of liposomes. Msp1 and TA proteins are co-reconstituted into liposomes, and the ATP-dependent translocation of TA proteins is confirmed through affinity purification.
Protocol
1. Liposome Preparation
- Combine chloroform stocks of lipids in appropriate ratios to mimic the outer mitochondrial membrane.
- Prepare 25 mg of lipid mixture. We use a previously established mixture of lipids that mimic mitochondrial membranes, consisting of a 48:28:10:10:4 molar ratio of chicken egg phosphatidylcholine (PC), chicken egg phosphatidyl ethanolamine (PE), bovine liver phosphatidyl inositol (PI), synthetic 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), and synthetic 1',3'-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (TOCL). Sample calculations are shown in Table 1.
- Bring all lipid stocks to room temperature before opening as this will limit condensation. As most labs do not have a precise way to measure the concentration of the lipids, any water absorbed by the chloroform stock will change the concentration of the lipid stock and thus the ratio of lipids used in the assay.
- As lipid stocks come in glass ampules, transfer the required amount of lipid to a glass vial using a 1 mL syringe. Add 2 mg of dithiothreitol (DTT) to the vial to prevent lipid oxidation. Work quickly as evaporation of chloroform will change the concentration of the lipid.
- Transfer any remaining lipid to a separate glass vial and fit it with a PFTE septa. Add 2 mg of DTT to the vial, wrap with parafilm, and store at -20 °C to prevent lipid oxidation. Try to use the lipids within 3 months of transfer to the vials. To prevent potential contamination of the chloroform stocks by marker runoff, transfer the stickers from the original ampules to the glass vials rather than label with marker.
- Evaporate chloroform under a very gentle stream of nitrogen while spinning the glass vial continuously by hand in a fume hood, essentially acting as a manual rotovap. Spin the vial at a consistent speed (20-40 rpm) by hand to keep the lipids moving. The goal is to evaporate all the chloroform and get an even coating of lipids over the entire glass vial.
CAUTION: Chloroform is neurotoxic, and this step should be performed in the fume hood.- Attach a fresh Pasteur pipette to the nitrogen tube. Do not let any of the lipids splash out of the vial or onto the Pasteur pipette. Aim the tip at the bottom of the vial so the air bounces off the bottom and pushes the lipids up towards the center of vial. Get an even coating over the entire vial while avoiding any accumulation in the corners or by the cap. This whole process takes about 5 minutes.
- As the mixture thickens into a "bead" of lipids, guide it into the center of the vial by changing the angle of the vial. Once the bead starts to become smaller, turn up the nitrogen stream slightly to disperse the bead, ensuring that none of the lipids blow out of the vial.
- Remove any remaining chloroform under vacuum.
- Put the glass vial on a house vacuum or diaphragm vacuum pump for 1 hour to remove the majority of residual chloroform. These vacuums are generally not strong enough to remove all of the chloroform, but they can tolerate small amounts of solvent better than rotary-vain vacuums.
- Put vial on a strong vacuum (<1 mTorr) for 12-16 h to remove all residual chloroform. Be sure to avoid bumping of the vial during this process.
- Resuspend the lipids in 1.25 mL of liposome buffer (50 mM HEPES KOH pH 7.5, 15% glycerol, 1 mM DTT). As we started with 25 mg of lipids, this results in a concentration of 20 mg/mL. Fully resuspend the lipids with no visible chunks. If the lipids pooled in the corner of the vial, this can be a lengthy process.
- Vortex the vial vigorously until the sample is milky smooth. We find that if the chloroform evaporation was done properly the night before, this process takes about 5-10 minutes.
- To ensure complete resuspension of the lipids, rotate on a wheel at room temperature for 3 hours at ~80 rpm. Remove vial from the wheel once every hour for 1 minute of vortexing to ensure even mixing.
- Transfer lipids carefully to a clean 1.5 mL microcentrifuge tube. Perform 5 freeze-thaw cycles using liquid nitrogen to freeze and a 30 °C heat block to thaw. This step helps to convert multilamellar vesicles to unilamellar vesicles.
- Extrude the lipids.
- During the freeze-thaw cycles, prepare the mini-extruder. Assemble the mini-extruder with 10 mm filter supports and a polycarbonate membrane of the desired pore size (we use 200 nm). The size of the filter will affect the size of the liposome, which will affect the concentration of proteins required for the reconstitution (Step 2.3).
- Place the mini-extruder onto a hot plate and bring the extruder temperature up to 60 °C.
- Draw up the lipids into a 1 mL gas-tight glass syringe and carefully place into one end of the mini-extruder. Place the empty gas-tight syringe into the other side of the mini-extruder. Allow the lipids to equilibrate to the extruder stand temperature for 5-10 minutes.
- Transfer the lipids to the alternate syringe by gently pushing the plunger of the filled syringe. Push the solution from the alternate syringe into the original syringe. Repeat this back-and-forth process 15 times, so that at the 15th pass the lipids end in the alternate syringe. Monitor the volume in each pass to make sure that there are no leaks.
- Prepare single-use aliquots of the lipids, flash freeze in liquid nitrogen and store at -80 °C. The liposomes are stable at -80 °C for several months. The reconstitutions require 10 μL of liposomes at a time (Step 2.3.4), so it is convenient to prepare 10 μL or 20 μL aliquots.
2. Reconstitution of Msp1 and Model TA protein
- Prepare the reconstitution buffer: 50 mM HEPES pH 7.5, 200 mM potassium acetate, 7 mM magnesium acetate, 2 mM DTT, 10% sucrose, 0.01% sodium azide, 0.2-0.8% Deoxy Big Chaps (DBC).
- Optimize reconstitution conditions for the new batch of liposomes. The concentration of DBC and biobeads required for optimal reconstitution varies depending on the batch of liposomes used. To limit prep-to-prep variability, use the same lot of DBC for all experiments. When changing lots of DBC, repeat the optimization process.
- Set up a series of reconstitutions with different concentrations of DBC (0.2% – 0.8%) and biobeads (25 mg – 100 mg) each time a new batch of liposomes is prepared. It is important to not drop the DBC below the critical micelle concentration (CMC) of ~0.12%. Once conditions are optimized, we recommend collecting all data using the same liposome prep and reconstitution conditions.
- Assay the effectiveness of the various reconstitution conditions by using the extraction assay described in Step 3.
- Prepare biobeads at a final concentration of 250 mg/mL.
- Weigh out 2.7 g of dried biobeads and resuspend in a 50 mL centrifuge tube of 100% methanol (about 45 mL) to wet the beads. Initially wet the biobeads in methanol to prevent air from being trapped in the pores of the beads. Once in methanol, keep the biobeads wet as any air trapped by the biobeads will alter their ability to absorb detergent.
- Remove methanol by washing the beads 8x with about 45 mL of ultrapure water (18.2 mΩ), hereafter referred to as ddH2O. Pellet beads by spinning at 3,200 x g for 1 minute. Decant the liquid and resuspend in ddH2O.
- After washing, resuspend in 10 ml of ddH2O with 0.02% sodium azide and store at 4 °C. Biobeads can be stored at 4 °C for several months. This stock is 250 mg/mL as it is assumed ~0.2 g is lost during the wash steps.
- Calculate the size of the liposome and the desired number of molecules of TA protein and Msp1 per liposome. This will determine the concentration of Msp1 and TA protein required for the reconstitution.
- First, calculate the number of lipid molecules per unilamellar liposome (Ntotal) using the equation
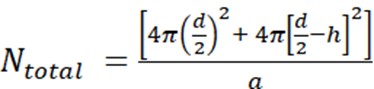
where d is the diameter of the liposome, h is the thickness of the bilayer, and a is the lipid headgroup area.- Measure the liposome diameter by DLS. In our example, a value of 70 nm for the liposome diameter (d) was obtained.
- Use a value of 5 nm for h and 0.71 nm2 for a, which is the headgroup size for phosphatidylcholine. In this particular situation, Ntotal is 37,610.
- Next, calculate the molar concentration of lipid MLipid using the average molecular weight of the lipids in the mixture. In this example, the concentration of lipids is 20 mg/mL (Step 1.4) and the average molecular weight of lipids is 810 g/mol (Table 1). This results in a value of 0.0247 M for MLipid.
- Next, calculate the molar concentration of liposomes, MLiposome, using the equation
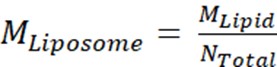
where MLipid is the molar concentration of lipid from Step 2.3.2, and Ntotal is the total number of lipids per liposome calculated in Step 2.3.1. In this example, the 20 mg/mL stock concentration of liposomes is approximately 660 nM. - Calculate the amount of Msp1 and TA protein required for a 100 μL reconstitution reaction.
NOTE: The final concentration of lipids in the reconstitution is 2 mg/mL, which is a 10x dilution of the liposome stock. This gives a final liposome concentration of 66 nM. The final concentration of Msp1 is 792 nM, which gives an average of 12 total copies (2 functional hexamers) per liposome. The final concentration of TA protein is 660 nM, which gives an average of 10 copies per liposome.
- First, calculate the number of lipid molecules per unilamellar liposome (Ntotal) using the equation
- In a PCR tube, mix together purified Msp1, TA protein, and liposomes in reconstitution buffer. The order of addition is buffer, proteins, and liposomes last. The total volume is 100 μL. Allow the mixture to sit on ice for 10 minutes. Purify the Msp1 and TA protein.
- Use the well-characterized model TA protein His-Flag-Sumo-Sec22 as a positive control substrate when first establishing the assay. This construct has a His-tag for easy purification, 3x-Flag tag for detection by western blot, a Sumo domain for increased solubility, and the TMD of the ER-TA protein Sec22 for reconstitution and recognition by Msp1.
- Ensure that stock solutions of both Msp1 and the TA protein are approximately 100 μM to minimize the effect of N-Dodecyl β-D-maltoside (DDM) from the protein purification on the reconstitution. Purified Msp1 is in 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 0.05% DDM whereas the purified TA protein is in 50 mM Tris pH 7.4, 150 mM sodium chloride, 10 mM magnesium chloride, 5 mM β-mercaptoethanol, 10% glycerol, 0.1% DDM. Stock solutions of approximately 100 μM TA protein and Msp1 monomer ensures that these components will make up < 5% of the final reconstitution volume, resulting in a dilution of DDM below the CMC.
- Add the desired amount of biobeads to the sample to remove detergent.
- Cut the tip of a p200 pipette tip to about 1/8th inch in diameter so that beads can fit through the tip. Vortex the biobeads tube thoroughly to obtain a uniform mixture and quickly remove the lid and pipette up the volume before the biobeads settle. Transfer the biobeads to an empty PCR tube.
- When the reconstitution has finished its 10-minute incubation on ice, use an uncut pipette tip to remove all of the liquid from the biobeads. Then transfer the 100 μL reconstitution into the tube with the biobeads. This must be done quickly so that the biobeads do not trap air, which will cause the beads to float.
- Allow the reconstitution to rotate on a wheel at ~80 rpm for 16 hours at 4 °C.
- Remove reconstituted material from biobeads. Do a quick spin in a picofuge to pellet the biobeads, and then use an uncut pipet tip to transfer the reconstituted material to a clean PCR tube. Repeat this process 1-2 times until there are no biobeads left in the sample. Keep reconstitution on ice.
- Pre-clear the reconstituted material to remove any proteins which failed to reconstitute into the liposomes.
- Prepare Extraction Buffer: 50 mM HEPES pH 7.5, 200 mM potassium acetate, 7 mM magnesium acetate, 2 mM DTT, 100 nM calcium chloride.
- Equilibrate the glutathione spin columns with Extraction Buffer according to the manufacturer’s directions. This typically involves 3 rounds of washing with 400 μL of buffer and then centrifuging at 700 x g for 2 minutes at room temperature to remove the buffer.
- Add 5 μM of each chaperone (GST-SGTA and GST-Calmodulin) to the reconstituted material. These chaperones will bind to the TMD of any proteins which failed to reconstitute into the liposomes. Purification of chaperones was described previously.
NOTE: These chaperones are commercially available, but we prefer to purify in-house to control for cost and quality. Both proteins are in 20 mM Tris pH 7.5, 100 mM sodium chloride, and 0.1 mM TCEP, and have a stock concentration around 160 μM. SGTA recognizes substrates with a highly hydrophobic TMD whereas Calmodulin binds TMDs with moderate hydrophobicity. Together, this chaperone cocktail can recognize a broad range of substrates. - Add 100 μL of extraction buffer to the reconstituted material, bringing the volume up to 200 μL. Add this to the equilibrated glutathione spin columns. Note that the glutathione spin columns provide the highest sample recovery when the pre-clearing volume is 200 – 400 μL.
- Plug the spin columns and rotate at ~80 rpm at 4 °C for 30 minutes to allow chaperones to bind to resin.
- Spin the columns at 700 x g for 2 minutes at room temperature. The flow through is pre-cleared material that is depleted of aggregated proteins. Keep material on ice and proceed directly with extraction assay.
3. Extraction Assay
- Prepare tubes for SDS PAGE analysis. Each reaction will have 4 tubes: INPUT (I), FLOW THROUGH (FT), WASH (W), and ELUTE (E).
- Add 45 μL of ddH2O sample to the INPUT tube, 40 μL of ddH2O to the FLOW THROUGH tube, and 0 μL to the WASH and ELUTE tubes.
NOTE: The best signal-to-noise ratio in this assay is obtained when the WASH and ELUTE samples are 5x concentrated relative to the INPUT and FLOW THROUGH samples. Due to the dilutions during the assay, this requires taking 5 μL of sample for the INPUT sample, 10 μL of sample for the FLOW THROUGH sample, and 50 μL of sample for the WASH and ELUTE samples. - Add 16.6 μL of 4x SDS PAGE Loading Buffer to each tube. The total volume of each sample is 50 μL before SDS PAGE Loading Buffer. The final volume is 66.6 μL (50 μL sample + 16.6 μL of 4x SDS PAGE Loading Buffer).
- Add 45 μL of ddH2O sample to the INPUT tube, 40 μL of ddH2O to the FLOW THROUGH tube, and 0 μL to the WASH and ELUTE tubes.
- Assemble the extraction assay.
- Prepare the extraction reaction containing 60 μL of pre-cleared proteoliposomes, 5 μM of GST-SGTA, 5 μM of GST-Calmodulin, and 2 mM ATP. Combine all reagents except ATP, which is used to initiate the reaction. Bring to a final volume of 200 μL with Extraction Buffer.
NOTE: As 60 μL of sample are used for each extraction assay, one reconstitution can be used for three different extraction assays. Perform positive and negative controls (+ATP and -ATP) on material from the same reconstitution. - Pre-warm extraction assay in 30 °C heat block for 2 minutes.
- Prepare the extraction reaction containing 60 μL of pre-cleared proteoliposomes, 5 μM of GST-SGTA, 5 μM of GST-Calmodulin, and 2 mM ATP. Combine all reagents except ATP, which is used to initiate the reaction. Bring to a final volume of 200 μL with Extraction Buffer.
- Initiate the extraction assay by adding ATP to final concentration of 2 mM and start timer.
- Give a 5-second spin in a picofuge to mix ATP into the reaction. Incubate reaction at 30 °C for 30 minutes.
- During the incubation, take 5 μL of the reaction and add to the INPUT tube. The timing of this is flexible.
- During this incubation period, equilibrate one glutathione spin column for each sample in the extraction assay.
- Perform pull-down on chaperones to isolate extracted material.
- Once the 30-minute incubation is finished, add 200 μl of extraction buffer to the tube to bring total volume to 400 μL. Add to equilibrated glutathione resin and allow to bind on wheel at 4° C for 30 minutes.
- Spin the columns at 700 x g for 2 minutes at room temperature to collect the flow through. Take 10 μL for the FLOW THROUGH tube. This sample contains substrates which are still integrated in the lipid bilayer.
- Wash resin twice with 400 μL of extraction buffer, discarding the flow through. On the third wash, keep the flow through and take 50 μL for the WASH tube.
- Prepare 5 mL of Elution Buffer by adding reduced glutathione to a final concentration of 5 mM in Extraction Buffer. Prepare this buffer fresh each time.
- Add 200 μL of Elution Buffer to the spin column. Incubate at room temperature for 5 minutes. Spin at 700 x g for 2 minutes at room temperature to elute. Keep the flow through. Repeat the process a second time so that the total elution volume is 400 μL.
- Take 50 μL of sample from the Elution sample and add it to the ELUTE tube.
| Lipid | Mole % | MW | Avg. MW | µmol in 25 mg | mg in 25 mg | Chlor stock (mg/mL) | µL for 25 mg |
| PC | 48% | 770 | 369.6 | 14.82 | 11.41 | 25 | 456.4 |
| PE | 28% | 746 | 208.88 | 8.64 | 6.45 | 25 | 258.0 |
| PI | 10% | 902 | 90.2 | 3.09 | 2.78 | 10 | 278.5 |
| DOPS | 10% | 810 | 81 | 3.09 | 2.50 | 10 | 250.1 |
| TOCL | 4% | 1502 | 60.08 | 1.23 | 1.85 | 25 | 74.2 |
| Conc. Corr Avg MW | 809.76 | ||||||
| µmol in 25 mg | 30.87 | ||||||
Table 1: Sample calculations for liposome preparation. The key goals of this table are to calculate the concentration corrected average molecular weight of the lipid mixture and the volume of each lipid stock required to make the liposomes. MW and stock concentration come from product labels. Lipids and mole % are chosen by user.
Divulgations
The authors have nothing to disclose.
Materials
| Biobeads | Bio-Rad | 1523920 | |
| Bovine liver phosphatidyl inositol | Avanti | 840042C | PI |
| Chicken egg phosphatidyl choline | Avanti | 840051C | PC |
| Chicken egg phosphatidyl ethanolamine | Avanti | 840021C | PE |
| Filter supports | Avanti | 610014 | |
| Glass vial | VWR | 60910L-1 | |
| Glutathione spin column | Thermo Fisher | PI16103 | |
| Mini-Extruder | Avanti | 610020 | |
| Synthetic 1, 2-dioleoyl-sn-glycero-3-phospho-L-serine | Avanti | 840035C | DOPS |
| Synthetic 1', 3'-bis[12-dioleoyl-snglycero-3-phospho]-glycerol | Avanti | 710335C | TOCL |
| Syringe 1 mL | Norm-Ject 53548-001 | ||
| Syringe 1 mL gas-tight | Avanti 610017 |

