Evaluating the Intracellular Distribution of Virus-Derived Peptide-Modified Antibody Conjugates
Abstract
Source: Beaudoin, S. et al., Initial Evaluation of Antibody-conjugates Modified with Viral-derived Peptides for Increasing Cellular Accumulation and Improving Tumor Targeting. J. Vis. Exp. (2018)
This video demonstrates the process of assessing the intracellular distribution of virus-derived peptide-modified antibody conjugates using confocal microscopy. The viral-derived nuclear localization sequence directs antibody-conjugates within the cytoplasm, confirmed through immunostaining and fluorescence measurement.
Protocol
1. Antibody Peptide Conjugation
NOTE: Cholic acid linked to NLS (ChAcNLS) can be synthesized at any commercial peptide manufacturer or university-affiliated peptide synthesis service platform.
- In a 1.7 mL microcentrifuge tube, prepare a 10 mg/mL (~1 mg total antibody) solution of 7G3 in phosphate buffered saline (PBS), pH 7.6.
- Dissolve sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC) in dimethyl sulfoxide (DMSO) at a concentration of 5-10 mM. Vortexing the solution thoroughly works best in order to achieve complete dissolution.
- Add to the tube containing 7G3 the dissolved sulfur-SMCC solution (typically between 5-20 μL depending on the molar ratio of sulfo-SMCC-to-7G3 desired). Testing sulfo-SMCC-to-7G3 ratios of 10-, 25-, and 50-to-1 is recommended. Incubate at room temperature for 1 h.
- Purify and concentrate the maleimide-derivatized 7G3 by using an ultrafiltration device and buffer exchange in PBS, pH 7.0. Centrifuge at 8,000 x g for approximately 5 min. Discard flow through and fill with PBS, pH 7.0, and centrifuge again.
- Perform this 7-8 times to exchange the buffer completely. Recover the concentrated 7G3 protein by placing the filter device upside down in a clean microcentrifuge tube. Centrifuge for 2 min at 1,000 x g. Recovery volume should be approximately 0.3 mL.
- In the tube containing the maleimide-activated 7G3, add a 2-fold molar excess of ChAcNLS relative to the sulfo-SMCC-to-7G3 ratio used in the previous step and incubate overnight at 4 °C. For example, if a 10-to-1 sulfo-SMCC-to-7G3 ratio was used, then use a 20-to-1 ChAcNLS-to-7G3 ratio. The total volume should not be significantly changed.
NOTE: Although ChAcNLS does not cause precipitation during the conjugation process this is a possibility that could occur with other peptides. Observe the tube during the reaction for signs of precipitation, which is most likely caused when peptides are hydrophobic. In these cases, it may be worth revisiting the peptide of interest in order to design changes to provide increased hydrophilicity. - Concentrate the 7G3-ChAcNLS using an ultrafiltration device to the desired concentration and buffer exchange in PBS pH 7.4 (see step 1.4). The recovery yield of total protein should be ≥ 75% of the original starting material.
- Perform sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) to evaluate the constructed 7G3-ChAcNLS candidates using a 12% polyacrylamide gel (provides sufficient separation of the ChAcNLS conjugated heavy and light chains from nonconjugated chains). Mix 10 μg of 7G3-ChAcNLS in PAGE loading buffer containing 2 μL β-mercaptoethanol and load into well.
- Set the appropriate voltage (120 V for 1 h for a 12% gel) and run the electrophoresis. Stop the electrophoresis when the Coomassie dye front reaches the bottom of the gel.
NOTE: Do not let the Coomassie dye front run off the gel. - Remove gel from electrophoresis apparatus and rinse with destaining solution containing 10% v/v of glacial acetic acid and 20% v/v methanol.
- Take a photo of the gel and with a ruler and measure the distance (cm) migrated for the standards and 7G3 conjugates. Calculate the retention factor (Rf) value, which is the distance migrated divided by the gel length (from where protein is loaded to the Coomassie migration front). Plot the molecular weight (MW) in log against the Rf values from each protein standard and extrapolate the size of the 7G3 conjugates.
- Calculate the number of peptides per antibody by dividing the difference in MW between 7G3 conjugates and unmodified 7G3 by 1768.5 g/mol (MW of ChAcNLS).
- Take digital images of the Coomassie-stained gel and perform densitometry analysis. At this point, selection of a lead candidate can be based on the AC with the maximum number of ChAcNLS molecules that does not contain significant aggregate species.
2. Confocal Microscopy for Intracellular Accumulation Evaluation
NOTE: It is important to test the cell selectivity of intracellular accumulation of the peptide-modified mAbs prior to developing formulations with a payload of interest. Because Tat has also been shown to have a propensity for nonspecific cell penetration, it is worth first analyzing intracellular accumulation and cell selectivity prior to undertaking costly development steps with expensive payloads. For this reason, Procedure 1 should also modify isotype specific irrelevant control mAbs. For the rest of the protocol, we will work with ACs modified with 10 ChAcNLS molecules per antibody. A schematic including key steps for Procedures 2 and 3 is described in Figure 1.
CAUTION: This step of the protocol involves the handling and manipulation of paraformaldehyde. Please follow manufacturer instructions when handling.
- Treat target TF-1a cells with 200 nM of 7G3-ChAcNLS including modified control mAbs. Incubate 5 x 106 antigen-positive cells with the conjugates for 1 h at 37 °C.
- After 1 h, remove supernatant and wash with 1 mL 3x in ice-cold PBS. Add fresh media and incubate for an additional hour at 37 °C.
- After the additional hour, remove supernatant and wash 3x in 1 mL ice-cold PBS. Add 0.5 mL of PBS containing 0.25% trypsin and ethylenediaminetetraacetic acid (EDTA) at 37 °C for 3 up to 30 min.
NOTE: During initial pilot testing it is recommended to not trypsinize cells in order to determine the differences in AC intracellular accumulation. This protocol promotes the use of trypsin, and we demonstrate how this improves assessment of intracellular accumulation efficiency of different AC candidates.- Test the different times for incubation by visually checking for all cells being detached. In addition, incubate cell aliquots with viability staining solutions and perform flow cytometric analysis as previously described.
- Neutralize trypsin with 1.5 mL of cell culture RPMI 1640 media/10% fetal bovine serum. Centrifuge cells at 500 x g for 5 min, remove supernatant, and wash 3x in 0.5 mL ice-cold PBS.
- Fix cells in 0.5 mL PBS containing 1% paraformaldehyde and 1% sucrose on ice for 30 min. Wash cells 3x in 0.5 mL ice-cold PBS and centrifuge at 250 x g for 5 min. Permeabilize cells in 0.5 mL PBS containing 0.15% Triton X-100 for 5 min on ice. Wash cells in ice-cold PBS and repeat centrifugation.
- Suspend cells in 0.1 mL PBS containing 2 μg/mL (or the manufacturer recommended concentration) of an anti-murine (isotype specific) Fc secondary polyclonal antibody conjugated to AlexaFluor 647 for 1 h at room temperature in the dark.
- Centrifuge at 250 x g for 5 min and wash cells 3x in 0.5 mL ice-cold PBS. Suspend cells in 0.5 mL PBS.
PAUSE: Cells can be stored in the refrigerator.
NOTE: It is essential to select a secondary antibody that recognizes the correct isotype of the mAb of interest. AF647 is selected because of its far-infrared emission, which does not interfere with propidium iodide (PI) staining of the nucleus.
- Centrifuge at 250 x g for 5 min and wash cells 3x in 0.5 mL ice-cold PBS. Suspend cells in 0.5 mL PBS.
- Add PI to a concentration of 10 μg/mL to the container with cells. Next, mount 1 x 105 cells onto glass slides using mounting media and cover with a glass coverslip.
- Examine cells with a Plan Apo 60X oil immersion objective NA 1.42 on an inverted laser scanning confocal microscope. Detect PI fluorescence using the 488 nm argon laser and the spectral scanning prism set for 600 – 650 nm. For AF647 fluorescence use the 633 nm helium-neon laser and the spectral scanning prism set for 650 – 700 nm. Collect fluorescence emissions from PI and AF647 sequentially.
NOTE: Use the same setting throughout the evaluation and comparison of the cells. However, you will have to adjust settings for trypsinized cells compared to non-trypsinized cells. - Use microscope software to acquire images. Collect images using serial horizontal optical sections of 1,024 x 1,024 pixels with 2X line averaging taken at 0.5 μm intervals through the entire cell thickness. Present the images as stacked z-projections from four consecutive slices at the maximum fluorescence intensity.
- Analyze cells with microscope software. Record the cellular distribution pattern of the conjugates. Specifically, evaluate whether intracellular fluorescence in the cytoplasm is grouped and near the cell surface or diffuse and homogeneous. Also evaluate the relative fluorescence intensity per cell.
Representative Results
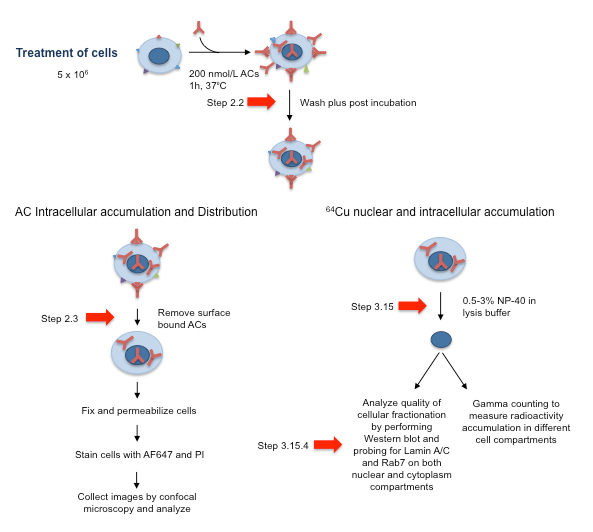
Figure 1: Schematic of AC Cell Treatment Approach and Subsequent Processing for Analysis by Confocal Microscopy and for Analysis of the Cell Fractionation Quality. Red arrows correspond to essential steps 2.2 and 2.3.
Divulgations
The authors have nothing to disclose.
Materials
| Sulfo-SMCC | Thermo Scientific | 22122 | There are many homo- and hetero-bifunctional maleimide crosslinkers to choose from. |
| Amicon Ultra-0.5 mL Centrifugal Filters | EMD Millipore | UFC505096 | There are pack sizes of 8, 24, and 96. Choose according to your needs. |
| Precision Plus Protein Kaleidoscope Standards | BioRad | 1610375EDU | Mulicolor recombinant proteins from 10-250 kDa. |
| Trypsin-EDTA (0.25%), phenol red | Thermo Scientific | 25200056 | 100 or 500 mL volumes to choose from. |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 647 conjugate | Thermo Scientific | A-21235 | 1-10 μg/mL recommended |
| NuPAGE LDS Sample Buffer (4x) | Thermo Scientific | NP0007 | |
| 2-Mercaptoethanol | Sigma Aldrich | M3148-25ML | |
| TF-1a cells | ATCC | ATCC CRL-2003 | |
| RPMI 1640 medium | ATCC | ATCC 30-2001 | |
| FluoView FV1000 Confocal Microscope | Olympus | ||
| Fluoview Software | Olympus | www.olympus-lifescience.com |

