Microfluidic On-chip Capture-cycloaddition Reaction to Reversibly Immobilize Small Molecules or Multi-component Structures for Biosensor Applications
Summary
We present a method for rapid, reversible immobilization of small molecules and functionalized nanoparticle assemblies for Surface Plasmon Resonance (SPR) studies, using sequential on-chip bioorthogonal cycloaddition chemistry and antibody-antigen capture.
Abstract
Methods for rapid surface immobilization of bioactive small molecules with control over orientation and immobilization density are highly desirable for biosensor and microarray applications. In this Study, we use a highly efficient covalent bioorthogonal [4+2] cycloaddition reaction between trans-cyclooctene (TCO) and 1,2,4,5-tetrazine (Tz) to enable the microfluidic immobilization of TCO/Tz-derivatized molecules. We monitor the process in real-time under continuous flow conditions using surface plasmon resonance (SPR). To enable reversible immobilization and extend the experimental range of the sensor surface, we combine a non-covalent antigen-antibody capture component with the cycloaddition reaction. By alternately presenting TCO or Tz moieties to the sensor surface, multiple capture-cycloaddition processes are now possible on one sensor surface for on-chip assembly and interaction studies of a variety of multi-component structures. We illustrate this method with two different immobilization experiments on a biosensor chip; a small molecule, AP1497 that binds FK506-binding protein 12 (FKBP12); and the same small molecule as part of an immobilized and in situ-functionalized nanoparticle.
Introduction
Efficient conjugation reactions are valuable tools for attaching bioactive molecules to surfaces for a variety of biotechnology applications. Recently, the very fast bioorthogonal [4+2] cycloaddition reaction between trans-cyclooctene (TCO) and 1,2,4,5-tetrazine (Tz) has been used to label cell surfaces, subcellular structures, antibodies and nanoparticles.1-7 Here, we use the [4+2] cycloaddition reaction in combination with antigen/antibody capture (GST/anti-GST) for reversible on-chip synthesis of multi-component structures for Surface Plasmon Resonance (SPR) interaction analysis and monitor the process in real-time (Figure 1).8,9 Notably, the capture-cycloaddition strategy enables surface regeneration using an established protocol.8 As a consequence, assembly of stable sensor surfaces with control over ligand orientation and density for variety of new assay formats is now possible. Using this strategy we demonstrate the immobilization of TCO/Tz-derivatized small molecules and characterize the cycloaddition rates in a variety of buffer conditions. We chose the well-known interaction between FKBP12 and a molecule AP1497 that binds FKBP1210-12 as an example to verify that the capture-cycloaddition strategy preserves the ability of the small molecule to interact with its target when either directly attached to immobilized GST antigens or to immobilized nanoparticles (NPs).
This method offers several benefits. First, the reversible immobilization of small molecules on sensor chips is now possible. Second, TCO/Tz immobilization of small molecules also enables label-free interaction studies that reverse the orientation of canonical SPR studies, and may provide a complementary view of a binding interaction. Third, this method enables the microfluidic synthesis of targeted nanoparticles, and immediate evaluation of their binding properties. This promises to improve the efficiency of evaluating or screening targeted nanoparticles, and also decrease the amounts of nanoparticles required.13-15 Fourth, this approach can measure the reaction kinetics of bioorthogonal cycloaddition reactions in real-time under continuous flow. Finally, the TCO/Tz immobilization chemistry is robust in the presence of serum. Taken together, we anticipate that this versatile approach will broadly facilitate construction of stable sensor surfaces for a wide variety of microfluidic studies with relevance to in vitro and in vivo cellular applications.
Protocol
1. Preparation of GST and Nanoparticle (NP) Conjugates
- GST-TCO preparation:
- Add 8 μl of TCO-NHS solution (50 mM in DMSO) to 100 μl of GST (1 mg/ml in PBS) and shake mixture at RT for 1 hr.
- Remove excess reagent using a Zeba spin desalting column. The recovered filtrate containing the GST-TCO conjugate is stored at 4 °C prior to use.
- GST-Tz preparation:
- Add 6 μl of Tz-NHS solution (25 mM in DMF) to 75 μl of GST (1 mg/ml in PBS) and shake mixture at RT for 1 hr.
- Dilute the reaction mixture with 25 μl of PBS and purify using a spin desalting column. The filtrate containing the GST-Tz conjugate is stored at 4 °C prior to use.
- NP-TCO preparation:
- Add 100 μl of TCO-NHS solution (50 mM in DMSO) to 150 μl of aminated nanoparticle (NP-NH2, 8.7 mg Fe/ml in PBS, 3 nm iron core ~8,000 Fe/NP) and shake mixture at RT for 1 hr.
- Remove excess reagent by gel filtration using a NAP-10 column eluted with PBS buffer. Collect the colored band containing the NP-TCO product.
- Concentrate the filtrate to a final volume of ~150 μl using a centrifugal filter device (100 k MWCO).
- Add 50 μl of succinic anhydride, (0.1 M in DMSO) to the NP-TCO solution and shake at RT for 1 hr (anhydride reacts with any remaining amines to form terminal carboxylic acids. This prevents nonspecific interactions with the dextran surface).
- Purify the NP-TCO product using a NAP-10 column eluting with PBS. Store the solution of NP-TCO at 4 °C prior to use.
2. Surface Preparation
All surface plasmon resonance assays are performed on a Biacore T100 instrument (GE Healthcare) at 25 °C using a CM5 sensor chip and PBS-P as the running buffer unless otherwise noted. Biacore Control and Evaluation software supplied with the instrument are employed for setting up experiments and analyzing data. Two modes of operation, application wizards and manual run will be used for surface preparation and monitoring on-chip capture-cycloaddition. The method builder mode will be used for setting up reverse orientation binding experiments and for measuring cycloaddition reaction rates. Data are double reference subtracted and kinetic analyses are performed using a 1:1 Langmuir binding model.
- Use the amine immobilization wizard template and select immobilization on Flow cells 1 (reference) and 2 (detection). Modify amine method to activate surface carboxylic groups by injection of a 1:1 solution of 0.4 M EDC: 0.1 M NHS for 480 sec at a flow rate of 10 μl/min. Set the injection of ethanolamine to quench remaining activated esters to 420 sec contact time at a flow rate of 10 μl/min.
- Input ligand name, anti-GST (18 μg/ml in acetate buffer, pH 5.0) and set to inject for a contact time of 420 sec at a flow rate of 10 μl/min.
- Place required solution vials in reagent rack 2 making sure to match the positions with the content list. Save and start immobilization.
Note: Running the immobilization wizard in this manner results in anti-GST immobilization levels in the range of ~14,000 to 17,000 RU. - Edit the immobilization wizard to only inject GST ligand (20 μg/ml) solution for 420 sec at 5 μl/min over reference Flow cell 1.
3. Monitoring On-chip Capture-cycloaddition of Functionalized Molecules
- Monitoring on-chip capture-cycloaddition in real-time (Figure 2).
- Place GST-TCO (20 μg/ml), Tz-BnNH2 (10 μM) and regeneration (10 mM glycine, pH = 2.0) solution vials in reagent rack 2. Select the manual run method, set the flow rate to 5 μl/min and flow path to Flow cell 2.
- Inject solution of GST-TCO for 420 sec.
- Inject solution of Tz-BnNH2 for 600 sec.
- Regenerate the surface with two 30 sec injections of regeneration solution.
- Monitoring on-chip assembly of multi-component structures in real-time (Figure 3):
- Place GST-Tz (20 μg/ml), NP-TCO (100 μg Fe/ml), Tz-BnNH2 (10 μM) and regeneration (10 mM glycine, pH = 2.0) solution vials in reagent rack 2. Select the instrument manual run method, set the flow rate to 5 μl/min and the flow path to Flow cell 2.
- Inject a solution of GST-Tz for 60 sec.
- Inject a solution of NP-TCO for 60 sec.
- Inject a solution of Tz-BnNH2 for 60 sec.
- Regenerate the surface with two 30 sec injections of regeneration solution.
4. Monitoring Immobilization Density and Determination of Cycloaddition Rates
- Characterization of small molecule cycloaddition reaction rates (Figure 4).
- Select new method and input general parameters. In the assay steps panel create a new step and name it Sample. Set the Purpose and Connect base settings to Sample.
- In the Cycle Types panel Create a new cycle step and insert the following commands; Capture, Sample, Regeneration 1 and Regeneration 2.
- Select the Capture command; input ligand name (GST-TCO, 20 μg/ml), contact time 120 sec at 5 μl/min and set flow path to: Second. Select the Sample command; input 180 sec contact time, 60 sec dissociation time, 30 μl/min flow rate and set flow path to: Both. Select the Regeneration 1 command; input regeneration solution name (10 mM glycine-HCl pH 2.0), contact time of 30 sec at 30 μl/min and set flow path to: Second. Repeat the same for Regeneration 2 command.
- Select Setup Run and set flow path to: 2-1. Select next and fill sample list with analyte solution (Tz-BnNH2) and concentration series (0.3-10 μM, 1:2 dilution). Select next to rack position panel. Place solution vials in reagent rack 2 and dilution series in 96-well plate making sure to match the positions with the content list. Save method template and start binding assay. Binding data and kinetic analysis are shown in Figure 4.
- Characterization of NP cycloaddition reaction rates (Figure 4).
- Open the saved method template from above and change the following parameters. Select the Capture panel; change ligand name to GST-Tz, 20 μg/ml. Select the Sample panel; change contact time to 120 sec contact time and dissociation time to 120 sec.
- Select the sample list; change analyte to NP-TCO and concentration series (7 to 224 nM, 1:2 dilution). Place solution vials in reagent rack 2 and dilution series in 96-well plate making sure to match the positions with the content list. Save method template and start binding assay. Binding data and kinetic analysis are shown in Figure 4.
5. Measuring the Binding of FKBP12 to Immobilized AP1497
Reversed-orientation binding studies employ FKBP12 as the analyte and compound AP1497 as immobilized ligand (Figure 5). The general method for this assay is set up as follows using the method builder tool:
- Select new method and input general parameters. In the assay steps panel create and name Capture, Sample and Regeneration steps. Select corresponding Purpose and Connect base settings.
- Select Cycle Types panel and create 3 cycle steps: Capture, Sample and Regeneration.
- Insert the Capture command twice under Capture cycle step. Select the Capture 1 panel and select capture solution as variable, set contact time to 300 sec at a flow rate of 5 μl/min and set flow path to: Second. Select the Capture 2 panel and select capture solution as variable, set contact time to 250 sec at a flow rate of 5 μl/min and set flow path to: Second.
- Insert the Sample command under the Sample cycle step. Select the Sample panel; set contact time to 60 sec, dissociation time to 200 sec at a flow rate of 30 μl/min and set flow path to: Both.
- Insert the Regeneration command twice under the Regeneration cycle step. Select the Regeneration 1 panel; input regeneration solution name (10 mM glycine-HCl pH 2.0), contact time of 30 sec at 30 μl/min and set flow path to: Second. Repeat the same for the Regeneration 2 panel.
- Select Setup Run and set flow path to: 2-1. Select next and input capture solution names (GST-TCO and AP1497-Tz), sample name (FKBP12), concentration series (0.020 to 5 μg/ml, 1:2 dilution) and MW (13,000). Place solution vials in reagent rack 2 and dilution series in 96-well plate making sure to match the positions with the content list. Save method template and start binding assay. Kinetic analysis data are shown in Figure 5.
6. Measuring the Binding of FKBP12 to AP1497 Attached to Immobilized NPs
The general method for nanoparticle immobilization, small molecule derivatization and FKBP12 binding assay is set-up as follows using the method builder tool:
- Open the saved method template from above and modify. Insert an additional Capture command under Capture cycle step. Select the Capture 1 panel; deselect capture solution as variable and name capture solution 1 (GST-Tz). Set contact time to 60 sec at a flow rate of 5 μl/min and set flow path to Second. Select the Capture 2 panel; deselect capture solution as variable and name capture solution 2 (NP-TCO). Set contact time to 90 sec at a flow rate of 5 μl/min and set flow path to second. Select the Capture 3 panel; deselect capture solution as variable and name capture solution 3 (AP1497-Tz). Set contact time to 180 sec at a flow rate of 5 μl/min and set flow path to Second.
- Select Setup Run and set flow path to: 2-1. Select next twice. Place solution vials in reagent rack 2 and dilution series in 96-well plate making sure to match the positions with the content list. Save method template and start binding assay. Kinetic analysis data are shown in Figure 5.
Representative Results
Data and figures have been adapted from reference 8.
Efficient reversible immobilization of bioactive small molecules with control over orientation and density plays a key role in development of new biosensor applications. Using the fast bioorthogonal reaction between TCO and Tz, we describe a method for the stepwise assembly and regeneration of ligand surfaces with retention of biological activity. Figure 2 shows the real-time monitoring of Tz-BnNH2 immobilization. A solution of GST-TCO is injected over a pre-immobilized anti-GST surface resulting in ~400 RU rise in response. A second injection with Tz-BnNH2 shows a fast ~15 RU rise in response. No dissociation of the derivatized antigen is observed after switching to running buffer providing evidence for structure stability. The surface is regenerated in the last step of the cycle to enable multiple capture-cycloaddition cycles. Using this procedure and replacing the Tz-BnNH2 moiety with the AP1497-Tz molecule generates a bioactive surface used for interaction studies with its target FKBP12 (Figure 5). Disruption of the antibody/antigen interaction (as shown in Figure 2) regenerates the anti-GST surface, allowing a new molecular assembly to be built. In this case, a GST-Tz injection (capture of ~800 RU) is followed with an injection of NP-TCO (cycloaddition ~600 RU), effectively immobilizing the nanoparticle to the sensor surface (Figure 3). Unreacted TCO groups on the NP are available for cycloaddition with injected Tz-BnNH2 (~22 RU). Alternatively, AP1497-Tz is used to monitor functionalized nanoparticle-FKBP12 interactions (Figure 5). No dissociation of the multi-component structures (GST-Tz/NP-TCO/Tz-BnNH2) are observed providing evidence for structure stability and bioactivity (AP1497-Tz/FKBP12 binding).
With capabilities for real-time immobilization (cycloaddition) reaction monitoring and surface regeneration, the straightforward characterization of the association rate ka (cycloaddition rate) is accomplished as shown in Figure 4. Knowledge of reaction kinetics provides guidelines for controlling immobilization density (contact time and concentration), an important parameter for biosensor assays. Injecting increasing concentrations of Tz-BnNH2 or NP-TCO in duplicate over GST-TCO or GST-TZ surfaces, respectively, generates the binding data (red lines). The binding data for two sequential analyte samples of the same concentration over the same surface are nearly superimposable reflecting minimal loss of antibody binding capacity due to multiple cycles of capture-cycloaddition and regeneration. The Evaluation software provides the best-fit kinetic analyses (black lines) for the cycloaddition reaction rates ka shown in Figure 4.
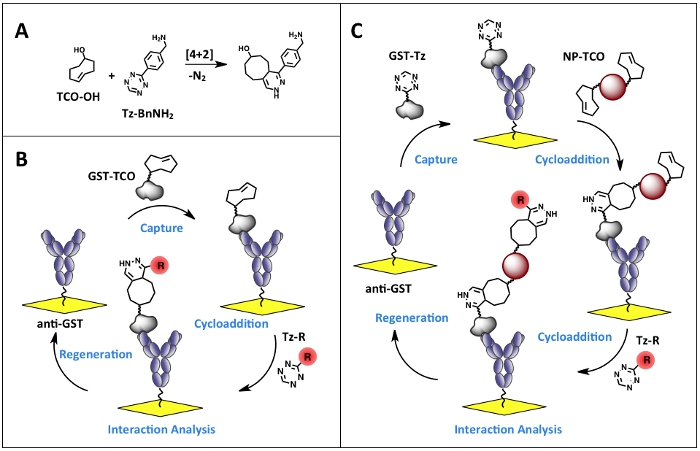
Figure 1. Bioorthogonal conjugation strategy for the reversible immobilization of derivatized molecules. A. [4+2] cycloaddition reaction between trans-cyclooctene (TCO) and 1,2,4,5-tetrazine (Tz) moieties to give the 1,4-dihydropyridazine adduct. B. Experimental scheme for reversible immobilization of Tz/TCO tagged molecules. C. Experimental scheme for reversible nanoparticle immobilization and functionalization. Click here to view larger figure.
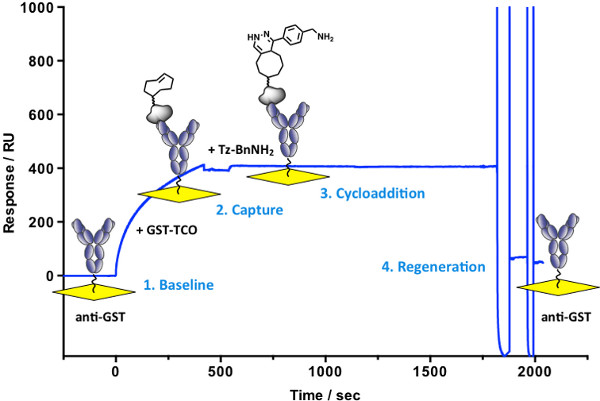
Figure 2. Real-time monitoring of small molecule immobilization. 1) Baseline: pre-immobilized anti-GST. 2) Capture of injected GST-TCO by anti-GST antibody (t = 0-420 sec), followed by injection of running buffer (t = 420-540 sec) showing persistence of interaction. 3) Cycloaddition between captured GST-TCO and tetrazine (Tz-BnNH2; ~15 RU rise in response; t = 540-1,140 sec). No signal decay occurs despite switching the mobile phase to running buffer (t = 1,140-1,820 sec). 4) Regeneration of the anti-GST surface with 2 short injections of 10 mM glycine, pH 2.1 (t = 1,820-2,000 sec).
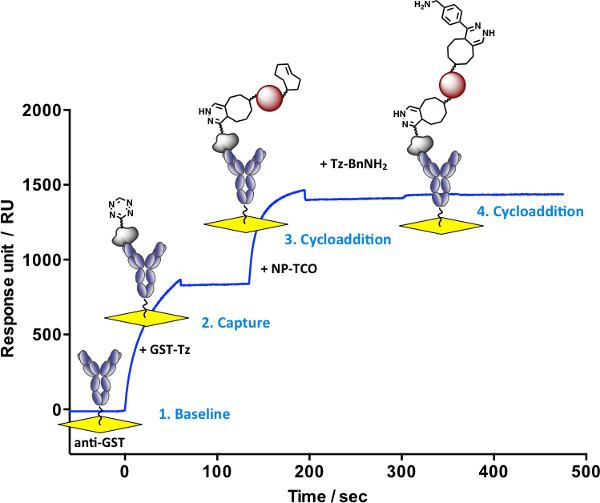
Figure 3. Real-time monitoring of nanoparticle immobilization and functionalization. 1) Baseline. 2) Capture of injected GST-Tz (t = 0-60 sec). 3) Cycloaddition between captured GST-Tz and injected NP-TCO (t = 135-195 sec), followed by injection of running buffer with no decay in signal (t = 195-300 sec). 4) Cycloaddition between immobilized NP-TCO and injected Tz-BnNH2 (~22 RU rise in response, t = 300-360 sec). There is no signal decay despite switching the mobile phase to running buffer (t = 360-480 sec) providing evidence for multi-component stability.
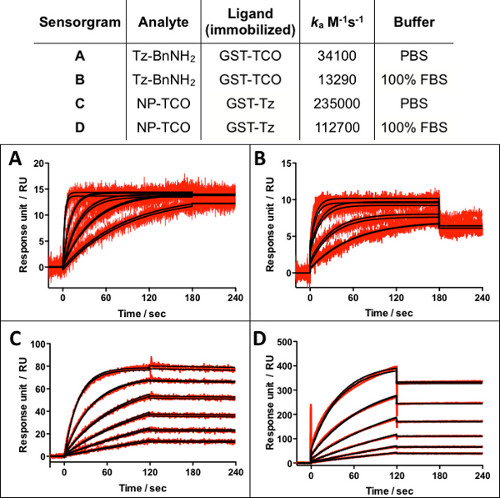
Figure 4. Representative sensorgrams showing binding data (red lines) and kinetic analyses (black lines) for the cycloaddition reaction between Tz and TCO tagged molecules in the presence or absence of 100% FBS. Table summarizes association (cycloaddition) rate constants, ka.
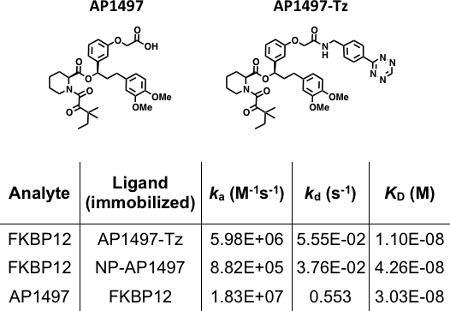
Figure 5. SPR studies of small molecule/protein interactions in varying configurations. (Top) Synthetic derivatives of FK506: AP1497 and AP1497-Tz. The table shows kinetic and equilibrium rate constants derived from binding data. Data from binding experiments where the protein is immobilized (i.e. traditional SPR experiments) are included for comparison.
Discussion
The capture-cycloaddition method described here enables rapid, reversible immobilization of modified nanoparticles and small molecules for label-free chip-based interaction and kinetic studies. The immobilization protocol can be performed in minutes requiring <10 μM concentrations of small-molecule ligands. By modulating ligand concentration and contact time immobilization densities can be closely controlled. Our data show that on-chip bioorthogonal reactions preserve the ability of in situ functionalized nanoparticles or immobilized small molecules to interact with their targets. We have also characterized bimolecular cycloaddition rates between two small molecules, (TCO and Tz) with low molecular weights. We envision that similar assays can be performed to measure and compare rates of other fast bioorthogonal reactions.
Reversible, continuous flow on-chip nanoparticle immobilization and in-situ functionalization provides rapid access to a variety of modified nanoparticles for screening and interaction analysis all in the same experiment.13-15 Compared to conventional nanoparticle synthesis, which requires different steps for purification, immobilization and screening, our method greatly reduces input quantities of biomaterials, solvents and reagents, while simultaneously decreasing environmental concerns dealing with nanoparticle use and disposal.16 Importantly, nanoparticle functionalization reactions are robust to the presence of serum concentrations typically used for cell culture experiments, and even to extremely high serum concentrations. This feature can facilitate structure-activity relationships for nanoparticle design in the presence of serum (which can affect the protein corona and surface properties of the nanoparticle).17,18
In most SPR assays, where protein-small molecule interactions are studied, the protein (ligand) is immobilized through an affinity tag (GST, biotin, etc.) or through covalent amide linkages. The analyte is then passed over the ligand where binding results in surface mass variations, which are detected as refractive index changes. Reversing the canonical SPR orientation and immobilizing the small molecule in a convenient and reversible manner provides facile access to study direct binding of soluble macromolecular targets and clarifies phase-specific (immobilized vs. soluble) artifacts.19,20 Assay formats of this type are especially valuable for inhibition and competition studies.21 Reversal of orientation can also be advantageous under several circumstances, i.e. a) with analytes of low molecular weight, considering that binding signal is proportional to surface mass change b) aggregation and non-specificity of hydrophobic analytes22 is eliminated making data analysis straightforward c) characterization of high affinity binders can be challenging due to long residence times (slow off-rates, especially when dealing with covalent inhibitors)23 and the need for surface regeneration conditions in preparation for the next analysis cycle d) regeneration agents may have degrading effects on the structure or function of the immobilized protein.
By incorporating the bioorthogonal capture-cycloaddition method as part of our immobilization strategy, we circumvent some of the difficulties associated with conventional small molecule immobilization techniques. These require much higher ligand concentrations since electrostatic attractions leading to surface pre-concentrations typical for protein immobilization protocols are ineffective with small molecules. Accordingly, increasing concentrations of ligands leads to increasing ratios of organic co-solvent for ligand dissolution. Both of these conditions are not compatible with micro-fluidic flow systems and as a consequence, conventional immobilizations must be performed outside the instrument where real-time monitoring is not possible.24 Lastly, in the event of a successful conventional immobilization, covalent surface modification prevents surface regeneration and limits the experimental range of biosensor surfaces resulting in increased investments in time, effort and costs.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We acknowledge funding from NIH (NHLBI Contract No. HHSN268201000044C to R.W., S.H. and S.Y.S.).
Materials
| Reagent | |||
| Sensor Chip CM5 | GE Healthcare | BR-1005-30 | |
| Amine coupling kit | GE Healthcare | BR-1000-50 | |
| GST capture kit | GE Healthcare | BR-1002-23 | |
| NAP-10 Columns | GE Healthcare | 17-0854-01 | |
| GST, lyophilized in 1X PBS | Genscript | Z02039 | 1 mg/ml |
| rhFKBP12 | R&D Systems | 3777-FK | |
| Surfactant P-20 | GE Healthcare | BR-1000-54 | |
| Glycine 2.0 | GE Healthcare | BR-1003-55 | |
| Zeba spin desalting column | Thermo | 89882 | 7 K MWCO |
| Amicon Ultra 4 | Fisher | UFC810096 | 100 K centrifugal filter |
| TCO-OH | Ref. 8 | Synthesized in-house | |
| TCO-NHS | Ref. 8 | Synthesized in-house, *Commercially available from Click Chemistry Tools # 1016-25 | |
| Tz-BnNH2 | Ref. 8 | Synthesized in-house | |
| Tz-NHS | Ref. 8 | 764701 | Synthesized in-house, *Commercially available from Sigma Aldrich # 764701 |
| NP-NH2 = CLIO-NH2 | Ref. 8 | Synthesized in-house | |
| AP1497, AP1497-Tz | Ref. 8 | Synthesized in-house | |
| Equipment | |||
| SPR Biosensor | GE Healthcare | Biacore T100 |
References
- Devaraj, N. K., Upadhyay, R., Haun, J. B., Hilderbrand, S. A., Weissleder, R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew Chem Int Ed Engl. 48, 7013-7016 (2009).
- Seitchik, J. L., et al. Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. J Am Chem Soc. 134, 2898-2901 (2012).
- Haun, J. B., Devaraj, N. K., Marinelli, B. S., Lee, H., Weissleder, R. Probing intracellular biomarkers and mediators of cell activation using nanosensors and bioorthogonal chemistry. ACS Nano. 5, 3204-3213 (2011).
- Budin, G., Yang, K. S., Reiner, T., Weissleder, R. Bioorthogonal probes for polo-like kinase 1 imaging and quantification. Angew Chem Int Ed Engl. 50, 9378-9381 (2011).
- Liu, D. S., Tangpeerachaikul, A., Selvaraj, R., Taylor, M. T., Fox, J. M., Ting, A. Y. Diels-Alder cycloaddition for fluorophore targeting to specific proteins inside living cells. J Am Chem Soc. 134, 792-795 (2012).
- Haun, J. B., Devaraj, N. K., Hilderbrand, S. A., Lee, H., Weissleder, R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 5, 660-665 (2010).
- Liong, M., et al. Specific pathogen detection using bioorthogonal chemistry and diagnostic magnetic resonance. Bioconjug Chem. 22, 2390-2394 (2011).
- Tassa, C., et al. On-chip bioorthogonal chemistry enables immobilization of in situ modified nanoparticles and small molecules for label-free monitoring of protein binding and reaction kinetics. Lab Chip. 12, 3103-3110 (2012).
- Pol, E. The importance of correct protein concentration for kinetics and affinity determination in structure-function analysis. J Vis Exp. (37), e1746 (2010).
- MacBeath, G., Schreiber, S. L. Printing proteins as microarrays for high-throughput function determination. Science. 289, 1760-1763 (2000).
- Ong, S. E., et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 106, 4617-4622 (2009).
- Tassa, C., et al. Binding affinity and kinetic analysis of targeted small molecule-modified nanoparticles. Bioconjug Chem. 21, 14-19 (2010).
- Weissleder, R., Kelly, K., Sun, E. Y., Shtatland, T., Josephson, L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 23, 1418-1423 (2005).
- Yuan, J., Oliver, R., Aguilar, M. I., Wu, Y. Surface plasmon resonance assay for chloramphenicol. Anal Chem. 80, 8329-8333 (2008).
- Myung, J. H., Gajjar, K. A., Saric, J., Eddington, D. T., Hong, S. Dendrimer-mediated multivalent binding for the enhanced capture of tumor cells. Angew Chem Int Ed Engl. 50, 11769-11772 (2011).
- Keelan, J. A. Nanotoxicology: nanoparticles versus the placenta. Nat Nanotechnol. 6, 263-264 (2011).
- Lundqvist, M., Stigler, J., Elia, G., Lynch, I., Cedervall, T., Dawson, K. A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci U S A. 105, 14265-14270 (2008).
- Lundqvist, M., et al. The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 5, 7503-7509 (2011).
- Mammen, M., Choi, S. -. K., Whitesides, G. M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed Engl. 37, 2754-2899 (1998).
- Kausaite-Minkstimiene, A., Ramanaviciene, A., Kirlyte, J., Ramanavicius, A. Comparative study of random and oriented antibody immobilization techniques on the binding capacity of immunosensor. Anal Chem. 82, 6401-6408 (2010).
- Arkin, M. R., Wells, J. A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 3, 301-317 (2004).
- Giannetti, A. M., Koch, B. D., Browner, M. F. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J Med Chem. 51, 574-580 (2008).
- Kimple, A. J., Willard, F. S., Giguere, P. M., Johnston, C. A., Mocanu, V., Siderovski, D. P. The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Galpha-interaction face. Biochim Biophys Acta. 1774, 1213-1220 (2007).
- GE Healthcare. . Biacore Sensor Surface Handbook BR-1005-71. , (2005).

