Segmentation and Measurement of Fat Volumes in Murine Obesity Models Using X-ray Computed Tomography
Summary
Fat content analysis is routinely conducted in studies utilizing murine obesity models. Emerging methods in small animal CT imaging and analysis are providing for longitudinal detail rich fat content analysis. Here we detail step by step procedures for performing small animal CT imaging, analysis, and visualization.
Abstract
Obesity is associated with increased morbidity and mortality as well as reduced metrics in quality of life.1 Both environmental and genetic factors are associated with obesity, though the precise underlying mechanisms that contribute to the disease are currently being delineated.2,3 Several small animal models of obesity have been developed and are employed in a variety of studies.4 A critical component to these experiments involves the collection of regional and/or total animal fat content data under varied conditions.
Traditional experimental methods available for measuring fat content in small animal models of obesity include invasive (e.g. ex vivo measurement of fat deposits) and non-invasive (e.g. Dual Energy X-ray Absorptiometry (DEXA), or Magnetic Resonance (MR)) protocols, each of which presents relative trade-offs. Current invasive methods for measuring fat content may provide details for organ and region specific fat distribution, but sacrificing the subjects will preclude longitudinal assessments. Conversely, current non-invasive strategies provide limited details for organ and region specific fat distribution, but enable valuable longitudinal assessment. With the advent of dedicated small animal X-ray computed tomography (CT) systems and customized analytical procedures, both organ and region specific analysis of fat distribution and longitudinal profiling may be possible. Recent reports have validated the use of CT for in vivo longitudinal imaging of adiposity in living mice.5,6 Here we provide a modified method that allows for fat/total volume measurement, analysis and visualization utilizing the Carestream Molecular Imaging Albira CT system in conjunction with PMOD and Volview software packages.
Protocol
1. Animals
- For the results reported below, three C57BL/6J mice and four B6.V-Lepob/J mice were obtained from Jackson Laboratories (Bar Harbor, Main, USA). B6.V-Lepob/J mice represent one of the earliest models of obesity and remain actively researched. B6.V-Lepob/J mice exhibit a phenotype characterized by increased adipocyte size and number and can weigh up three times more than wild type mice.7,8 Here, B6.V-Lepob/J mice were employed as a positive control for an obese phenotype to illustrate the feasibility of the Albira CT system for CT based fat content measurements.
- Imaging was performed when the animals reached approximately 12 weeks old. (Obesity in B6.V-Lepob/J mice is manifest after 4 weeks of age).
- Mice were anesthetized by Isofluorane (2.5% flow rate) and kept under at 2.5% via a nose-cone setup for imaging. Animals were positioned prone in the standard rat bed (M2M Imaging Inc. Cleveland, OH) in supplied with the Albira image station. Limbs were positioned lateral from the torso for a uniform CT acquisition.
- After image acquisition was completed, mice were removed from the nose cone and returned to a recovery cage until ambulatory.
2. Image Acquisition and Reconstruction
- Image acquisitions are performed using the Albira CT system (Carestream Molecular Imaging, Woodbridge, CT). Mice were anesthetized by Isofluorane (2.5% flow rate) and kept under at 2.5% via a nose-cone setup for imaging. Acquisitions were performed to scan a bed of 115 mm length, using 600 projections. The X-ray source was set to a current of 200 μA and voltage of 45 kVp, and used a 0.5 mm Al filter to harden the beam.
- Approximate radiation deep dose equivalent for CT settings was 220 mSv, and shallow dose equivalent was 357.4 mSv. These doses are over 20-fold lower than reported LD50 values.9
- Images are reconstructed using the FBP (Filtered Back Projection) algorithm via the Albira Suite 5.0 Reconstructor using “Standard” parameters. These combined acquisition and reconstruction settings produce a final image with 125 μm isotropic voxels, deemed sufficient for whole animal analysis. For detailed region specific analysis, a reconstruction with 35 μm isotropic voxels may be selected for a final resolution of 90 μm.
3. Image Analysis
Image analysis is performed using the PMOD (PMOD Technologies LTD, Zurich, Switzerland) analysis software. Images are segmented in PMOD according to tissue density-first for total volume and then for fat volume.
3.1 Images may be reduced for analysis to minimize computational demands.
- To reduce, navigate to the main View tab.
- Select Tools > Reduce.
- Select X by 2, Y by 2, and Z by 2.
- Check Replace.
- Select Run.
The message: “Bounding box will change” displays once the reduction is complete.
3.2 Images can be masked to eliminate bed and nose-cone elements for subsequent volume-of-interest (VOI) analysis.
- To mask, navigate to Planes, Layouts, Rotations, Mirror, 3D Markers > Planes & Layouts.
- Select Show plane Z.
- Scroll to the nose cone in the Z plane.
- Select the main VOI tab > Draw vertices.
- Draw a region of interest (ROI) around the animal nose-excluding the bed and nose cone.
- Select Copy Actual ROI.
- Move to the next slice, and paste ROI from buffer through the relevant planes of nose.
- Use Edit Group of Vertices to adjust the ROIs as necessary.
- Select Delete ROI at the first planes beyond the nose cones.
- Generate a new VOI to encompass the animal girth (excluding the animal bed) at the first planes beyond the nose cones.
- Navigate to VOI Tools > Masking & Algebra.
- Enter -1000 in the dialogue box provided.
- Select the Mask voxels outside the selected VOIs button.
The message: “Irreversible data operation. Do you want to continue?” displays.
- Select Yes.
- Navigate to Planes, Layouts, Rotations, Mirror, 3D Markers > Planes & Layouts.
- Select Show all planes.
- Examine the VOI for integrity.
- Select Save.
- Save as Analyze.
- Change the file name prefix.
3.3 First, segment the image for total animal volume:
- Select Tools > External.
- Select the Segmentation check box.
- Enter a range of -300 à +3500 (density range reference derived from abdominal fat-region density range).
- Select Run Segmentation.
- Examine the integrity of the segmentation.
- Select OK.
- Select Remove ROI.
- Select VOI statistics.
Reported statistics represent the total volume.
- Record the reported volume.
3.4 Next, segment the image for fat volume:
- Return to the non-segmented masked image for fat-volume segmentation.
- To load the saved Masked data file, check the Analyze box in the Load window).
- Select Tools > External.
- Select the Segmentation check box.
- Enter a range of -200 à -50.
- Select Run Segmentation.
- Examine the integrity of the segmentation.
- Select OK.
- Select VOI statistics.
The reported statistics represent the fat volume.
- Record the reported volume.
- Select Save.
- Save as Analyze.
- Change the file name prefix.
Optional: If skin/peripheral density remains, the “Erosion and Dilation” protocol below may be performed to eliminate these regions for VOI analysis.
- Select Tools > External.
- Select the Morphological check box. The Morphological view displays.
- Select Erosion.
- Select OK.
- Select Tools > External.
- Select the Morphological check box. The Morphological view displays.
- Select Dilation.
- Select OK.
4. Visualization of CT Images
4.1 VolView v3.2 (Kitware, Clifton Park, NY, USA) was utilized to create rendered 3D visual displays of segmented images.
- Open the CT data set in Analyze format.
- Use the default settings in the pop-up window.
- Open the plug-ins menu.
- Under Utility, select Merge volumes.
- Uncheck Rescale Components.
- Click Assign Second Input.
- Choose the segmented fat data for the second input.
- Use the default settings in the pop-up window.
- Click Apply plug-in.
- Double-click Volume view window for a larger view of the subject mouse.
4.2 Return to the Color/Opacity tab. The component drop-down box refers to which data set is currently being edited. Two sliders are located at the bottom of the tab and determine the relative brightness of each component data set within the overlay, using values 0 à 1. For component 1, the CT, we prefer to use a grayscale color scheme. To change the color:
- In the Scalar Color Mapping section, double-click on one of the color sliders.
- To remove a slider, drag it out of the box.
- To add a new slider, click anywhere within the slider area.
- Remove one of the sliders.
- Make the left color slider black (scalar value (S)= -19000).
- Make right color slider white ((S) = 15000).
- From the Scalar Opacity mapping box, create a new point by clicking within the box. This will give a total of three points in the window.
- For the middle point, change the (S) à ~-3000, and the Opacity (O) value to 0.
- Select the third point at the right of the window.
- Change (S) to about 32000, and O à .25.
- The first point can be anywhere at left, just as long as the Opacity value is set to 0.
- Change to component two, which should edit the look of the fat.
- Change each of the color sliders to red by double-clicking and sliding the Hue (H) slider to the left to false-color map the fat to red. Very little else should be needed to adjust the look of the fat.
4.3 To create a three-panel rotation movie displaying the CT, fat, and overlay:
- Click and the drag the subject mouse into an upright position with the back facing you.
- Under Component Weights, set the value of Component two à 0 to display only the CT scan.
- Click Review > Camera.
- Select a number of frames for the rotation movie (for the present case, we chose “36“).
- Change the X rotation value à 360 degrees.
- Select Create.
- In the pop-up dialog box, create a new folder named CT, and save the file in tiff format, which will output a series of rotation images.
- Repeat this step for the fat image, as well as for the overlaid fat/CT image, saving them into individual folders each time.
4.4 ImageJ v 1.43u was used to generate a rotation movie file using the VolView output images.
- In ImageJ, select File > Import > Image Sequence.
- Select the first image in the CT folder. The software will automatically detect the other files and open them as a stack.
- Repeat to open the fat and overlay sequences.
- Open the ROI manager under Analyze > Tools > ROI Manager.
- Draw an ROI around the subject mouse, excluding needless background pixels.
- In the ROI manager, click Add to add the ROI.
- Select a different image sequence.
- In the ROI manager, click the ROI to apply it to the picture. In this fashion, each of the cropped stacks will match up perfectly.
- When the ROI is on all the stacks, right-click within the ROI.
- Select Duplicate.
- Select Check the Duplicate Stack Box to separate the ROI from the rest of the image.
- Close the larger image stacks.
- Repeat this procedure for all three image sequences.
- Go to Image > Stacks > Tools > Combine to combine the stacks together.
- Select CT for Stack 1.
- Select Fat for Stack 2.
- Repeat, and select Combined Stacks for Stack 1 and Overlay for Stack 2.
- There is now a three-panel, image-rotation stack that can be previewed by selecting Play in the lower-left corner of the image window.
- To save the movie as an AVI, select File > Save as… > AVI…
- Click Save.
5. Representative Results
Results for three WT (C57BL/6J) mice and four obese (B6.V-Lepob/J) mice are reported here as a representative example of fat/total volume ratio measurements employing the Albira CT system. Figure 1 below provides a representative display created with VolView v3.2 for the segmentation (i.e. total volume and fat volume) of obese mice CT images.
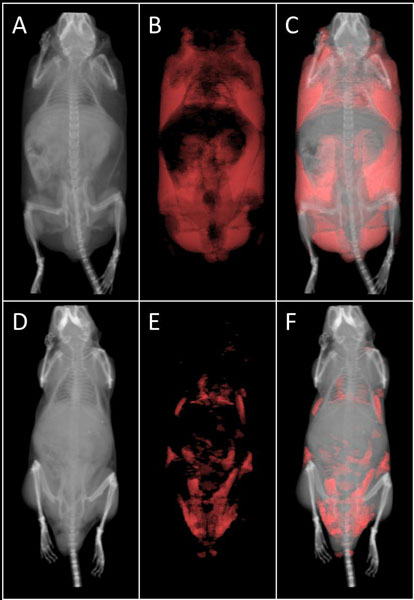
Figure 1. Representative CT images segmented for fat. (A) Obese mouse (B6.V-Lepob/J) CT total volume in grayscale, (B) fat volume in red, and (C) image fusion. (D) WT mouse (C57BL/6J) CT total volume in grayscale, (E) fat volume in red, and (F) image fusion.
Total volumes, fat volumes, and calculated fat/total volume ratios are reported below in Table 1 for each WT mouse and each obese mouse. The averaged fat/total volume ratio for the WT group and the obese group was 0.09 and 0.42 respectively (Figure 2). The fat/total volume ratios for the WT mice versus the obese mice was found to differ significantly (p = 0.001).
| WT (C57BL/6J) | Total (cm3) | Fat (cm3) | Fat/Total Ratio | Obese (B6.V-Lepob/) | Total (cm3) | Fat (cm3) | Fat/Total Ratio |
| Animal 1 | 28.79 | 3.00 | 0.10 | Animal 1 | 66.25 | 26.75 | 0.40 |
| Animal 2 | 33.25 | 3.05 | 0.09 | Animal 2 | 61.15 | 26.31 | 0.43 |
| Animal 3 | 30.30 | 2.63 | 0.09 | Animal 3 | 64.19 | 25.7 | 0.40 |
| Animal 4 | 54.25 | 23.78 | 0.44 |
Table 1. Total volume, fat volume, and fat/total volume ratios for WT and obese mice. Total and fat volumes were derived from segmented images using PMOD VOI analysis.
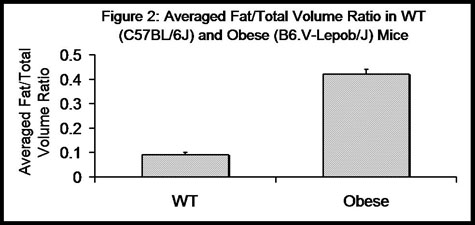
Figure 2. Averaged fat/total volume ratios for WT mice versus obese mice. Averaged fat/total volume ratios for WT (C57BL/6J) and obese (B6.V-Lepob/J) found to be 0.09 and 0.42 respectively are displayed. (Error bars = single standard deviation). WT versus obese fat/total volume ratios were found to differ significantly (p-value = 0.001).
Discussion
Here, utilizing B6.V-Lepob/J mice we have illustrated the feasibility of performing fat content measurements in a small animal model using the Albira CT system. These measurements are consistent with expectations for comparisons of intra-group and inter-group measurements. Firstly, representative results provided here highlight limited intra-group variability in measurements of fat/total volume ratios in both WT and obese mice groups using these procedures. Secondly, fat/total volume ratios for WT versus obese mice differ significantly. Finally, based on comparisons (not shown) with previous values reported for relative total fat mass and percent body fat for WT versus B6.V-Lepob/J mice, our measurements for fat/total volume ratios for WT versus B6.V-Lepob/J mice fall within an expected range,7, 8.
The methods detailed here could be applied or adapted to other models and/or study objectives. Modifications in reconstruction parameters may be required to achieve specific objectives. For example, Judex et al. (2010) reported that 50 μm resolution images were required for some region specific analysis. One cm isotropic volumes of an image may be selected for 35 μm reconstructions in the Albira 5.0 Suite Reconstructor using the “HR” reconstruction option. Once the Albira CT system has been utilized for region and organ specific fat content measurements the full benefits (i.e. simultaneous region and organ specific fat volume measurements and longitudinal measurements) of CT based fat content analysis may be realized for the Albira CT system.
Conclusions:
Here we provide a detailed, step by step method for the measurement of fat content in living mice using X-ray CT imaging. We acquired our CT data sets using an Albira image station, and performed subsequent segmentation and analysis using the PMOD software suite. Finally, we provide instructions to enable facile rendering and visualization of the fat tissue distribution within the whole animal.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We warmly thank the Notre Dame Integrated Imaging Facility (NDIIF) and Carestream Health for financial support for this project.
References
- Clarke, P. J., O’Malley, P. M., Schulenberg, J. E., Johnston, L. D. Midlife and Socioeconomic Consequences of Persistent Overweight Across Early Adulthood: Findings From a National Survey of American Adults (1986-2008). Am. J. Epidemiol. , (2008).
- Brockman, G., Bevova, M. Using Mouse Models to Dissect the Genetic of Obesity. TRENDS in Genetics. 18, 367-376 (2002).
- Bray, G. A. Progress in Understanding the Genetics of Obesity. Journal of Nutrition. 127, 940S-942S (1997).
- Carroll, L. Mouse Models of Obesity. Clinics in Dermatology. 22, 345-349 (2004).
- Judex, S., Luu, Y. K., Ozcivici, E., Adler, B., Lublinsky, S., Rubin, C. T. Quantification of Adiposity in Small Animal Rodents using Micro-CT. Methods. 50, 14 (2010).
- Luu, Y. K., Lublinsky, S., Ozcivici, E., Capilla, E., Pessin, J. E., Rubin, C. T., Judex, S. In Vivo Quantificaiton of Subcutaneous and Visceral Adiposity by Micro Computed Tomography in a Small Animal Model. Med. Eng. Phys. 31, 34-41 (2009).
- Medina-Gomez, G., Gray, S. L., Yetukuri, L., Shimomura, K., Virtue, S., Campbell, M., Curtis, R. K., Jimenez-Linan, M., Blount, M., Yeo, G. S., Lopez, M., Seppänen-Laakso, T., Ashcroft, F. M., Oresic, M., Vidal-Puig, A. PPAR gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PLoS Genet. 3, e64 (2007).
- Sakkou, M., Wiedmer, P., Anlag, K., Hamm, A., Seuntjens, E., Ettwiller, L., Tschöp, M. H., Treier, . M.A role for brain-specific homeobox factor bsx in the control of hyperphagia and locomotory behavior. Cell Metab. 5, 450-463 (2007).
- Patchen, M. L., MacVittie, T. J., Souza, L. M. Postirradiation treatment with granulocyte colony-stimulating factor and preirradiation WR-2721 administration synergize to enhance hemopoietic reconstitution and increase survival. International Journal of Radiation Oncology. 22, 773-779 (1992).

