A Polyaniline-based Sensor of Nucleic Acids
Summary
Nucleic acids are common analytes for assessing biological systems; however, bias from enzymatic manipulation can cause concern. Here a method is described for label-free detection of nucleic acids using polyaniline. This sensitive, cost-effective sensor technology can distinguish single nucleotide differences between molecules.
Abstract
Detection of nucleic acids is at the center of diagnostic technologies used in research and the clinic. Standard approaches used in these technologies rely on enzymatic modification that can introduce bias and artifacts. A critical element of next generation detection platforms will be direct molecular sensing, thereby avoiding a need for amplification or labels. Advanced nanomaterials may provide the suitable chemical modalities to realize label-free sensors. Conjugated polymers are ideal for biological sensing, possessing properties compatible with biomolecules and exhibit high sensitivity to localized environmental changes. In this article, a method is presented for detecting nucleic acids using the electroconductive polymer polyaniline. Simple DNA “probe” oligonucleotides complementary to target nucleic acids are attached electrostatically to the polymer, creating a sensor system that can differentiate single nucleotide differences in target molecules. Outside the specific and unbiased nature of this technology, it is highly cost effective.
Introduction
Conjugated polymers provide many options for molecular sensors. This includes fluorescence, electronic, and colorimetric responses1. There have been many efforts to incorporate conjugated polymers in nucleic acid sensors. However, most systems require secondary detection, limiting sensing options2. Recently, we reported a conjugated polymer-based sensor platform built on polyaniline (PANI) that exploits properties of this polymer, creating a label-free system3. PANI is an extensively conjugated electro-active polymer with properties such as fluorescence and resistance that are suitable for measuring biological systems4. The excitons within the structure are not localized leading to mobility of the positive charge between monomeric subunits. This provides a flexible scaffold of positive charges that can interact with the negatively charged backbone of DNA5,6. Importantly, electrostatically attached DNA is orientated such that nitrogenous bases can participate in base pairing. Association with DNA alters the electronic properties of PANI, an effect that can be enhanced by UV irradiation (Figure 1)3. Using this system, oligonucleotides complementary to target nucleic acids can be immobilized on PANI. Multiple studies have demonstrated that upon hybridization electrostatically adsorbed oligonucleotides dissociate from PANI or other cationic matrices due to conformational changes caused by the switch to a double-stranded DNA structure3,5,7.
In a sensor system where probe attachment modulates conjugated polymer properties, hybridization events can be transduced without labels or enzymatic modification of probes or target nucleic acids. Conjugated polymers offer great flexibility in detection methods, one of which is fluorescence. Through monitoring PANI fluorescence, concentrations of target nucleic acids as low as 10-11 M (10 pM) can be detected3. Detection is rapid, occurring within 15 minutes of hybridization, and specific where a single mismatch in a target molecule can be differentiated3.
Fabrication of PANI-sensors is straightforward. High molecular weight PANI can be generated that is well-dispersed in water using standard synthesis procedures involving aniline monomer, surfactant, and controlled addition of an oxidant. Yield can be very high and unreacted oxidant removed by washing with water, ensuring no further PANI growth. PANI-probe association occurs spontaneously upon mixture, and complex formation is enhanced by mild UV exposure. Hybridization can be carried out immediately, and the changes in PANI fluorescence assayed following a short incubation. The simplicity of this technology makes it highly accessible to many laboratories.
Protocol
1. Processable PANI Synthesis
- Dissolve aniline (1 ml, 11 mmol) completely in 60 ml of chloroform in a 250 ml round-bottom flask. Stir at 600 rpm for 5 min and cool to 0-5 °C with ice. This usually takes 15-20 min (Figure 2A).
- Add sodium dodecyl benzene sulphonate (NaDBS) (7.44 g, 21 mmol) to the aniline solution in a round-bottomed flask while stirring at 600 rpm.
- Dissolve ammonium persulphate (APS) (3.072 g, 13.5 mmol) in 20 ml water and add all of it drop-wise over 30 min to avoid overheating the reaction.
- Carry out the reaction at 0-5 °C for 24 hr, and allow it to reach room temperature for another 24 hr.
- Observe the reaction mixture initially turn milky white after 15 min, then dark brown after 2 hr, and finally to dark green after 24 hr (Figure 2B-F).
- Filter the PANI-NaDBS solution with a Buchner funnel. Mix with 80 ml chloroform and 120 ml water in a separation funnel (Figure 2G).
- Incubate the solution for 24 hr at room temperature and collect the dark green PANI from the separation funnel, leaving unreacted NaDBS and APS in the aqueous supernatant.
2. PANI-probe Mixing and UV Irradiation
- Dilute PANI solution 10x with chloroform-water (1:3 v/v) and mix 200 µl of diluted PANI with 6.4 µmol of probe DNA oligonucleotides by gentle rocking for 15 min in a microfuge tube.
- Irradiate the PANI-DNA solution with 1,200 µJ/cm2 of UV in a crosslinker for 2 min. It is critical that UV exposure is limited to the indicated amount. Extended exposure to UV compromises the fluorescence change in PANI, likely due to covalent cross-linking of PANI and DNA.
- Pellet complexes by centrifugation at 17,000 x g for 6 min, and wash with phosphate buffered saline (PBS). Pellet again, and re-suspend in PBS.
3. Hybridization of PANI-probe
- Add 8 µl of 100 µM complementary DNA oligonucleotides or target nucleic acids to 200 µl of PANI-probe complexes.
- Perform hybridization by rocking solution mixture for 15 min at 40 °C.
- Pellet the PANI complexes by centrifugation at 17,000 x g for 6 min. Wash with PBS and re-suspend in water.
4. Emission Steady State Fluorescence Measurement
- Add PANI from different treatments into a 96 well microplate and measure emission fluorescence in the 270-850 nm range by excitation at 250 nm. An emission peak for PANI should be observed around 500 nm.
5. Fluorescence Microscopy Measurement of Hybridized Duplex
- Drop coat PANI on a borosilicate glass coverslip and dry at 40 °C for 48 hr.
- Add probe (8 µl of 100 µM) on a dried PANI film and irradiate it with UV light (1,200 µJ/cm2) for 2 min.
- Wash the PANI-probe film with PBS and dry at 40 °C for 48 hr.
- Perform hybridization for 15 min by adding target nucleic acids. This could be a biological sample or a control target oligonucleotide (8 µl of 100 µM). Follow with a PBS wash.
- Obtain the fluorescent images at 40X magnification, with a 500 nm long pass filter.
Representative Results
Figure 2A captures the reaction setup at the start of the polymerization process, i.e., before APS addition. Micelle formation is the initial step in the reaction process-PANI synthesis occurs at the micellar interface. Figure 2B shows a milky solution after 5 min. 30 min after APS is added the reaction turns to a slightly brown color. Figure 2C shows the color change associated with the formation of oligomers. Figure 2D shows a dark brown color after 4 hr, indicating a high concentration of short chain PANI, consistent with a very slow reaction. Finally after 48 hr a dark green solution of high molecular weight PANI can be observed that is dispersed in chloroform-water (Figure 2E). Figure 2F shows a well-dispersed, homogenous PANI solution in the separating funnel with no phase separation.
Sensing nucleic acids can be carried out in emulsion (Figure 2) or on the surface of drop-coated films (Figure 3). The basal fluorescence of PANI films is relatively low (Figure 3A). Probe DNA is added over the film, and allowed to dry at room temperature. When the expected DNA-PANI complex forms, an intense increase in fluorescence can be observed (Figure 3B). Electron influx from the negatively charged DNA causes higher electron density in PANI molecules, causing the fluorescence increase. After hybridization induced dissociation of the probe-target duplex, PANI fluorescence returns to basal intensity (Figure 3C). Dissociation is caused by changes in the DNA backbone upon hybridization that compromise PANI-probe interaction. Sensing nucleic acids with this technology can also be performed directly in emulsion (Figure 3D). A benefit of using the emulsion-based system is compatibility with plate reader instruments. This permits a highly precise measurement of PANI fluorescence, and avoids difficulties caused by irregularities in film coating. A PANI-based sensor in emulsion format is highly specific. The introduction of a single mismatch (mm) in the target oligo fails to restore basal PANI fluorescence (Figure 3D).
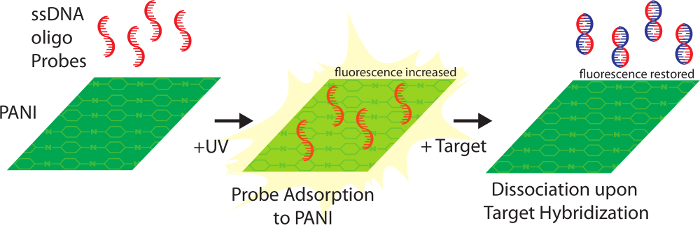
Figure 1. Attachment of probes mediated by UV exposure increases PANI fluorescence. Hybridization of target nucleic acids results in detachment from PANI, and reversion of the polymer to basal fluorescence. Please click here to view a larger version of this figure.
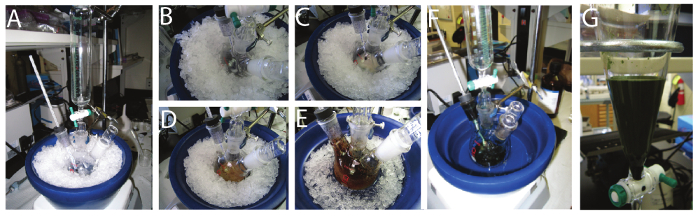
Figure 2. Synthesis of processable PANI. (A) Start of polymerization reaction. (B) After 5 min when APS is slowly added. (C) 30 min after APS addition. (D) After 1 hr. (E) After 4 hr. (F) Green product after 48 hr of reaction. Please click here to view a larger version of this figure.
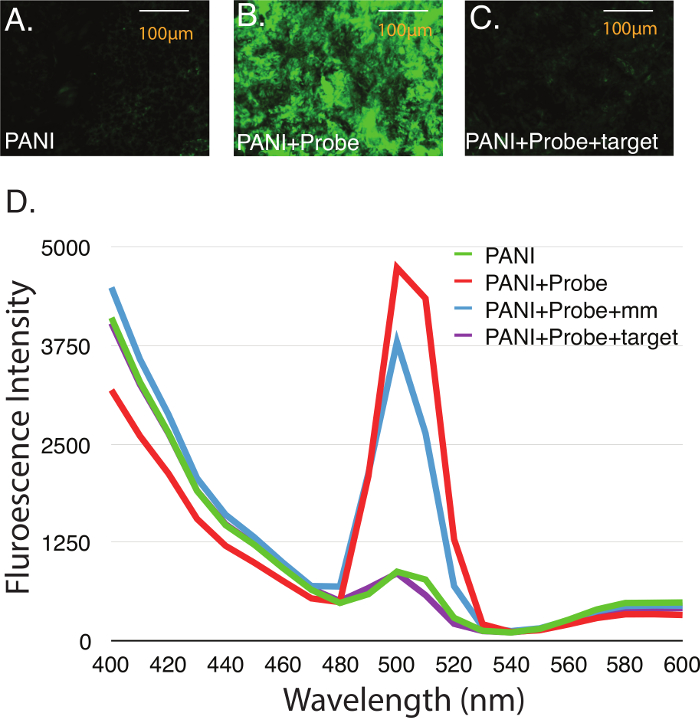
Figure 3. Use of PANI Fluorescence to detect nucleic acid hybridization. (A) PANI film created through drop-coating on borosilicate glass, followed by drying for 48 hr at 40 °C. (B) PANI fluorescence after probe immobilization. (C) Following hybridization of target DNA with PANI-immobilized probes. (D) Fluorescence at 500 nm of PANI emulsion before and after hybridization with either complementary or single mismatch (mm) target oligonucleotides. Please click here to view a larger version of this figure.
Discussion
A PANI-based sensor of nucleic acids requires solubilization of the polymer in water in order to interact with DNA and RNA. The dispersion of PANI in water is accomplished using surfactants, forming micelles as previously reported8. In addition to the NaDBS used here other anionic surfactants like dodecyl ester of 4-sulfophthalic acid, nonionic surfactants like nonyl phenol ethoxylate, or cationic surfactants like cetyltrimethyl ammonium bromide could also be used for synthesis of processable PANI9,10. The synthesis described here begins with a fixed proportion of the anionic surfactant NaDBS added to the monomer aniline at a 2:1 molar ratio. The surfactant NaDBS has a high hydrophile-lipophile balance (HLB) allowing polymerization of aniline monomer emulsified in chloroform-water by establishing equilibrium between the micellar polymeric oil droplets and water. The oxidant APS is then added at a 1:1.5 molar ratio to generate micelles of growing PANI, where the inner core acts as a nucleating agent. This leads to head to tail attachment of monomers, while also keeping PANI in dynamic equilibrium with the surrounding solvent11,12. The reaction proceeds in a controlled manner until high molecular weight polymers are formed13. This protocol will generate particles that range from 50 nm to 1,500 nm3. Critical to establishing a reaction where PANI grows slowly yet remains soluble is the slow addition of the oxidant APS, low temperature, and the absence of strong protonic acids. Some optimization of reactant proportions may be necessary to generate well-dispersed PANI. PANI synthesized by this method is stable for more than one year as an emulsion. Excessive dilution should be avoided as the HLB balance of the dispersion may be disrupted, causing PANI to precipitate. High speed mixing can help to keep the PANI in solution. Perfect solubility is not desirable, as it would preclude pellet formation, which is an important criterion for hybridization procedures in an emulsion-based sensor.
After generating dispersed PANI, DNA probe oligos are attached to the polymer matrix through electrostatic interaction. Interaction of PANI imine groups and the phosphates of the backbone of DNA occur spontaneously. Non-specific PANI-nucleic acid interactions complicate analyte sensing in complex biological samples14. To achieve association between PANI and probe DNA that can be distinguished from spontaneous interactions, the complex is irradiated with UV. The creation of polar moieties induced by UV-triggered charge localization increases wettability of the polymer surface15. Optimal exposure is approximately 2 min, and can be confirmed if PANI fluorescence at 500 nm is elevated roughly 5X (Figure 3). Over-exposure of the complex to UV can lead to crosslinking between PANI and DNA, which unlike electrostatic interactions, does not cause an elevation of PANI fluorescence.
The process of generating sensors is easy and quick. The polymerization reaction is completed after 48 hours followed by 24 hours to separate from unreacted oxidant and NaDBS. Synthesis will need to be carried out infrequently as the quantity of stock PANI produced by the protocol in this article is very large relative to the amount used in a sensor. PANI emulsion at high concentrations is stable; however, any dilution that occurs during probe attachment can lead to precipitation of PANI, compromising sensor performance. The immediate use of PANI-probe complexes to detect targets will avoid this issue. Together, creation of complexes and hybridization can be done in under an hour. PANI-probe films are more stable and can be used after long-term storage. A limitation of this sensor technology is that it is not highly amenable to multiplexing. Each detection event must be carried out in a separate tube. Films may provide a better format for multiplexing as different probes could be spotted on different regions of a film. A sample could be applied over the spot array, and the fluorescence at each spot used to evaluate expression of a different gene.
The detection mechanism described here provides several advantages over other systems. Reports of detecting nucleic acids through a dissociation mechanism involving PANI rely on fluorophores attached to probe DNA oligos5. Other strategies use covalent attachment of probes, which fail to distinguish single base-pair differences in target nucleic acids16-17. The omission of labeling or other secondary modes of detection in this technology creates opportunities to use other properties of electro-active PANI. The sensor response thus could be extended to the measurement of electrochemical properties altered by the attachment-detachment dynamics of the sensor. Labs that adopt this technique will find the sensor system to be cost effective in addition to fast and straightforward.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors have nothing to disclose.
Materials
| Aniline | Fisher Scientific | A7401-500 | ACS, liquid, refrigerated |
| Ammonium peroxydisulfate | Fisher Scientific | A682-500 | ACS, crystalline |
| Sodium dodecylbenzene sulfonate | Pfaltz & Bauer | D56340 | 95% solid |
| Chloroform | Fisher Scientific | MCX 10601 | Liquid |
| DNA primers | MWG operon | n/a | custom DNA sequence ~20bps |
| Microplate | USA Scientific | 1402-9800 | 96 well, polypropylene as it is unreactive to chloroform |
| Microplate Adhesive Film | USA Scientific | 2920-0000 | Reduces well-to-well contamination, sample spillage and evaporation |
| Microscope Cover Glass | Fisher Scientific | 12-544-D | PANI coated on UV irradiated cover glass |
| UV crosslinker | UVP | HL-2000 | Energy: X100 μJ/cm2; Time: 2min |
| Hybridization Oven | VWR | 01014705 T | Temperature: 400C; with rocking for 15 min |
| Glass Apparatus | Fisher Scientific | Three necked round bottom flask for reaction; dropping funnel, stoppers, condenser, separating funnel | |
| Microscope | Leica Microsystems | Leica IMC S80 | Magnification 20X; Pseudo color 536 nm; Exposure 86 ms; Gain 1.0X; Gamma 1.6 |
| Microplate Reader | Molecular Devices | 89429-536 |
References
- Hahm, J. I. Functional polymers in protein detection platforms: optical, electrochemical, electrical, mass-sensitive, and magnetic biosensors. Sensors (Basel). 11 (3), 3327-3355 (2011).
- Rahman, M. M., Li, X. B., Lopa, N. S., Ahn, S. J., Lee, J. J. Electrochemical DNA hybridization sensors based on conducting polymers. Sensors (Basel). 15 (2), 3801-3829 (2015).
- Sengupta, P. P., et al. Utilizing Intrinsic Properties of Polyaniline to Detect Nucleic Acid Hybridization through UV-Enhanced Electrostatic Interaction. Biomacromolecules. 16 (10), 3217-3225 (2015).
- Song, E., Choi, J. -. W. Conducting Polyaniline Nanowire and Its Applications in Chemiresistive Sensing. Nanomaterials. 3 (3), 498 (2013).
- Liu, S., et al. Polyaniline nanofibres for fluorescent nucleic acid detection. Nanoscale. 3 (3), 967-969 (2011).
- Oliveira Brett, A. M., Chiorcea, A. -. M. Atomic Force Microscopy of DNA Immobilized onto a Highly Oriented Pyrolytic Graphite Electrode Surface. Langmuir. 19 (9), 3830-3839 (2003).
- Zhang, Y., et al. Poly(m-Phenylenediamine) Nanospheres and Nanorods: Selective Synthesis and Their Application for Multiplex Nucleic Acid Detection. PLoS ONE. 6 (6), e20569 (2011).
- Namgoong, H., Woo, D. J., Lee, S. -. H. Micro-chemical structure of polyaniline synthesized by self-stabilized dispersion polymerization. Macromol Res. 15 (7), 633-639 (2007).
- John, A., Palaniappan, S., Djurado, D., Pron, A. One-step preparation of solution processable conducting polyaniline by inverted emulsion polymerization using didecyl ester of 4-sulfophthalic acid as multifunctional dopant. J Polym Sci A: Polym Chem. 46 (3), 1051-1057 (2008).
- El-Dib, F. I., Sayed, W. M., Ahmed, S. M., Elkodary, M. Synthesis of polyaniline nanostructures in micellar solutions. J Appl Polym Sci. 124 (4), 3200-3207 (2012).
- Tsotcheva, D., Tsanov, T., Terlemezyan, L., Vassilev, S. Structural Investigations of Polyaniline Prepared in the Presence of Dodecylbenzenesulfonic Acid. J Therm Anal Calorim. 63 (1), 133-141 (2001).
- Jia, W., et al. Polyaniline-DBSA/organophilic clay nanocomposites: synthesis and characterization. Synthetic Met. 128 (1), 115-120 (2002).
- Kim, B. -. J., Oh, S. -. G., Han, M. -. G., Im, S. -. S. Preparation of Polyaniline Nanoparticles in Micellar Solutions as Polymerization Medium. Langmuir. 16 (14), 5841-5845 (2000).
- Scales, C. W., et al. Corona-Stabilized Interpolyelectrolyte Complexes of siRNA with Nonimmunogenic, Hydrophilic/Cationic Block Copolymers Prepared by Aqueous RAFT Polymerization†. Macromolecules. 39 (20), 6871-6881 (2006).
- Kadashchuk, A., et al. Localized trions in conjugated polymers. Phys Rev B. 76 (23), 235205 (2007).
- Chang, H., Yuan, Y., Shi, N., Guan, Y. Electrochemical DNA Biosensor Based on Conducting Polyaniline nanotube Array. Anal. Chem. 79, 5111-5115 (2007).
- Zhu, N., Chang, Z., He, P., Fang, Y. Electrochemically fabricated polyaniline nanowire-modified electrode for voltammetric detection of DNA hybridization. Eletrochim. Acta. 51, 3758-3762 (2006).

