Analyzing the Effects of Stromal Cells on the Recruitment of Leukocytes from Flow
Summary
The ability of inflamed endothelium to recruit leukocytes from flow is regulated by mesenchymal stromal cells. We describe two in vitro models incorporating primary human cells that can be used to assess neutrophil recruitment from flow and examine the role that mesenchymal stromal cells play in regulating this process.
Abstract
Stromal cells regulate the recruitment of circulating leukocytes during inflammation through cross-talk with neighboring endothelial cells. Here we describe two in vitro “vascular” models for studying the recruitment of circulating neutrophils from flow by inflamed endothelial cells. A major advantage of these models is the ability to analyze each step in the leukocyte adhesion cascade in order, as would occur in vivo. We also describe how both models can be adapted to study the role of stromal cells, in this case mesenchymal stem cells (MSC), in regulating leukocyte recruitment.
Primary endothelial cells were cultured alone or together with human MSC in direct contact on Ibidi microslides or on opposite sides of a Transwell filter for 24 hr. Cultures were stimulated with tumor necrosis factor alpha (TNFα) for 4 hr and incorporated into a flow-based adhesion assay. A bolus of neutrophils was perfused over the endothelium for 4 min. The capture of flowing neutrophils and their interactions with the endothelium was visualized by phase-contrast microscopy.
In both models, cytokine-stimulation increased endothelial recruitment of flowing neutrophils in a dose-dependent manner. Analysis of the behavior of recruited neutrophils showed a dose-dependent decrease in rolling and a dose-dependent increase in transmigration through the endothelium. In co-culture, MSC suppressed neutrophil adhesion to TNFα-stimulated endothelium.
Our flow based-adhesion models mimic the initial phases of leukocyte recruitment from the circulation. In addition to leukocytes, they can be used to examine the recruitment of other cell types, such as therapeutically administered MSC or circulating tumor cells. Our multi-layered co-culture models have shown that MSC communicate with endothelium to modify their response to pro-inflammatory cytokines, altering the recruitment of neutrophils. Further research using such models is required to fully understand how stromal cells from different tissues and conditions (inflammatory disorders or cancer) influence the recruitment of leukocytes during inflammation.
Introduction
Inflammation is a protective response to microbial infection or tissue injury that requires tight regulation of leukocyte entry into and exit from the inflamed tissue to allow resolution1,2. Cross-talk between endothelial cells (EC) that line blood vessels, circulating leukocytes and tissue-resident stromal cells is essential for coordinating this process3. However, uncontrolled recruitment of leukocytes and their ineffective clearance underpin the development of chronic inflammatory diseases4. Our current understanding of leukocyte recruitment in health and disease is incomplete and more robust models are needed to analyze this process.
The mechanisms supporting the recruitment of leukocytes from blood through vascular EC in post-capillary venules have been well described1,2,5. Circulating leukocytes are captured by specialized receptors (e.g., VCAM-1, E-selectin, P-selectin) which are up-regulated on inflamed endothelium. These transient interactions allow leukocytes to interact with surface bound chemokines and lipid-derived mediators (either endothelial or stromal in origin) that activate integrins expressed by leukocytes6-11. This in turn stabilizes adhesion and drives migration across the endothelium and into the tissue12-15. Within tissue, recruited leukocytes are subjected to stromal-derived agents that influence their motility, function and survival16,17. Growing evidence strongly suggests that signals received at each stage of the recruitment process conditions leukocytes for the next. However, our understanding of leukocyte recruitment remains incomplete and very little is known about the components shaping leukocyte movement within tissue.
In Birmingham we have developed several in vitro “vascular” models to study the recruitment of leukocytes from flow9,18,19. We now understand that vascular EC act as immediate regulators of leukocyte recruitment responding to changes in their local microenvironment. Specifically, tissue-resident stromal cells can actively regulate the inflammatory response, in part by conversing with neighboring vascular EC to influence their role in recruitment3. We have previously shown that various stromal cells modulate the ability of EC to support adhesion and migration of leukocytes in a tissue-specific manner, and that these effects become altered in chronic diseases13,16,20,21. Thus, stromal cells establish tissue ‘address-codes’ that define the context of each inflammatory response22. More recently, we have demonstrated that bone-marrow derived MSC (BMMSC) potently down-regulate the response of EC to cytokines, leading to a reduction in the recruitment of both neutrophils and lymphocytes23.
The mechanisms governing recruitment elucidated in vitro have largely used assays incorporating a single cell type (e.g., EC) or protein in isolation. However, these studies do not take into consideration the effects of the local tissue environment (i.e., the presence of stromal cells) on recruitment of leukocytes and their subsequent migration into the tissue. Here we describe two flow-based methods in which stromal cells, specifically mesenchymal stem cells (MSC), are co-cultured with EC23. Such models allow us to examine the effect of stromal cell on endothelial responses, in particular their ability to support leukocyte recruitment from flow.
Protocol
1. Isolation and Culture of Primary Human Endothelial Cells and Mesenchymal Stem Cells
- Isolation and culture of human umbilical vein endothelial cells (HUVEC):
- Put umbilical cord on a tray with paper towels and spray with 70% ethanol. Place in a tissue culture hood. Identify the vein and cannulate at both ends. Place a cable tie around the cannulated end to secure it.
- Wash out venous blood with PBS using a syringe. Fill syringe with air and pass through the vein to remove and discard the residual PBS.
- Thaw 10 mg/ml collagenase type Ia and dilute 1:10 in PBS (with calcium and magnesium chloride) to a final concentration of 1 mg/ml. Pass collagenase solution into the vein until both cannulae are filled. Close the clamps on cannulae at both ends.
- Cover the tray with tissue and spray with 70% ethanol. Place the cord into an incubator for 20 min at 37 °C and 5% CO2.
- Take the cord out of the incubator and tighten cable ties. Massage the cord gently for 1 min. Flush out the cell suspension with 10 ml PBS and collect in a 50 ml centrifuge tube.
- Fill syringe with air and pass through the vein to remove any residual PBS twice, collecting the PBS in a 50 ml centrifuge tube used in step 1.1.5. Centrifuge at 400 x g for 5 min at RT.
- Aspirate supernatant and resuspend pellet in 1 ml complete EC medium. EC medium consists of M199 supplemented with 35 µg/ml gentamicin sulphate, 10 ng/ml human epidermal growth factor, 1 µg/ml hydrocortisone, 2.5 μg/ml amphotericin B, and 20% fetal calf serum.
- Add 4 ml of EC medium and the EC suspension to a 25 cm2 flask. Change the medium on the following day and then every 2 days.
- EC exhibit a cobblestone-like morphology (Figure 1Ai). For adhesion assays, EC are generally seeded when they reach 100% confluence (see Section 1.4).
- Isolation of Wharton’s jelly mesenchymal stem cells (WJMSC) from human umbilical cords:
- Isolate Wharton’s jelly-derived MSC (WJMSC) from fresh umbilical cords or from cords that have already been used to isolate EC. Cut umbilical cord into 5 cm long pieces. Cut each piece longitudinally to reveal the blood vessels (2 arteries [white, rigid] and 1 vein [yellow, distended]).
- Using sterile scissors and forceps remove the blood vessels and discard. Cut all the tissue into 2 – 3 mm3 pieces. Using forceps place 2 – 3 mm3 pieces into a 50 ml centrifuge tube.
- Thaw 100 mg/ml stock collagenase type II and dilute 1:100 in 10 ml PBS to a final concentration of 1 mg/ml. Thaw 20,000 U/ml stock hyaluronidase and dilute 1:400 to a final concentration of 50 U/ml in the collagenase solution.
- Add enzymatic cocktail to the centrifuge tube containing the tissue fragments. Incubate the tissue fragments for 5 hr at 37 °C on a slow rotator.
- Dilute the cell suspension 1:5 in PBS. Place a 100 µm pore filter into a new 50 ml centrifuge tube. Pour the cell suspension onto the 100 µm pore filter.
NOTE: Remaining tissue fragments will be retained on the filter and cells will be collected in the 50ml centrifuge tube. - Discard the filter. Centrifuge the cell suspension at 400 x g for 10 min at RT. Aspirate supernatant and resuspend WJMSC pellet in 12 ml complete WJMSC culture medium (DMEM low glucose, 10% FCS and 100 U/ml penicillin + 100 µg/ml streptomycin mix).
- Seed all cells in a 75 cm2 tissue culture flask. Change the medium after 24 hr with 12 ml complete WJMSC culture medium. Replace medium every 2 – 3 days. Cells should reach 70 – 80% confluence within 2 weeks. Passage when WJMSC reach 70 – 80% confluence (see Section 1.4).
- Expansion of bone marrow-derived MSC (BMMSC):
- Isolate human bone marrow-derived MSC (BMMSC) as previously described24. Add 10 ml pre-warmed MSC growth medium (MSCGM) into a 15 ml centrifuge tube.
- Thaw a vial of p2 BMMSC by placing in a 37 °C water bath for 2 min. Add the BMMSC suspension to the centrifuge tube containing MSCGM. Mix well by pipetting.
- Centrifuge at 400 x g for 5 min at RT. Aspirate supernatant completely.
- Resuspend cells in 1 ml MSCGM and count the cells using a haemocytometer or a digital cell counter such as a cellometer.
- Seed cells into 75 cm2 culture flasks at a density of 5,000 – 6,000 cells per cm2 in 12 ml MSCGM. Change the medium after 24 hr with 12 ml MSCGM. Feed cells with 12 ml MSCGM every 2 – 3 days.
- Passage when BMMSC reach 70 – 80% confluence (see Section 1.4). Cells exhibit a fibroblastic morphology (Figure 1Aii).
- Detachment of EC and MSC:
- Aspirate medium from 25 cm2 culture flasks. Add 2 ml of 0.02% EDTA for approximately 2 min. Aspirate EDTA and add 2 ml trypsin (2.5 mg/ml). View under the microscope until the cells become round.
- Tap the flask to detach the cells. Inactivate the trypsin by adding 8 ml culture medium (dependent on cell type; EC medium for HUVEC, DMEM LG for WJMSC and MSCGM for BMMSC) to the culture flask and transfer suspension to a 15 ml centrifuge tube.
- Centrifuge at 400 x g for 5 min at RT. Aspirate supernatant and resuspend the pellet as described below.
- For passaging of MSC, resuspend pellet in 3 ml culture medium. Add 11 ml culture medium into three separate 75 cm2 culture flasks. Add 1 ml cell suspension to each culture flask (1:3 split). Passage WJMSC and BMMSC 3 times (p3) before use in co-culture assays.
- For seeding EC or MSC in adhesion assays – see Sections 2 and 3.
- Freezing MSC:
- At passage 3 detach MSC as described in Section 1.4. Aspirate supernatant and resuspend in 3 ml ice-cold CryoSFM. Pipette 1 ml aliquots of cell suspension into 1.5 ml ice-cold cryovials. Put cryovials in a freezing container.
- Store the container at -80 °C O/N. Transfer to liquid nitrogen. Thaw vial of MSC (follow steps 1.3.1 – 1.3.6). Resuspend MSC in 5 ml culture medium (choose appropriate medium for WJMSC or BMMSC) and seed into a 25 cm2 flask.
2. Establishing Endothelial-mesenchymal Stem Cell Co-cultures on Ibidi Microslides
- Trypsinize a confluent 25 cm2 flask of EC (~1.5 x 106 cells; as Section 1.4). Resuspend EC in 380 µl MSCGM (1 x 25 cm2 flask will seed two 6-channel Ibidi microslides (~1.25 x 105/channel), for adhesion assays all cell types are cultured in MSCGM). Add 30 µl of EC suspension to each channel (this will cover the growth area through capillary action). Incubate Ibidi microslide at 37 °C and 5% CO2 for 1 hr.
- Add 140 µl MSCGM to each channel and then aspirate it off. Repeat twice for a total of 3 washes. Add 140 µl MSCGM and place in the incubator at 37 °C and 5% CO2 for 24 hr.
- For co-cultures detach MSC (Section 1.4). Perform a cell count using a haemocytometer or a cellometer. Adjust the concentration of MSC to 1.5 x 105 cells/ml.
- Aspirate excess medium from the Ibidi channels (leaving only the growth area of the channel in medium). Add 30µl of MSC suspension to the Ibidi channels. Aspirate the medium that was ejected from the growth area of the channel and add another 30 µl of MSC suspension. Repeat once more and then place the Ibidi microslide in the incubator at 37 °C and 5% CO2 for 1 hr.
- Add 140 µl MSCGM to each channel and then aspirate it off. Repeat twice for a total of 3 washes. Add a final volume of 140 µl MSCGM to each channel and place in the incubator at 37 °C and 5% CO2 for 24 hr.
- Thaw 1 x 105 U/ml stock TNFα and dilute 1:1,000 in MSCGM to a final concentration of 100 U/ml (equivalent to ~10 ng/ml). Perform a serial dilution by diluting 100 U/ml TNFα by 1:10 in MSCGM to obtain 10 U/ml. Dilute 10 U/ml TNFα by 1:10 in MSCGM to obtain 1 U/ml.
- Treat channels with TNFα at 37 °C for 4 hr prior to the assay. Temperature fluctuations during the cytokine treatment will alter the patterns of neutrophil recruitment observed. Add fresh MSCGM to untreated channels.
3. Establishing Endothelial-Mesenchymal Stem Cell Co-cultures on Filters
- Detach WJ or BMMSC as described in Section 1.4. Resuspend the pellet in 1 ml MSCGM. Perform a cell count cells using a haemocytometer or a cellometer.
- Adjust volume so that the final concentration is 5 x 105 MSC in 500 µl of MSCGM.
- Using sterile forceps invert 6-well, 0.4 µm PET Transwell filters and place in a sterile box.
- Seed 5 x 105 MSC onto the outer surface of the filters. Incubate filters at 37 °C and 5% CO2 for 1 hr.
- Collect media from the outer surface of the filter. Count the number of non-adherent MSC in the media. Re-invert filters using sterile forceps and place in a matching 6-well plate containing 3 ml MSCGM.
- Add 2 ml of MSCGM onto the inner surface of the filter (Figure 1B). Place in an incubator at 37 °C and 5% CO2 for 24 hr. Trypsinize a confluent 25 cm2 flask of EC (Figure 1Ai; as Section 1.4).
- Resuspend EC in 8 ml MSCGM (1 x 25cm2 flask will seed four 6-well filters; ~5 x 105 EC/filter). Aspirate medium from the top and bottom of the porous filters. Add 3 ml fresh MSCGM into the lower chamber (underneath the filter). Add 2 ml EC suspension to the inner surface of each filter. Incubate for 1 hr at 37 °C and 5% CO2.
- Aspirate medium to wash off non-adherent EC and replace with fresh MSCGM. Set up parallel EC mono-culture filters by seeding cells on the inner surface without first seeding MSC. Incubate O/N at 37 °C and 5% CO2.
- Check that the EC monolayer is confluent and contains no gaps. Sub-confluent monolayers cannot be used for adhesion assays (Figure 1B).
- Treat the upper and lower chambers of the filters with 1, 10, or 100 U/ml TNFα (equivalent to ~10 ng/ml) at 37 °C for 4 hr prior to the assay (described in Section 2).
4. Isolation of Leukocytes
- Take venous blood from healthy volunteers and aliquot immediately into EDTA tubes. Invert tubes gently to mix.
- Layer 2.5 ml Histopaque 1077 onto 2.5 ml Histopaque 1119 in a 10 ml round bottomed tube. Layer 5 ml whole blood onto the Histopaque gradient. Centrifuge at 800 x g for 40 min at RT.
- Harvest peripheral polymorphonuclear neutrophils (PMN) at the interface of Histopaque 1077 and 1119 (above the erythrocyte layer). Place in a 10 ml round bottomed tube and make up to 10 ml with PBSA. Gently invert tube and centrifuge at 400 x g for 5 min at RT.
- Dilute 7.5% BSA solution 1:50 in 100 ml PBS (with calcium and magnesium chloride) to a final concentration of 0.15% (w/v; PBSA). Aspirate supernatant and resuspend in 10 ml PBSA. Centrifuge at 400 x g for 5 min at RT.
- Resuspend in 1 ml PBSA. Take a 20 µl aliquot of the cell suspension and add to 380 µl PBSA (1:20 dilution). Count cells using a haemocytometer or a cellometer. Dilute to the required concentration (1 x 106/ml for Transwell filters and Ibidi microslide) in PBSA. Maintain neutrophil suspension at RT until the assay.
5. Assembling the Flow System
- Set up the flow system as shown in Figure 1C. Turn on the heater and set to 37 °C. Attach a 20 ml syringe (remove the plunger) and a 5 ml syringe to a 3-way tap. Attach the tap to the Perspex chamber using Micropore tape.
- Measure and cut a long piece of silicon 2/4 mm (thick) tubing that is approximately the distance between the valve and the 3-way tap. Cut an 8 – 10 mm long piece of 1/3 mm (thin) tubing and insert into one end of the thick tubing. Attach the thick tubing side onto the 3-way tap.
- Connect the thin tubing end onto a port of the electronic 3-way microvalve. This is the “wash reservoir”. Cut a 6 – 8 mm long piece of thick and thin tubing.
- Insert the thin tubing into one end of the thick tubing. Attach the thick tubing end onto a 2 ml syringe (remove the plunger).
- Connect the thin tubing end of the 2 ml syringe onto a port on the electronic microvalve to make the “sample reservoir”. Connect the valve to the filter flow chamber by measuring and cutting a long piece of thin tubing that is the distance between the microvalve and the center of the microscope stage.
- For the microslide model, attach an 8 – 10 mm piece of thick tubing to the end of the thin tubing. Place an L shaped connector to the end of the thick tubing. This will connect to the microslide. For the filter model, attach an 8 – 10 mm piece of Portex Blue Line Manometer connecting tubing to the end of the thin tubing.
- Place the thin tubing end onto the microvalve. This is the common output for the wash and sample reservoirs. Fill reservoirs with PBSA. Prime tubing by flowing PBSA through them to remove any air bubbles.
- Attach Manometer tubing to a 29 mm (50 ml) glass syringe. Prime the syringe by filling with 10 ml PBSA. Invert the syringe so that the end connected to the tubing faces upwards and push out all air bubbles. Refill with 5 ml PBSA.
- For the microslide model only, attach a 10 – 12 mm piece of thick tubing to the end of the Manometer tubing leading to the glass syringe. Place an L shaped connector onto the end of the thick tubing. Place the glass syringe into a syringe pump for infusion/withdrawal.
- Calculate the refill flow rate (Q) required to generate the desired wall shear stress (τw in Pascal, Pa) of 0.1 Pa (Transwell filters) or 0.05 Pa (Ibidi microslides) using the following formulae:
γw = (6.Q) / (w.h2)
τ = n.γ
Where w = internal width and h = internal depth of the flow channel. n = viscosity of the flowing solution; PBSA is n = 0.7 mPa.s. For the parallel plate filter flow chamber, the width (w) is 4 mm and the depth (h) is 0.133 mm. The depth of the chamber can vary slightly due to differences in the thickness of the Parafilm gasket used. For the Ibidi microslide, the width is 3.8 mm and the depth is 0.4 mm.
NOTE: Due to differences in the dimensions of the flow channel, and capture dynamics we use different shear stresses for the microslide model compared to the filter model6.
6. Setting Up the Parallel Plate Flow Chamber Incorporating Filters
- Cut a piece of Parafilm (the same size as the glass coverslip) using a metal template. Cut out a 20 x 4 mm slot in the Parafilm (to create the flow channel) using a metal template. Use the gasket to mark the flow channel on the glass coverslip.
- Align the edges of the 6-well filter on the glass coverslip. Ensure that the filter covers the flow channel markings. Carefully cut out the filter using a type 10A scalpel.
- Carefully cover the filter with a Parafilm gasket, ensuring that the flow channel slot is in the middle of the filter (Figure 1D). Use a piece of clean tissue and push out any bubbles.
- Place the coverslip in the recess of the bottom plate of the Perspex flow chamber. Position the top Perspex plate over the top of the gasket and screw the plates together (Figure 1D).
- Turn the 3-way tap to allow wash buffer (PBSA) to flow through the valve. Connect Manometer tubing to the inlet port of the top Persex plate. Run PBSA through the flow channel to allow bubbles to pass through.
- Connect the Manometer tubing from the syringe pump into the outlet port of the Perspex plate. Set the syringe pump to refill and press run. Clean any PBSA that has dripped onto the upper plate of the chamber.
- Place the chamber on the stage of an invert phase-contrast microscope. Adjust focus to visualize the EC above the filters (Figure 1B).
7. Setting Up Microslides for Flow
- Place the microslide onto the stage of an invert phase contrast microscope. Connect the L shaped connector into the inlet port of a channel. Run PBSA through the flow channel.
- Place the L shaped connector from the syringe pump into the outlet port of the channel. Set the syringe pump to refill and press run. Clean any PBSA that has dripped onto the microslide. Adjust focus to visualize the EC monolayer.
8. Perfusion of Leukocytes Over Endothelial Cells
- Put 2 ml of purified neutrophils into the sample reservoir and leave to warm for 2 min. Wash the endothelium with PBSA for 2 min.
- Turn the valve ON to perfuse neutrophils over endothelium. Deliver the neutrophil bolus for 4 min. Turn the valve OFF and perfuse PBSA from the wash reservoir for the remainder of the experiment.
- Ensure that air bubbles do not pass through the flow channel at any point during the assay as this will disrupt the EC monolayer and cause detachment of adherent neutrophils.
9. Recording Neutrophil Capture and Behavior
- Record neutrophil recruitment either during neutrophil flow or post-perfusion.
- Make all digital recordings of at least 5 – 10 fields in the center of the flow channel. Identify the center of the channel by moving the objective to the edge of the channel at the inlet port and identifying the middle of the port.
- For recording during neutrophil flow, take images of a single field every 10 sec for 1 min. Move along the channel and record another field for 1 min. Repeat for duration of bolus.
- For recording post-perfusion, make 10 sec recording of 5 – 10 fields down the center of the flow channel for assessing leukocyte behavior (typically 2 min after the end of the neutrophil bolus). Take images every second within the 10 sec interval. This allows sufficient time for capture from flow and the behavior of adherent neutrophils to be analyzed.
- Record a single field containing at least 10 transmigrated neutrophils for 5 min, taking images every 30 sec. This can be used to calculate the velocity of migrated cells (either above or underneath the endothelium).
- Record another series of 10 sec fields (typically 9 min post-perfusion). This allows neutrophils time to migrate through the endothelial monolayer.
- Stop the syringe pump and remove the tubing. Disassemble the flow chamber and rinse the sample reservoir and tubing. Repeat for subsequent filters/microslide channels.
10. Analysis of Leukocyte Recruitment and Behavior
- Count the number of neutrophils in each field during the 10 sec recordings at the 2 min time point. All cells must be present in the first second field in order to be counted. Cells that are partially in the field on 2 sides (e.g., top/right hand border) of the field of view are included in the counts, as long as they remain in the field for the full 10 sec.
- Calculate the mean number of adherent neutrophil per field. Measure the length and width of the recorded field. Calculate the area of the field. Calculate how many fields there are in 1 mm2. Multiply the mean neutrophil count by the number of fields in 1 mm2.
- Calculate the total number of neutrophils perfused by multiplying the amount of neutrophils perfused (e.g., 2 x 106/ml*Q [e.g., 0.0999 ml/min for parallel plate flow chamber]) by the duration of the bolus (e.g., 4 min).
- Divide the neutrophil count/mm2 by the total number of neutrophils perfused to determine the total number of cells that have adhered (adherent cells/mm2 / 106 perfused).
- Assess whether the adherent neutrophils are rolling, firmly adherent or transmigrated (Supplementary Video 1 and 2).
- A rolling neutrophil is phase bright and will slowly move along the endothelial monolayer (1 – 10 µm/sec; Supplementary Video 1).
- Firmly adherent cells are phase bright and bound to the EC surface, either remaining stationary (i.e., not moving during recording) or have undergone shape change and are migrating over the EC surface (Figure 1Ei).
- A transmigrated neutrophil is phase dark and below the EC layer (Figure 1Eii).
- Calculate the percentage of adherent cells that are rolling, stationary and transmigrated. Alternatively, neutrophil behavior can be expressed as total cell numbers that exhibit the different behaviors by applying the same formula used to calculate total adhesion (as described in steps 10.6 – 10.7).
- Mark the leading edge of a rolling neutrophil. Mark the leading edge of the same cell at the end of the 10 sec sequence. Draw a line between the 2 points and measure the distance that the cell has travelled.
- Divide this value by the duration of the recording in which the cell is rolling (i.e., 10 sec). Try and select neutrophils that are in the field for the entire 10 sec interval.
- To calculate the velocity of neutrophils migrating over the surface (shaped changed phase bright) or underneath the endothelium (phase dark) use the 5 min recording (Supplementary Video 2).
- Draw an outline of migrated cells at the beginning of the sequence and track their movements throughout the sequence. Make note of the X and Y positions of the centroid at each 1 min interval for each cell. Subtract values of the X and Y position from the first image of the sequence from the values in the second image. This is based on Pythagoras’ theorem.
- Subtract the values from the second image from the third image. Do this for all images in the sequence. Square the X and Y values and add them together.
- Square root the resulting value. Calculate the velocity for each cell by averaging the velocities calculated at each minute interval. Track 10 migrated neutrophils and calculate the mean velocity.
Representative Results
Initially, we analyzed the effect of stimulating EC with TNFα on the recruitment of neutrophils from flow using the Ibidi microslide model (Section 7 – 9). In the absence of TNFα, little if any neutrophils adhered to the endothelial monolayer (Figure 2A). This was expected, as untreated/resting EC do not express the necessary adhesion molecules (selectins) or chemokines to support binding25,26. In contrast, cytokine-stimulation significantly increased neutrophil adhesion to the endothelium in a dose-dependent manner (Figure 2A). Adhesion normally remains stable over the course of the assay. Binding of leukocytes to untreated endothelium indicates that either the EC are activated (i.e., contaminated with LPS during the culture process) and/or the neutrophils were activated during the isolation process. Indeed, LPS has been shown to increase the expression of E-selectin, ICAM-1 and VCAM-125-27 on the surface of EC, allowing them to bind neutrophils.
When analyzing the behavior of the recruited neutrophils we typically observe a dose-dependent decrease in the percentage of neutrophils rolling (Figure 2B) with a concomitant dose dependent increase in the percentage of neutrophils migrating through the endothelial monolayer (Figure 2C). At the lower doses of TNFα-stimulation (1 U/ml) a larger proportion of neutrophils appear phase bright indicating that they are attached to the apical surface of the endothelium (Figure 2B). In contrast, at 10 and 100 U/ml (higher doses) approximately 40% of the recruited neutrophils appear phase dark at 2 min indicating that these cells have migrated through the endothelial monolayer and are underneath the endothelium (Figure 2C). Neutrophils are able to migrate through the EC within 1 – 2 min, with transmigration reaching maximal levels at ~10min post-perfusion28. Here we observed an increase in neutrophil transmigration from 40% at 2 min to 60% by 9 min post perfusion (Figure 2C). We observed no effect of TNFα concentration on the velocity of rolling (~3 µm/sec) or migrating (~10 – 12 µm/min) neutrophils.
In the filter based model, TNFα-stimulation increased neutrophil adhesion in a dose-dependent manner similar to that seen using the Ibidi microslide model (Figure 3A). In terms of behavior, neutrophil rolling was unaffected by TNFα dose (Figure 3B), whilst a dose-dependent increase in percentage transmigration was observed (Figure 3C). In this series of experiments we observed no significant effect of time on neutrophil transmigration (Figure 3C).
We provide methods on how to generate two different co-culture constructs, each of which is devised to answer specific questions. In the Ibidi microslide model, EC and MSC are cultured in a single monolayer in direct contact with one another. This model is useful for examining the effect of therapeutic injection of MSC into the blood and their subsequent integration into the EC monolayer. In contrast in the filter-based model, EC and MSC are cultured on opposite sides of the filter in close proximity but not necessarily in direct contact. This more closely resembles tissue, with endothelial cells forming a monolayer representing the blood vessel, and MSC residing in the subendothelial compartment. This allows us to examine the effects of tissue-resident MSC on the response of EC to cytokine stimulation.
Based on our experiences, we observe that maximal neutrophil recruitment and transendothelial migration occurs when EC are stimulated with 100 U/ml TNFα. As such, we have used this concentration to examine the effect of MSC co-culture on endothelial recruitment of neutrophils from flow. Here, we present data for BMMSC in co-culture with EC using the microslide and filter-based models however, other types of MSC can also be examined e.g., WJMSC. In both models the presence of BMMSC significantly reduced neutrophil adhesion to the EC when compared to EC cultured alone (Figure 4A). Co-culture had no effect on the behavior of recruited neutrophils, with similar levels of rolling and transmigration observed on EC cultured alone or with MSC (Figure 4B and C). Thus, MSC can modify the EC response to cytokine stimulation, which suppresses their ability to support neutrophil recruitment from flow.
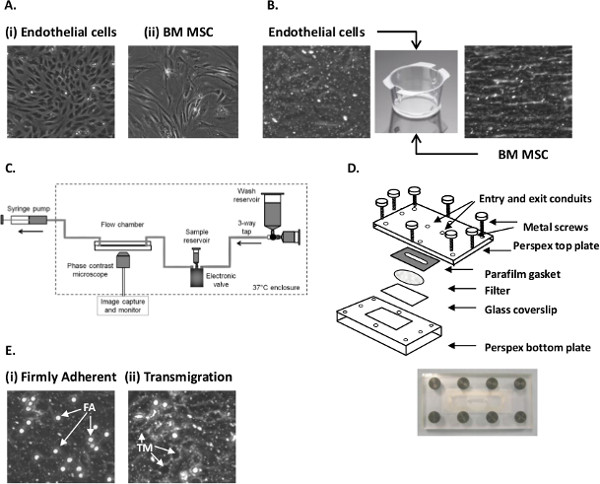
Figure 1. Establishing EC-MSC co-culture and analyzing neutrophil recruitment using a flow-based adhesion assay. (A) Micrograph of (i) primary EC and (ii) passage 3 BMMSC grown on tissue culture flasks. (B) Micrograph of EC and MSC cultured on 6-well Transwell filter inserts. (C) Diagram of the perfusion system used to generate flow. (D) Schematic representation of the parallel plate filter flow chamber. (E) Micrograph of (i) firmly adherent (FA) and (ii) transmigrated (TM) neutrophils following recruitment from flow to EC stimulated with 100 U/ml TNFα. Images C and D are taken from Figures 2 and 3 in Methods in Molecular Biology: T-cell Trafficking, 2010, pg 53-5428 with kind permission from Springer Science and Business Media. Please click here to view a larger version of this figure.
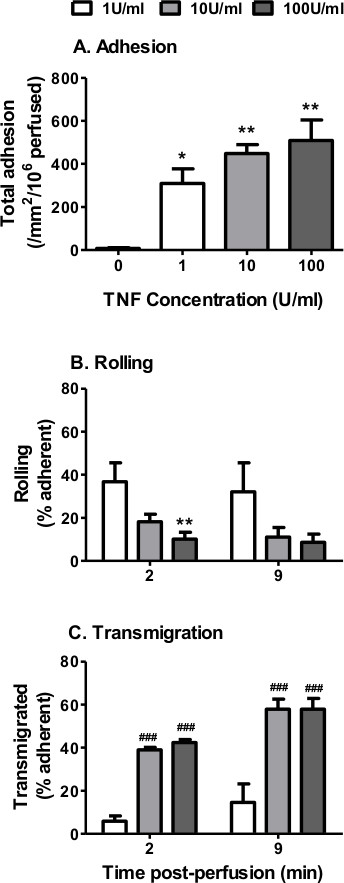
Figure 2. Neutrophil recruitment from flow to TNFα-stimulated EC using Ibidi microslides. EC were stimulated with increasing concentrations of TNFα (0 – 100 U/ml) for 4 hr. A 4 min bolus of neutrophils was perfused over the EC monolayer at 0.05 Pa. (A) Neutrophil adhesion assessed at 2 min. ANOVA showed a significant effect of TNFα treatment on neutrophil adhesion, p < 0.01. Neutrophil behavior was assessed at 2 and 9 min and expressed as a percentage of adherent cells that were (B) rolling or (C) transmigrated. ANOVA showed a significant effect of TNFα treatment on the behavior of the adherent neutrophils, p < 0.001. In C, ANOVA showed a significant of time on transmigrated neutrophils p < 0.01. Data are mean ± SEM from n = 3 experiments. *p < 0.05 and **p < 0.01 compared to the unstimulated (0 U/ml) EC control by Dunnett post-test. ##p < 0.01 and ###p < 0.001 compared to the 1 U/ml EC at the same time point by Bonferroni post-test.
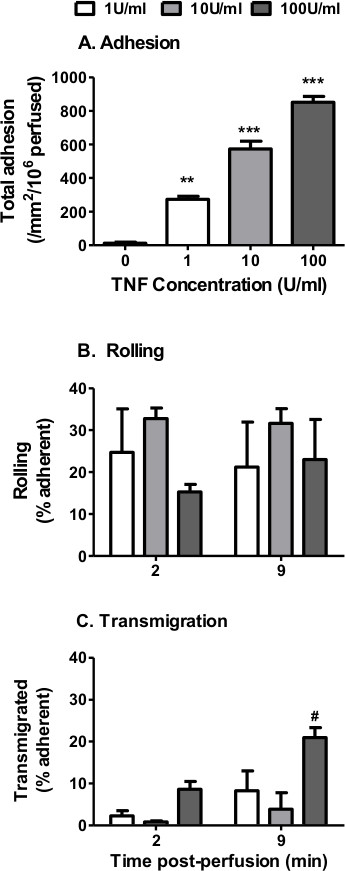
Figure 3. Neutrophil recruitment from flow to TNFα-stimulated EC using filter-based assay. EC were stimulated with increasing concentrations of TNFα (0 – 100 U/ml) for 4 hr. A 4 min bolus of neutrophils was perfused over the EC monolayer at 0.1 Pa. (A) Neutrophil adhesion assessed at 2 min. ANOVA showed a significant effect of TNFα treatment on neutrophil adhesion, p < 0.001. Neutrophil behavior was assessed at 2 and 9 min and expressed as a percentage of adherent cells that were (B) rolling or (C) transmigrated. In C, ANOVA showed a significant effect of time and cytokine treatment on neutrophil transmigration, p < 0.05. Data are mean ± SEM from n = 3 experiments. **p < 0.01 and ***p < 0.001 compared to the unstimulated (0 U/ml) EC control by Dunnett post-test. #p < 0.05 compared to the 1 U/ml EC at the same time point by Bonferroni post-test.
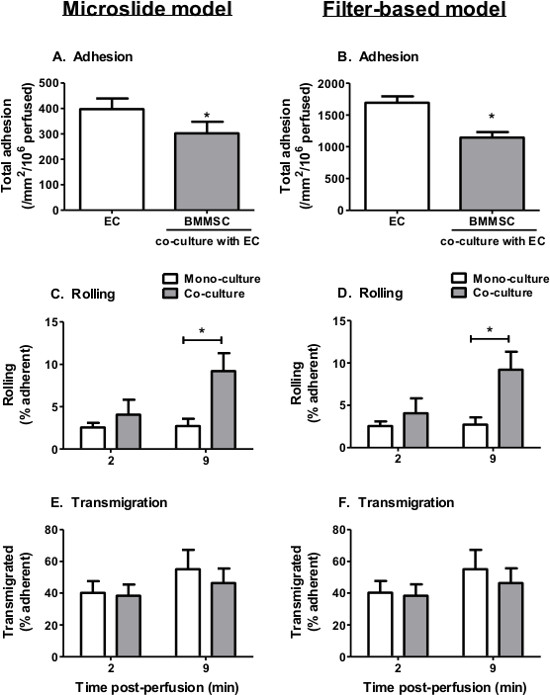
Figure 4. Neutrophil recruitment from flow to TNFα-stimulated EC-BMMSC co-cultures. BMMSC were co-cultured with EC for 24 hr prior to stimulation with 100 U/ml TNFα for 4 hr. A 4 min bolus of neutrophils was perfused over the EC monolayer at 0.05 Pa for (A, C, E) microslides and 0.1 Pa for (B, D, F) filters. (A-B) Neutrophil adhesion was assessed at 2 min. Neutrophil behavior was assessed at 2 and 9 min and expressed as a percentage of adherent cells that were (C-D) rolling or (E-F) transmigrated. In C and D, ANOVA showed a significant effect of culture conditions on neutrophil rolling, p < 0.05. In E and F, ANOVA showed a significant effect of time on neutrophil transmigration, p < 0.05. However, no significant differences were observed in transmigration between individual treatments by Bonferroni post-test. Data are mean ± SEM from n = 5 experiments. *p < 0.05 compared to the EC monoculture by paired t-test or Bonferroni post-test.
Supplementary Video 1. Analysis of neutrophil rolling velocities. EC cultured on a Transwell filter were stimulated with 100 U/ml TNFα for 4 hr. A bolus of neutrophils was perfused over the EC for 4min. Representative digitized sequence of a single 10 sec field taken 2 min post-perfusion. The change in position of a single rolling neutrophil from the beginning to the end of the 10 sec sequence can be used to calculate the velocity at which the neutrophil is rolling.
Supplementary Video 2. Analysis of neutrophil migration velocities. EC cultured on a Transwell filter were stimulated with 100 U/ml TNFα for 4 hr. A bolus of neutrophils was perfused over the EC for 4 min. Representative digitized sequence of a single 5 min field to track the movement of transmigrated neutrophils. This can be used to calculate the velocity.
Discussion
Here we describe two in vitro “vascular” models for studying the recruitment of circulating neutrophils by inflamed endothelium. A major advantage of these models is the ability to analyze each step in the leukocyte adhesion cascade in order, as would occur in vivo. We have previously observed a dose-dependent increase in neutrophil adhesion to and transmigration through TNFα-stimulated EC9,29. We also describe how both models can be adapted to study the effects of stromal cells on leukocyte recruitment. Here, MSC were co-cultured with EC in an Ibidi microslide or on opposite sides of a porous filter. This allows both cell types to communicate with one another, thereby modifying each other’s phenotype and response. We have shown here, and in previous studies23, that the presence of MSC suppressed neutrophil adhesion to TNFα-stimulated EC. This indicates that stromal cells modify the EC response to cytokines and subsequently alters the recruitment of circulating leukocytes.
In addition to the models described above, Cellix Biochips, Bioflux plates and Glyotech parallel plate flow chambers are commercially available flow channel systems that provide a surface for culturing endothelium and observing recruitment. In all systems, certain parameters should be considered whilst establishing endothelial cultures and performing the flow-based adhesion assays, some of which are highlighted below and in previous reports30,31. Any anti-inflammatory agents, such as hydrocortisone, that may affect the cytokine responses should be omitted from the medium for the duration of the culture and assay18,29. When using the Ibidi microslide ensure that there are no air bubbles present in the flow channel during the culture of the EC as these will disrupt the EC monolayer. The integrity of the endothelial monolayer should be confirmed prior to cytokine-treatment, as neutrophils will bind to the gaps in the monolayer where the BSA has coated the microslide/filter. It is also essential to ensure that EC are maintained at 37 °C throughout the cytokine-stimulation because TNFα is temperature sensitive and only has the maximal effect at 37 °C. For the flow assay itself, select an appropriate wash buffer, we typically use PBSA for short assays (less than 30 mins) and M199 medium supplemented with BSA (0.15%) for longer assays (1 – 48 hr)30,31. Finally ensure that there are no air bubbles present in the flow channel during the assay as this disrupts the flow rate, damages the EC monolayer and activates adherent neutrophils.
One of the major advantages of the multi-cellular in vitro models described here is their ability to replicate the in vivo interactions between EC and stromal cells. It is difficult to isolate the effects of specific stromal components in vivo and to modify them in a controlled manner. In our models, stromal cells can be manipulated to elucidate how they communicate with EC and influence the inflammatory process in health and disease. For example, using siRNA technology we have previously shown that production of IL-6 by MSC during co-culture was necessary for their immunosuppressive effects23. Each model can be used to address specific questions i.e., the effect of tissue-resident stromal cells (filter-based model) or therapeutically administered stromal cells (Ibidi microslides and other commercially available systems) on leukocyte recruitment. In both cases we have titrated different stromal cell types to ensure their viability and to establish a suitable ratio of stromal cell to EC for assessing effects on recruitment15,25. Similarly culture medium must be compatible with each cell type incorporated into the model. In our hands co-cultures are typically performed in the stromal cell medium11,18,23,31.
Using the filter-based model, we have previously shown that various stromal cells modulate the ability of EC to support adhesion and migration of leukocytes in a tissue-specific manner and that these effects become altered in chronic diseases3. This led to the concept that stromal cells establish tissue-specific “address codes” which actively regulate the recruitment of leukocytes to inflamed tissue24. These models are specifically designed to examine the initial stages of recruitment in great detail but are unable to study the subsequent migration within the subendothelial space (i.e., away from the endothelium into the tissue). Multi-cellular, multilayered 3D constructs such as a static collagen gel assay12,32 would be more appropriate for studying these latter phases of recruitment.
Our flow-based adhesion models are highly versatile. We have described their use in the context of neutrophil recruitment, but other leukocyte subsets can be investigated in a similar manner. We have also used the Ibidi microslide system to investigate the recruitment of circulating MSC by EC23, and whether this occurs through the same adhesion cascade reported for leukocytes. Likewise these models could be used to examine the recruitment and incorporation of metastatic tumor cell lines, and their subsequent effects on endothelial responses and leukocyte recruitment. Alternatively, the filter-based model could be adapted to incorporate different types of stromal cells (e.g., fibroblasts, podocytes, smooth muscle cells) from healthy tissues13,21 and sites of disease11,18,20. This would enable the study of tissue-specific regulatory pathways acting at the level of on EC and/or leukocytes. In all cases the disruption of normal regulatory processes in a range of disease conditions can be examined to identify key regulatory mediators (such as IL-6 and TGFβ) and potential new therapeutic targets. In the context of chronic inflammation these agents might be used to switch off the recruitment process, whilst in cancer biology one could imagine their use to turn on recruitment to target the tumor.
Divulgations
The authors have nothing to disclose.
Acknowledgements
Umbilical cords were collected with the assistance of the Birmingham Women’s Health Care NHS Trust. HMM was supported by an Arthritis Research UK Career Development Fellowship (19899) and Systems Science for Health, University of Birmingham (5212).
Materials
| Collagenase Type Ia | Sigma | C2674 | Dilute in 10ml PBS to get a final concentration of 10mg/ml. Store at -20°C in 1ml aliquots. |
| Dulbecco's PBS | Sigma | D8662 | With calcium and magnesium chloride. Keep sterile and store at room temperature. |
| 1X Medium M199 | Gibco | 31150-022 | Warm in 37 °C water bath before use. |
| Gentamicin sulphate | Sigma | G1397 | Store at 4°C. Add to M199 500ml bottle. |
| Human epidermal growth factor | Sigma | E9644 | Store at -20°C in 10µl aliquots. |
| Fetal calf serum (FCS) | Sigma | F9665 | FCS must be batch tested to ensure the growth and viability of isolated EC. Heat inactivate at 56°C. Store in 10ml aliquots at -20°C. |
| Amphotericin B | Gibco | 15290-026 | Potent and becomes toxic within a week so fresh complete HUVEC medium must be made up every week. Store at -20°C in 1ml aliquots. |
| Hydrocortisone | Sigma | H0135 | Stock is in ethanol. Store at -20°C in 10µl aliquots. |
| Collagenase Type II | Sigma | C6885 | Dilute stock in PBS to a final concentration of 100mg/ml. Store at -20°C in 100µl aliquots. |
| Hyaluronidase | Sigma | H3631 | Dilute stock in PBS to a final concentration of 20,000U/ml. Store at -20°C in 100µl aliquots. |
| 100µm cell strainer for 50ml centifuge tube | Scientific Lab Supplies (SLS) | 352360 | Other commercially available cell strainers (e.g. Greiner bio-one) can also be used. |
| DMEM low glucose | Biosera | LM-D1102/500 | Warm in 37 °C water bath before use. |
| Penicillin/Streptomycin mix | Sigma | P4333 | Store at -20°C in 1ml aliquots. |
| 25cm2 tissue culture flask | SLS | 353109 | |
| 75cm2 tissue culture flask | SLS | 353136 | |
| Bone marrow mesenchymal stem cells vial | Lonza | PT-2501 | Store in liquid nitrogen upon arrival. Cells are at passage 2 upon arrival but are designated passage 0. Exapand to passage 3 and store in liquid nitrogen for later use. |
| Mesenchymal stem cell growth medium (MSCGM) | Lonza | PT-3001 | Warm in 37 °C water bath before use. For Cell Tracker Green staining use medium without FCS. |
| EDTA (0.02%) solution | Sigma | E8008 | Store at 4°C. Warm in 37°C water bath before use. |
| Trypsin solution | Sigma | T4424 | Store at -20°C in 2ml aliquots. Thaw at room temperature and use immediately. |
| Cryovials | Greiner bio-one | 2019-02 | Keep on ice before adding before adding cell suspension. |
| Mr. Frosty Freezing Container | Nalgene | 5100-0001 | Store at room temperature. When adding cryovials with cells store at -80°C for 24h before transfrring cells to liquid nitrogen. |
| Ibidi u-Slide VI (0.4), T/C treated, sterile | Ibidi | IB-80606 | Alternative models include glass capillaries, Cellix Biochips (www.cellixltd.com), BioFlux Plates (www.fluxionbio.com/bioflux/) and GlycoTech parallel plate flow chambers (http://www.glycotech.com/apparatus/parallel.html). |
| Cell tracker green dye | Life technologies | C2925 | Store in 5µl aliquots at -20°C. Dilute in 5ml prewarmed (at 37°C) MSCGM. |
| Cell counting chambers | Nexcelom | SD-100 | Alternatively a haemocytometer can be used. |
| Cellometer auto T4 cell counter | Nexcelom | Auto T4-203-0238 | |
| Tumor necrosis factor α (TNFα) | R&D Systems | 210-TA-100 | Dilute stock in PBS to a final concentration of 100,000U/ml. Store at -80°C in 10µl aliquots. |
| 6-well, 0.4µm PET Transwell filters | SLS | 353090 | |
| K2-EDTA in 10ml tubes | Sarstedt | Store at room temperature. | |
| Histopaque 1119 | Sigma | 11191 | Store at 4°C. Warm to room temperature before use. |
| Histopaque 1077 | Sigma | 10771 | Store at 4°C. Warm to room temperature before use. |
| 10ml round bottomed tube | Appleton Woods | SC211 142 AS | |
| 7.5% BSA Fraction V solution | Life technologies | 15260-037 | Store at 4°C. |
| 20ml Plastipak syringes | BD falcon | 300613 | |
| 5ml Plastipak syringes | BD falcon | 302187 | |
| 2ml Plastipak syringes | BD falcon | 300185 | |
| 3M hypo-allergenic surgical tape 9m x 2.5cm | Micropore | 1530-1 | Use to secure the syringe tap onto the wall of the perspex chamber. |
| Silicon rubber tubing, internal diameter/external diameter (ID/OD) of 1/3mm (thin tubing) | Fisher Scientific | FB68854 | Cut silicon tubing to the appropriate size. All tubing leading directly to the electronic microvalve must be thin. |
| Silicon rubber tubing ID/OD of 2/4mm (thick tubing) | Fisher Scientific | FB68855 | |
| Portex Blue Line Manometer tubing | Smiths | 200/495/200 | Tubing leading to the syringe pump. |
| 3-way stopcock | BOC Ohmeda AB | ||
| Glass 50ml syringe for pump | Popper Micromate | 550962 | Must be primed prior to use by removing any air bubbles. |
| Glass coverslip | Raymond A Lamb | 26x76mm coverslips made to order. Lot number 2440980. | |
| Parafilm gasket | American National Can Company | Cut a 26x76mm piece of parafilm using an aluminium template and cut a 20x4mm slot into it using a scalpel 10a. Gasket thickness is approximately 133µm. | |
| Two perspex parallel plates | Wolfson Applied Technology Laboratory | Specially designed chamber consisting of parallel plates held together by 8 screws. The lower plate has a viewing slot cut out in the middle and a shallow recess milled to allow space for the coverslip, filter and gasket. The upper perspex plate has an inlet and outlet hole positioned over the flow channel. | |
| Electronic 3-way microvalve with min. dead space | Lee Products Ltd. | LFYA1226032H | Electronically connected to a 12 volt DC power supply. |
| Syringe pump for infusion/withdrawal (PHD2000) | Harvard Apparatus | 70-2001 | Set the diameter to 29mm and refill (flow) rate. |
| L-shaped connector | Labhut | LE876 | To attach to the inlet and outlet ports onto the Ibidi microslide channel. |
| Video camera | Qimaging | 01-QIC-F-M-12-C | Connected to a computer which enables digitall videos to be recorded. |
| Image-Pro Plus 7.0 | Media Cybernetics | 41N70000-61592 | For data analysis. Manually tag cells displaying the different behaviors. Track cells for analysis of rolling and migration velocities. |
| Refer to product datasheets for details on hazards of using the reagents described here. |
References
- Springer, T. A. Traffic signals on endothelium for lymphocyte recirculation and leukocyte emigration. Ann. Rev. Physiol. 57, 827-872 (1995).
- Ley, K., Laudanna, C., Cybulsky, M. I., Nourshargh, S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nature reviews. Immunology. 7 (9), 678-689 (2007).
- McGettrick, H. M., Butler, L. M., Buckley, C. D., Rainger, G. E., Nash, G. B. Tissue stroma as a regulator of leukocyte recruitment in inflammation. Journal of leukocyte biology. 91 (3), 385-400 (2012).
- Serhan, C. N., Savill, J. Resolution of inflammation: the beginning programs the end. Nature immunology. 6 (12), 1191-1197 (2005).
- Schmidt, S., Moser, M., Sperandio, M. The molecular basis of leukocyte recruitment and its deficiencies. Molecular immunology. 55, 49-58 (2013).
- Luu, N. T., Rainger, G. E., Nash, G. B. Differential Ability of Exogenous Chemotactic Agents to Disrupt Transendothelial Migration of Flowing Neutrophils. The Journal of Immunology. 164 (11), 5961-5969 (2000).
- Smith, C. W., Rothlein, R., et al. Recognition of an endothelial determinant for CD 18-dependent human neutrophil adherence and transendothelial migration. The Journal of clinical investigation. 82 (5), 1745-1756 (1988).
- Luscinskas, F. W., Brock, A. F., Arnaout, M. A., Gimbrone, M. A. Endothelial-leukocyte adhesion molecule-1-dependent and leukocyte (CD11/CD18)-dependent mechanisms contribute to polymorphonuclear leukocyte adhesion to cytokine-activated human vascular endothelium. J. Immunol. 142 (7), (1989).
- Bahra, P., Rainger, G. E., Wautier, J. L., Nguyet-Thin, L., Nash, G. B. Each step during transendothelial migration of flowing neutrophils is regulated by the stimulatory concentration of tumour necrosis factor-alpha. Cell adhesion and communication. 6 (6), 491-501 (1998).
- Piali, L., Weber, C., et al. The chemokine receptor CXCR3 mediates rapid and shear-resistant adhesion-induction of effector T lymphocytes by the chemokines IP10 and Mig. European journal of immunology. 28 (3), 961-972 (1998).
- McGettrick, H. M., Smith, E., et al. Fibroblasts from different sites may promote or inhibit recruitment of flowing lymphocytes by endothelial cells. European journal of immunology. 39 (1), 113-125 (2009).
- McGettrick, H. M., Hunter, K., Moss, P. a., Buckley, C. D., Rainger, G. E., Nash, G. B. Direct observations of the kinetics of migrating T cells suggest active retention by endothelial cells with continual bidirectional migration. Journal of leukocyte biology. 85 (1), 98-107 (2009).
- McGettrick, H. M., Buckley, C. D., Filer, A., Rainger, G. E., Nash, G. B. Stromal cells differentially regulate neutrophil and lymphocyte recruitment through the endothelium. Immunology. 131 (3), 357-370 (2010).
- Tull, S. P., Yates, C. M., et al. Omega-3 Fatty acids and inflammation: novel interactions reveal a new step in neutrophil recruitment. PLoS biology. 7 (8), e1000177 (2009).
- Ahmed, S. R., McGettrick, H. M. Prostaglandin D2 regulates CD4+ memory T cell trafficking across blood vascular endothelium and primes these cells for clearance across lymphatic endothelium. Journal of immunology. 187 (3), 1432-1439 (2011).
- Bradfield, P. F., Amft, N., et al. Rheumatoid fibroblast-like synoviocytes overexpress the chemokine stromal cell-derived factor 1 (CXCL12), which supports distinct patterns and rates of CD4+ and CD8+ T cell migration within synovial tissue. Arthritis and rheumatism. 48 (9), 2472-2482 (2003).
- Filer, A., Parsonage, G., et al. Differential survival of leukocyte subsets mediated by synovial, bone marrow, and skin fibroblasts: site-specific versus activation-dependent survival of T cells and neutrophils. Arthritis and rheumatism. 54 (7), 2096-2108 (2006).
- Lally, F., Smith, E., et al. A novel mechanism of neutrophil recruitment in a coculture model of the rheumatoid synovium. Arthritis and rheumatism. 52 (11), 3460-3490 (2005).
- Chakravorty, S. J., McGettrick, H. M., Butler, L. M., Buckley, C. D., Rainger, G. E., Nash, G. B. An in vitro. model for analysing neutrophil migration into and away from the sub-endothelial space: Roles of flow and CD31. Biorheology. 43 (1), 71-82 (2006).
- Rainger, G. E., Nash, G. B. Cellular Pathology of Atherosclerosis Smooth Muscle Cells Prime Cocultured Endothelial Cells for Enhanced Leukocyte Adhesion. Circulation Research. 88 (6), 615-622 (2001).
- Kuravi, S. J., McGettrick, H. M. Podocytes regulate neutrophil recruitment by glomerular endothelial cells via IL-6-mediated crosstalk. Journal of immunology. 193 (1), 234-243 (2004).
- Parsonage, G., Filer, A. D. A stromal address code defined by fibroblasts. Trends in immunology. 26 (3), 150-156 (2005).
- Luu, N. T., McGettrick, H. M. Crosstalk between mesenchymal stem cells and endothelial cells leads to downregulation of cytokine-induced leukocyte recruitment. Stem cells. 31 (12), 2690-2702 (2013).
- Bevilacqua, M. P., Nelson, R. M., Mannori, G., Cecconi, O. Endothelial-leukocyte adhesion molecules in human disease. Annual review of medicine. 45, 361-378 (1994).
- Stanness, K. A., Beatty, P. G., Ochs, H. D., Harlan, J. M. An endothelial cell surface factor(s) induced in vitro. 136 (12), 4548-4553 (1986).
- Burton, V. J., Butler, L. M. Delay of migrating leukocytes by the basement membrane deposited by endothelial cells in long-term culture. Experimental cell research. 317 (3), 276-292 (2011).
- Luu, N. T., Rainger, G. E., Buckley, C. D., Nash, G. B. CD31 Regulates Direction and Rate of Neutrophil Migration over and under Endothelial Cells. Journal of Vascular Research. 40 (5), 467-479 (2003).
- McGettrick, H. M., Buckley, C. D., Ed Rainger, G., Nash, G. B. Influence of stromal cells on lymphocyte adhesion and migration on endothelial cells. Methods in molecular biology. 616, 49-68 (2010).
- Butler, L. M., McGettrick, H. M., Nash, G. B. Static and dynamic assays of cell adhesion relevant to the vasculature. Methods in molecular biology. 467, 211-228 (2009).
- Jeffery, H. C., Buckley, C. D., Moss, P., Rainger, G. E., Nash, G. B., McGettrick, H. M. Analysis of the effects of stromal cells on the migration of lymphocytes into and through inflamed tissue using 3-D culture models. Journal of immunological methods. 400-401, 45-57 (2013).

