Improved Visualization and Quantitative Analysis of Drug Effects Using Micropatterned Cells
Summary
Adhesive micropatterns that normalize cellular architecture can be used to increase sensitivity in the detection of drug effects, improve reproducibility and simplify automated image acquisition and analysis. Such technology will benefit drug/siRNA screening assays, performed on conventional cell culture supports and consequently suffering from excessive cell-to-cell variability.
Abstract
To date, most HCA (High Content Analysis) studies are carried out with adherent cell lines grown on a homogenous substrate in tissue-culture treated micro-plates. Under these conditions, cells spread and divide in all directions resulting in an inherent variability in cell shape, morphology and behavior. The high cell-to-cell variance of the overall population impedes the success of HCA, especially for drug development. The ability of micropatterns to normalize the shape and internal polarity of every individual cell provides a tremendous opportunity for solving this critical bottleneck 1-2.
To facilitate access and use of the micropatterning technology, CYTOO has developed a range of ready to use micropatterns, available in coverslip and microwell formats. In this video article, we provide detailed protocols of all the procedures from cell seeding on CYTOOchip micropatterns, drug treatment, fixation and staining to automated acquisition, automated image processing and final data analysis. With this example, we illustrate how micropatterns can facilitate cell-based assays. Alterations of the cell cytoskeleton are difficult to quantify in cells cultured on homogenous substrates, but culturing cells on micropatterns results in a reproducible organization of the actin meshwork due to systematic positioning of the cell adhesion contacts in every cell. Such normalization of the intracellular architecture allows quantification of even small effects on the actin cytoskeleton as demonstrated in these set of protocols using blebbistatin, an inhibitor of the actin-myosin interaction.
Protocol
1. Cell Preparation and Seeding on Micropatterns
- Place 2 CYTOOchips in 2 independent wells of a 6 well plate ensuring that you read “CYTOO” in the lower right corner of the CYTOOchip.
Micropatterns can be fluorescently labeled with Cy3 or Cy5 dyes. CYTOOChips used for this experiment have Cy3 fluorescently labeled micropatterns.
CAUTION: Keep chips in the dark. - Collect HeLa cells (~80% confluent) by trypsinization. Check under a microscope that cells are properly dispersed by the trypsin treatment. Collect and centrifuge cells at 300g for 4 min, then gently resuspend the cell pellet. Count cells and dilute to a concentration of 15,000 cells/ mL in completed DMEM/F12 culture media.
- Dispense 4 mL (60,000 cells) into each well containing a CYTOOchip.
CAUTION: The plate should be moved as little as possible to avoid inducing any rotating movements in the medium as this will tend to concentrate cells at the center. - Let the cells sediment for 10 min under the hood then move them to the cell incubator. After 10-20 min, cells will start to adhere to the micropatterns. After 10 min, check regularly under the microscope.
- As soon as cells have attached, change the cell medium and gently flush the coverslip surface using the following procedure:
- Keeping the plate flat, gently aspirate the medium with a pipette from the side of the well.
CAUTION: Never let the chip dry out, this means never aspirate off all the liquid but leave enough to cover the CYTOOchip at all times. - Add 4 mL PBS and aspirate again first starting at the center of the chip then moving to the side of the well. Aspirate off the PBS as described without exposing the chip to air. Repeat 3-4 times.
- Check under the microscope for floating cells:
- If a large number of floating cells remain, repeat the washing procedure.
- If you see very few cells, aspirate off part of the PBS and replace it with 2 mL fresh medium. Aspirate and add 4 mL of fresh medium twice.
- Place the plate in a tissue culture incubator at 37°C with 5% CO2 for a minimum of three hours to allow cells to achieve full spreading.
NOTE: Using this method between 10-30% (2000-6500) micropatterns will be occupied by a single or multiple cells.
CAUTION: The time needed for cells to adhere before the washing step and the time needed for full spreading of cells on micropatterns will be specific to each cell line.
2. Blebbistatin Treatment
- Dissolve the blebbistatin in 100% DMSO to obtain a stock solution of 17 mM. Dilute the stock solution further to 5 mM with 100% DMSO. Make a final dilution in culture medium to obtain a final concentration of 5 μM blebbistatin in 0.1% DMSO.
- For control conditions, prepare 4 mL of medium containing 0.1% DMSO only.
- Aspirate the media on cells and replace it with the 4 mL medium prepared for treated and control conditions.
- Incubate cells for 1hr at 37°C.
3. Cell Fixation and Staining of Actin
- Solution preparation for cell fixation
Please see reagent section for the preparation of cytoskeleton buffer (CB) and PFA 5%. CB is particularly recommended for fluorescence labeling of actin filaments and microtubules.- Prepare 1xCB-sucrose: add 1 g of sucrose to 2 mL of CB.
Inclusion of sucrose helps preserve internal cell structures. - Prepare 4% PFA-CB-sucrose: Add 1 mL of 1xCB-sucrose to 4 mL of warm PFA 5%.
CAUTION: This solution can be kept at 4°C for a few days only.
- Prepare 1xCB-sucrose: add 1 g of sucrose to 2 mL of CB.
- PFA fixation
- Use forceps to pick up the CYTOOchip on the CYTOO logo and place it (without washing) in a well of a 6-well plate containing 2 mL of 4% PFA-CB-sucrose solution
- Incubate 10 min at RT
- Remove PFA-CB-sucrose and wash once with PBS
- Do a second wash with 2 mL of 100 mM NH4Cl for 10 min in order to quench residual PFA crosslinking activity
NOTE: PFA fixed slides can be stored in PBS for several days at 4°C before organelle staining for fluorescent imaging.
- Permeabilization of cell membrane with Triton X-100
Permeabilization of cells with detergent allows antibodies or other probes to penetrate the cell membrane and eliminates some of the cytosolic pool of cytoskeleton proteins that can otherwise reduce the contrast of cytoskeletal polymers making it difficult to analyse their localization.- Remove NH4Cl and add 2 mL/well of 0.1% Triton X-100-CB for 3 min at RT
- Wash twice with 2 mL PBS
- Actin staining
Fluorescent-phalloidin which binds to native actin only is used to stain actin filaments.- Add 2 mL of FITC-conjugated phalloidin diluted 1:2000 in PBS with 1.5% BSA
- Incubate 1 h at RT
- Wash once with 2 mL PBS
- Nuclei staining
- Add 2 mL of Hoechst (stain with 1μg/ mL in PBS)
- Incubate 3 min at RT
- Wash 2 times with 2 mL PBS
- Mounting of slide
- Pick up the CYTOOchips on the CYTOO logo and mount CYTOOchip onto a standard microscope slide using 25-30 μL of Mowiol or any other mounting solution.
4. Microscope Setup and Automated Image Acquisition
Numerous imaging software packages exist that control the microscope for fast automated image acquisition and display. This example uses Metamorph imaging software and automatic acquisitions are performed on a Nikon Eclipse Ti microscope equipped with a CCD Hamamatsu camera and an Intensilight mercury-fiber illuminator. Images are acquired at 20x magnification in three wavelengths corresponding to DAPI (nuclei), Cy-3 (micropatterns) and FITC-Phalloidin (actin). The number of micropatterns visualized per field will vary depending on the camera and objective setups. CYTOO micropatterns are positioned at defined intervals from each other (100 or 130 μm) across the chip greatly facilitating acquisition.
- Select 20x magnification
- Open Multidimensional Acquisition (in the Menu bar: Apps/Multidimensional Acquisition) and set up the different parameters of the experiment:
- Number of wavelengths: 3
- Wavelength types: DAPI, FITC, Cy3
- Set exposure time for each wavelength by first focusing on a field of cells
- Autofocus can be performed on each wavelength
- Select where to save data
- Select Cy3 wavelength and position the camera so that 12 micropatterns are visualized and centered in the camera field (4 columns and 3 rows of patterns).
- Create a journal running MultiDimensional Acquisition. The procedure for creating a journal is described in Figure 1.
- Open Scan stage function (in the Menu bar: Devices/Stage/Scan Stage). To acquire 24 images each containing 12 micropatterns at 20x magnification, enter 6 columns and 4 rows with -400 μm X step size and 300 μm Y step size. In Set Journal to Execute, upload the journal MDA.JNL. Then press Scan to start the automated acquisition of the whole block in the three wavelengths.
NOTE: The X and Y step sizes indicated here will vary according to the number of micropatterns present in a camera field. Depending on your stage set up, direction in X and/or Y may require negative values for movement in the correct direction.
5. Automated Image Processing and Analysis
Many image processing and analysis software packages exist. The procedures described below outline image processing and analysis steps performed by 2 custom-made macros written for the open source program ImageJ. The first macro CellRef creates a Reference Cell, the second Hypotenuse measures rigidity/collapse of the cell membrane. Both are available upon request through www.cytoo.com.
- Create image stacks for all three channels centered on micropatterns (macro CellRef).
Use of micropatterns avoids cell clustering and clumping, greatly simplifying the first step in automated image processing which is localization and segmentation of individual cells. The fluorescently labeled micropattern images are used to define and delimit regions centered on the micropatterns. These regions are then transposed onto the images acquired in other channels and cropped. Corresponding cropped images are then assembled into stacks for each wavelength. A RGB-stack is also created from the merge of the three channels (Figure 2). - Apply single cell selection filter (macro CellRef)
As indicated above, 10-30% (2000-6500) of the micropatterns are occupied by a single cell or multiple cells, while the remaining micropatterns are empty. To select images that show micropatterns occupied by just a single cell, the macro detects the number of nuclei per image and eliminates from all stacks (RGB and different wavelengths) those containing more than one nucleus or no nuclei (Figure 3). - Eliminate non-normalized and cells that are not fully stretched across the micropattern using cell area cut-off filter (macro CellRef)
To remove dividing cells that would have escaped the nuclei filter, another filter using the FITC channel for actin is applied. Cell area is calculated and those below a certain cut off (cell areas about 2 times less than interphase cell area) are removed from all stacks. The cell envelope area is determined by the size of the pattern. - Align cell images (macro CellRef)
From the previous steps, images containing single micropatterned cells were selected. Even though these images are centered on micropatterns, they are never all perfectly aligned to each other. To conclude the Image processing, an ImageJ plugin (MultiStackReg) is applied to realign all collected images and all channels based on the micropattern position. This step generates aligned stacks of individual images that can now be used for single cell analysis or creation of a Reference Cell (Figure 4) - Building a Reference Cell (macro CellRef)
Growing cells on micropatterns means that all the cells have similar shapes. This normalization permits a precise alignment and overlay of many cell images. Summing up the signal in each image over the whole stack allows one to visualize and measure an “average drug effect”. The final overall image or Reference Cell is a unique and useful tool for representation and image analysis and can only be obtained with micropatterned cells (Figure 4). In ImageJ, the Reference Cell is constructed by making a projection of the filtered and aligned images from a stack (Image>Stacks>Z-project). Several projection methods can be applied such as average or maximum intensity but usually a median projection is chosen which eliminates main outliers of the image stack. To facilitate examination of the results, a LUT (Look Up Tables) converting the monocolor image into a color-coded frequency map is applied. - Measuring and quantifying parameters of interest (macro Hypotenuse)
In this video article, L-micropatterns are used because the form organizes the actin cytoskeleton into a single major stress fiber. By highlighting actin in this way, blebbistatin’s impact on the cytoskeleton can be determined in many ways: measuring changes in the curvature of the actin stress fiber, staining intensity, length or thickness of the stress fiber, changes in morphometric features of the actin filaments or cell shape etc. These parameters can be measured either on individual cell images or on the final Reference Cell itself.
Here, the impact of 5 μM blebbistatin was characterized by counting the number of collapsed cells using a second ImageJ macro named Hypotenuse (Figure 7). Briefly, thresholded images of the L-micropattern are used to define a theoretical hypotenuse of the cell triangle shape. Next, thresholding of the actin stained images is performed in order to approximate actual cell shape. The area between the theoretical hypotenuse of the cell triangle and the actual concave cell membrane is then measured. When the cell has collapsed, an area underneath the theoretical hypotenuse appears. The percentage of collapsed cells is used to compare control and treated conditions.
6. Representative Results
Examples of what cells should look like on micropatterns immediately and several hours after the critical washing steps described in 1.6-1.8, are shown in Figure 5. After 15 min on CYTOOchips, round HeLa cells are observed at equal distances indicating that they have adhered to the fibronectin micropatterns (Figure 5a). Adhesion is complete when cells remain immobilized despite gentle shaking of the culture vessel. To minimize the number of micropatterns occupied by multiple cells, chips are then flushed with PBS to remove cells floating over the cytophobic areas. After several hours, cells that are fully stretched over the micropatterns will look like those shown in Figure 5b.
Typical single cell images obtained after incubation of cells with blebbistatin, fixation and staining of actin and nuclei are presented in Figure 6. After automated acquisition of multiple images and image processing using the ImageJ CellRef macro, a color-coded frequency map is generated that depicts the intensity of actin averaged over all cell images (Figure 6c and 6f). The comparison of the Reference Cell for nontreated and treated conditions shows the potential of this approach for easy observation of the phenotype under study and is especially useful for exploring cellular drug effects.
An example of one possible analysis of the effect of blebbistatin on cells is presented in Figure 7. The number of collapsed cells (ie. cells with a blank area existing under the theoretical hypotenuse) detected in 50 control cells and 50 cells treated with 5 μM blebbistatin as detected by the ImageJ Hypotenuse macro is shown. The increased number of collapsed cells in the blebbistatin treated population reflects relaxation of the stress fibers and hence tension at the membrane due to blebbistatin inhibition of actinomyosin contractile forces within the cell.
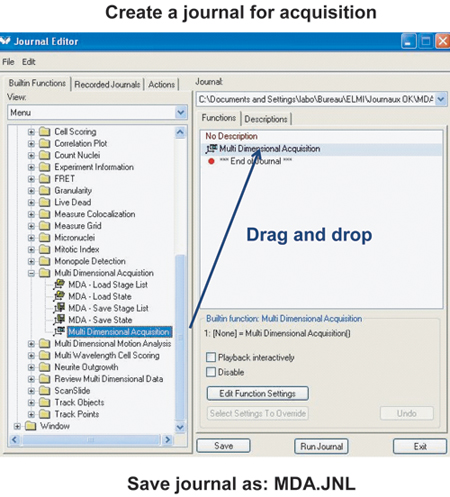
Figure 1. Procedure to create a new journal with Metamorph.
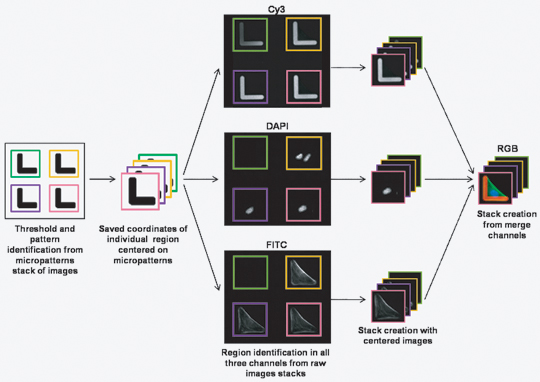
Figure 2. Flow chart of the automatic image processing procedure.
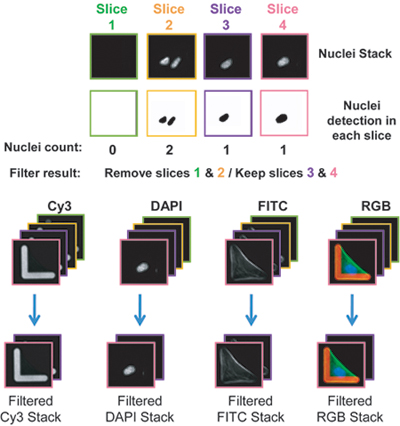
Figure 3. Filtering out of single cells.
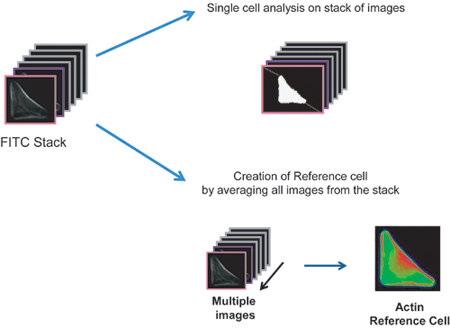
Figure 4. Building a Reference Cell.
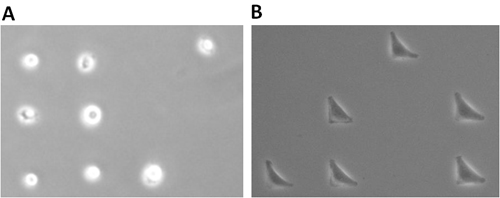
Figure 5. Cell seeding. A) Hela cells adhered to micropatterns after the flushing step (4x magnification) B) HeLa cells after full spreading on L micropatterns (10x magnification).
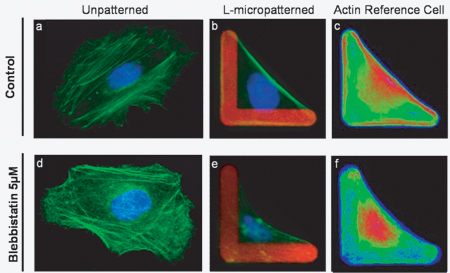
Figure 6. Comparison of nonpatterned and micropatterned cells in control and drug treated conditions. HeLa cells were seeded for 3 hours and treated with 5 μM blebbistatin for 1 hour (lower panel) or left untreated (upper panel). In control nonpatterned cells (a), actin assembles into multiple fibers which disassemble imperceptibly upon treatment with 5 μM blebbistatin (d). On L-micropatterns, control cells adopt a triangular shape (b) and develop a major actin fiber (stress fiber) between the 2 apices of the L. Compared to nonpatterned cells (d), blebbistatin induces a major phenotypic change on L-micropatterned cells (e). Direct visualization of drug effects and the comparison of control and treated conditions can be visualized rapidly with the Reference Cell presentation built from 20 cells. Similar to individual cells, a clear and intense stress fiber is present on the control Reference Cell (c), while blebbistatin treated cells collapse and the major stress fiber disappears (f).
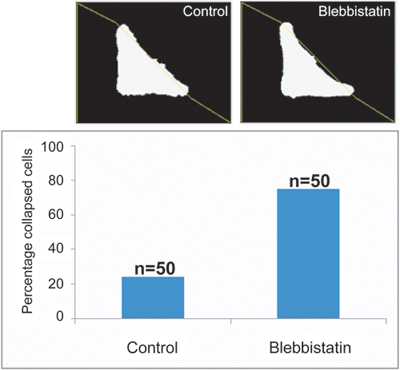
Figure 7. Example of an analysis of the effects of blebbistatin on cells using the macro Hypothenuse. While 25% of control cells were collapsed, this increased 3 fold to 75% in the 5 μM blebbistatin treated cells reflecting the weakened actinomyosin contractile network (n=50 cells).
Discussion
Choice of micropattern
While the effects of 5 μM blebbistatin are barely detectable on standard culture supports, efficient quantification is possible on micropatterns that concentrate actin into a single major stress fiber. This enhances visualization of the actin cytoskeleton aiding accurate quantification of blebbistatin effects on the cell. The choice of micropattern shape is critical in the design of the assay and is designed according to the phenotypic change to be highlighted in the study.
Conclusions
Micropatterns facilitate the visualization and quantification of drug effects, especially at low concentrations. The replacement of homogenous planar substrates with adhesive micropatterns for cell-based assays improves the accuracy, sensitivity and quality of data produced. In consequence, fewer cells are needed for analysis in order to achieve robust statistical outcomes 3. Adhesive micropatterns offer a real opportunity for improving functional studies and exploring phenotypic changes at lower drug concentrations in order to avoid inducing toxic or indirect drug effects. If adequate screening platforms are used, micropatterns can meet the demands of pharmaceutical companies for improving gene/drug screening applications. Some additional examples of recent publications that have exploited micropatterns for analyzing and quantifying cellular phenomenon are cited below 3-13.
Divulgations
The authors have nothing to disclose.
Materials
| Name of the reagent | Company | Catalogue number | Comments | |
| Cell culture | ||||
| CYTOOchips 20×20 L-M-FN | CYTOO | 10-114 | CYTOOchips are available for adsorption of the protein of your choice | |
| PBS 1x | Gibco | 14040-091 | ||
| Counting chamber (Kova slide) | Prolabo | 71287.01 | ||
| DMEM/F-12 + Glutamax-I | Gibco-Invitrogen | 25300-054 | ||
| FBS (f tal bovine serum) | PAA | 31331-028 | ||
| Penicillin & Streptomycin | PAA | A15-101 | ||
| 6 wells-plate | Dutscher | 353046 | ||
| centrifuge | Fisher Scientific | M65838 | ||
| Microscope | Nikon | |||
| Drugs, fixation and immunostaining | ||||
| Cytoskeleton Buffer 1X (CB) | Sigma | MES : M8250 KCl : I2636 MgCl : M2393 EGTA : E3889 |
10 mM MES pH 6.1, 138mM KCl, 3mM MgCl, 2mM EGTA |
|
| Sucrose | Sigma | S2395 | ||
| PFA 32 % | Electron Microscopy Sciences | 15714 | ||
| PFA 5% | Mix 10 mL of PFA 32% with 54 mL of CB 1.185X | |||
| Triton-X100 | Sigma | T9284 | ||
| PBS tablets | Gibco-Invitrogen | 18912-014 | ||
| Bovin serum albumin (BSA) | Sigma | A7806 | ||
| Phallodin-FITC | Sigma | P5282 | ||
| Hoescht | Invitrogen | H3570 | ||
| Blebbistatin | Euromedex | BN0640 | ||
| DMSO | Sigma | D8418 | ||
| NH4Cl 0.1M | Sigma | M11209 | ||
| Image Acquisition | ||||
| Epifluorescent microscope | Nikon | Eclipse Ti | ||
| CCD Camera | TBC | TBC | ||
| ImageJ Software | http://rsbweb.nih.gov/ij/ | |||
References
- Thery, M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 7, 947-953 (2005).
- Thery, M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci U S A. 103, 19771-19776 (2006).
- Schauer, K. Probabilistic density maps to study global endomembrane organization. Nat Methods. , (2010).
- Bischofs, I. B., Klein, F., Lehnert, D., Bastmeyer, M., Schwarz, U. S. Filamentous network mechanics and active contractility determine cell and tissue shape. Biophys J. 95, 3488-3496 (2008).
- Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M., Ingber, D. E. Geometric control of cell life and death. Science. 276, 1425-1428 (1997).
- Dupin, I., Camand, E., Etienne-Manneville, S. Classical cadherins control nucleus and centrosome position and cell polarity. J Cell Biol. 185, 779-786 (2009).
- Gomez, E. W., Chen, Q. K., Gjorevski, N., Nelson, C. M. Tissue geometry patterns epithelial-mesenchymal transition via intercellular mechanotransduction. J Cell Biochem. 110, 44-51 (2010).
- Kilian, K. A., Bugarija, B., Lahn, B. T., Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 107, 4872-4877 (2010).
- Kwon, M. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22, 2189-2203 (2008).
- Nelson, C. M. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A. 102, 11594-11599 (2005).
- Pouthas, F. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci. 121, 2406-2414 (2008).
- Thery, M., Jimenez-Dalmaroni, A., Racine, V., Bornens, M., Julicher, F. Experimental and theoretical study of mitotic spindle orientation. Nature. 447, 493-496 (2007).
- Thery, M., Pepin, A., Dressaire, E., Chen, Y., Bornens, M. Cell distribution of stress fibres in response to the geometry of the adhesive environment. Cell Motil Cytoskeleton. 63, 341-355 (2006).

