Spinal Cord Neurons Isolation and Culture from Neonatal Mice
Summary
This study presents a technique for the isolation of neurons from WT neonatal mice. It requires the careful dissection of the spinal cord from the neonatal mouse, followed by the separation of neurons from the spinal cord tissue through mechanical and enzymatic cleavage.
Abstract
We present a protocol for the isolation and culture of spinal cord neurons. The neurons are obtained from neonatal C57BL/6 mice and are isolated on postnatal day 1-3. A mouse litter, usually 4-10 pups born from one breeding pair, is gathered for one experiment, and spinal cords are collected individually from each mouse after euthanasia with isoflurane. The spinal column is dissected out and then the spinal cord is released from the column. The spinal cords are then minced to increase the surface area of delivery for an enzymatic protease that allows for the neurons and other cells to be released from the tissue. Trituration is then used to release the cells into solution. This solution is subsequently fractionated in a density gradient to separate the various cells in solution, allowing for neurons to be isolated. Approximately 1-2.5 x 106 neurons can be isolated from one litter group. The neurons are then seeded onto wells coated with adhesive factors that allow for proper growth and maturation. The neurons take approximately 7 days to reach maturity in the growth and culture medium and can be used thereafter for treatment and analysis.
Introduction
Understanding spinal cord pathology requires the use of various models, both on the macroscopic and microscopic levels. Large and small animal models1,2,3 are used for in vivo investigations of spinal cord disease and injury. While studying these issues in vivo has its merits, analysis of the spinal cord is limited to whole spinal cord homogenate or to tissue sections4. This creates some ambiguity when trying to isolate specific responses and targets in the spinal cord among its resident neurons and surrounding glia. The increasing availability of genetically manipulated mice allows for more detailed investigations of the biology at cellular and molecular levels. Thus, a neonatal mouse model is used here, allowing for the study of the unique properties and biology of spinal cord neurons in vitro.
The isolation and maintenance of neurons in vitro is not particularly straightforward. There is a relative abundance of techniques for neuron isolation from the cortical tissue of adult rodents that seem to result in a substantial number of isolated neurons (i.e. millions)5,6,7. In contrast, the yield of neurons from spinal cord tissue is lower8,9,10, in part due to the smaller mass of tissue. Furthermore, in mice, there is a relative paucity of techniques for the isolation of neonatal spinal cord neurons, and existing methods are limited by lower neuron yields (i.e. hundreds)9 or laborious and resource-heavy techniques requiring the isolation of embryonic mice10.
In this protocol, we use a technique that allows for the cost- and resource-effective isolation of a substantial number of neurons from the spinal cords of neonatal mice. As is common in previously published techniques, we use papain as an enzymatic protease, allowing for the release of neurons from the spinal cord tissue5,6 . In addition, we use a density gradient for refined cell separation, which has previously been shown to be effective6,10. While the medium in which the cells are incubated can vary, in our experience and as previously published11, supplementation with fresh B27 culture medium supplement has proven to be critical for neuron longevity. The neurons are typically viable for up to 10 days, allowing for treatment to be carried out.
Protocol
The care and treatment of animals in this procedure were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of Colorado.
1. Preparing Solutions
- Prepare and store all solutions at appropriate temperatures, as shown in Table 1.
2. Coating Wells and Slides
NOTE: Neurons do not adhere well to plastic or glass surfaces.
- One day prior to the isolation of the neurons, coat the wells of a sterile 24-well culture plate with 0.5 mL of Poly-D-Lysine (PDL; Table 2) and leave it in a laminar flow hood O/N.
NOTE: The preferred method is to use 24-well plates and to coat only the center wells, as evaporation from the peripheral wells tends to be more accelerated and to lead to inconsistent results. Additionally, glass slides can be placed inside the wells prior to coating. The neurons will attach to the coated glass slides and can be removed later for further microscopic imaging. - On the day of isolation, remove the PDL from the wells. Wash with sterile water for a few minutes before removing any remaining water and allowing it to dry for 1 h (see below).
- After drying, coat the slides with laminin (Table 2).
NOTE: Approximately 10 μL of laminin (10 µg/µL) is enough to coat each well of a sterile 24-well plate. This is mixed with 340 μL of medium (total volume: 350 μL) in order to cover the entire well. Let this sit for 2 h at RT in a culture hood and then aspirate prior to coating the cells.
3. Harvesting Spinal Cords
NOTE: All instruments should be autoclaved (135 °C and 30 psi for 4 x 7 min cycles) for sterility.
- Euthanize 1-3 day-old C57BL/6 mouse pups in a chamber with isoflurane. Wait 30 s after the cessation of movement and pinch the leg to confirm a lack of response.
- Separate the head from the body using scissors, with pup in the prone position.
- Stabilize the hind legs or tail and arms on the procedure table, with dorsal side facing the user.
- Cut the skin off using curved iris scissors.
- Cut the spinal cord from the lumbar region just above the hips and proceed to cut both sides of the thorax to separate it from the body.
NOTE: This requires the careful dissection of the spinal cord from visceral organs to avoid inadvertent damage to other organs (Figure 1a). - Wash sequentially for 10 s in 3 x 10 cm Petri dishes containing 5 mL of 0.2 μm filter-sterilized Phosphate-Buffered Saline (PBS) to remove excess tissue.
- Insert a 22 G needle and syringe filled with 5 mL of filter-sterilized PBS into the caudal end of the spinal column and flush cranially, allowing the cord to exit into a fourth Petri dish (Figure 1b).
- Collect the spinal cord in a 15 mL tube with 5 mL of HABG (Table 1) on ice. Use care to avoid crushing the spinal cord.
- Repeat steps 3.1-3.8 for each pup in the litter.
NOTE: Ideally, this process should take less than 30 min per spinal cord to ensure healthy neuron isolation.
4. Isolating Neurons
NOTE: The following step should be performed in a laminar flow hood. Familiarity with basic sterile technique is expected.
- Tissue Mincing
- Take the tube containing the spinal cords and shake lightly to suspend the tissue.
- Pour tissue from the tube into a 60 mm glass Petri dish and dice with a razor blade to create fine pieces ~0.5 mm in size.
- Transfer the tissue with a wide-bore pipette into a 15 mL tube containing 5 mL of HABG.
- Place it in a 30 °C water bath for 30 min to allow the cells to equilibrate at this temperature. Keep the cells on a shaker just enough to allow them to be suspended in the fluid.
NOTE: This step is done to avoid shocking the cells upon transfer from ice to the digestion medium. Keeping the cells at 30 °C helps to decrease cell death associated with an otherwise increased metabolism at 37 °C.
- Prepare the digestion medium (Table 1).
- Prepare the density gradient (Table 1).
- Prepare each of the 4 layers in 4 separate 15 mL tubes, as outlined in Table 1.
- Add 1 mL from each layer into a new 15 mL tube. Start with layer 1 at the bottom and sequentially add until reaching layer 4 at the top. Avoid disturbing the layers while adding.
- Wash the PDL-coated plates from step 2.2. Wash with sterile water for a few minutes before removing any remaining water and allowing them to dry for 1 h.
- Transfer the tissue to digestion medium.
- Remove the tissue-containing tube from the shaker water bath at 30 °C and allow it to settle for a few minutes.
- Remove the digestion medium tube from the 37 °C water bath and aspirate it into a leur-lock syringe.
- Aspirate off the excess HABG from the tissue-containing tube.
- Use a leur-lock 0.2 μm filter on the syringe to add digestion medium to the tissue-containing tube.
- Place the tube in a 30 °C water bath for 30 min. Keep the cells shaking just enough to allow them to be suspended in the fluid.
NOTE: It is important not to keep the cells in the digestion medium for too long or to let the temperature get too high, which could lead to excessive digestion and result in the tissue becoming suspended in a gelatinous mixture.
- During this period, coat the laminin as in step 2.3.
- Perform trituration (i.e. separating the cells from the tissue).
- Remove the tube from the shaking 30 °C water bath and allow it to settle for a few min.
- Aspirate excess digestion medium.
- Suspend the tissue in 2 mL of HABG.
- Using a narrow-bore pipette, triturate 10x for 45 s.
NOTE: This is probably the single most crucial step and can significantly decrease the yield if not done properly.- Aspirate the tissue into the pipette and immediately empty the contents back.
- Avoid introducing air, as it will significantly decrease the viable yield.
NOTE: The ideal pipette is a 9" glass pipette. The tip of the pipette should be fire polished to smooth out rough surfaces. It should then be siliconized by placement in a 1:20 solution of dichlorodimethylsilane (DMDCS) in chloroform and left O/N. The pipette should then be removed and allowed to air dry. Subsequently, it should be autoclaved for sterilization.
Caution: DMDCS and chloroform are highly flammable, and siliconizing should be carried out in a fume hood.
- Aspirate the top 2 mL of supernatant and place it into a new 15 mL tube labeled "collection."
- Repeat steps 4.7.3-4.7.5 two additional times (the cell collection tube should have 6 mL by the end).
- Slowly transfer the collection tube contents into the gradient tube prepared in step 4.2, avoiding the disruption of the gradient.
- At this point, remove the previously prepared neurobasal medium (Table 1) from the refrigerator and allow it to warm in a 37 °C bath.
- Purify the neurons.
- Centrifuge the gradient tube for 15 min at 800 x g and 22 °C.
- Collect the desired layer(s) with a pipette (Figure 2) and place in a new 15 mL tube. For the highest-purity neuron isolation (i.e. >90%), collect layer 3. For more yield with less purity (i.e. >70-80%), collect layers 2 & 3.
- Dilute out the density gradient by adding 5 mL of HABG to the newly collected layers.
- Centrifuge at 200 x g for 2 min at 22 °C.
- Discard the supernatant, re-suspend in 5 mL of HABG, and flick the pellet to suspend the cells.
- Centrifuge at for 2 min 200 x g at 22 °C.
- Discard the supernatant, resuspend in 3 mL of neurobasal medium, and flick the pellet to resuspend the cells.
- Count the cells.
- Take 10 μL of the solution, now with cells in neurobasal medium, and mix with 10 μL of Trypan blue.
- Place 10 μL of this mixture in a glass counting chamber.
- Using a standard glass counting chamber, count the number of cells in each of the four 4 x 4 quadrants. Add all of the cells counted (n), multiply by 2 (dilution factor), divide by 4 (number of quadrants counted), multiply by 3 (volume of neurobasal medium), and multiply by 104 to obtain the concentration of cells in cells/mL.
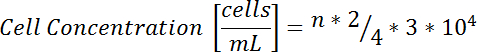
- Seed the cells on culture plates
- Dilute the cell suspension to 300,000 cells in 1 mL of neurobasal medium.
NOTE: Based on the concentration of cells obtained in the above steps, additional neurobasal medium is added to obtain a final concentration of 3*105 cells/mL. The equation is C1V1=C2V2, where C1 is the initial concentration of cells obtained from the harvest; V1 is 3 mL; and C2 is 3*105 cells/mL, as discussed above. The equation is solved for V2. Add the volume of neurobasal medium necessary to make the total volume of cells in solution equal to V2. - Shake gently to distribute the cells in solution and add 1 mL to each well in the coated 24-well plates.
- Dilute the cell suspension to 300,000 cells in 1 mL of neurobasal medium.
Representative Results
Using this technique, a single litter (4-10 pups) allows for the isolation of 1-2.5 106 neurons suitable for seeding onto culture plates. Typically, 4-8 wells are seeded at the concentration mentioned above (i.e. 300,000 cells/mL). Figure 3 demonstrates the appearance of neurons at this concentration after a week in culture at low- (a) and high- (b) magnification light microscopy. However, we have also been able to culture cells at concentrations as high as 500,000 cells/well and as low as 100,000 cells/well. Using the higher concentrations requires more medium and careful attention to environmental conditions, as nutrients can be depleted quickly, leading to a toxic acidic environment for the cells. With lower concentrations, cells tend not to reach full maturity, and an overgrowth of supporting cells (i.e. oligodendrocytes, microglia, and astrocytes) is observed.
The cells will typically start adhering to the surface within the first couple of hours (Figure 4a). Axons will begin to sprout within the first 24-48 h (Figure 4b-4c). Connections between various neurons in culture typically reach maturity at 7 days (Figure 4d), at which point experiments are typically carried out on the neurons.
Neurons are identifiable under light microscopy, as they have distinct axonal projections. Our isolations using layer 3 (Figure 2) yields ~80-90% neurons in culture. This was confirmed using the immunofluorescent staining of neuron-specific markers. In Figure 5c, the neuron cytoskeletal protein Microtubule-Associated Protein 2 (MAP2) is stained, showing outlines and axonal projections of the neurons after a week in culture. Similarly, in Figure 6c, the neuron-specific nuclear marker NeuN is stained, showing neuronal nuclei. These neuronal markers are combined with nucleus (DAPI, Figure 5a, 6a) and cytoplasm (GFP, Figure 5b, 6b) imaging, and the resulting images are merged (Figure 5d, 6d), detailing the relative abundance of neurons to other cells in culture. When using layers 2 and 3, the yield of neurons is slightly higher; however, there tend to be fewer neurons in the pure solution (~70%).
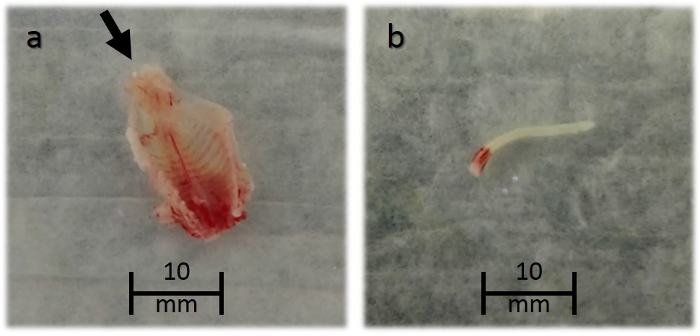
Figure 1: Spinal Column Dissected and Spinal Cord Released from a 3 day-old Neonatal Mouse. The arrow in (a) points towards the caudal end of the spine, where a needle is inserted to release the spinal cord. A released spinal cord (b) is shown submerged in PBS. Please click here to view a larger version of this figure.
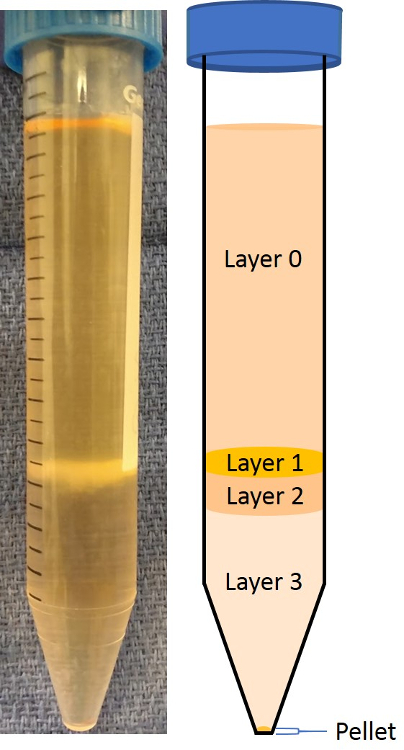
Figure 2: Density Gradient, with Cells Added-on after Centrifugation. The layers are outlined and better visualized on the cartoon depiction. The most superficial layer (0) is generally debris from the spinal cord tissue. Layer 1 is rich in oligodendrocytes and astrocytes. Layers 2 and 3 contain the majority of neurons. While layer 3 contains neurons with the highest purity, some supporting cells (i.e., astrocytes and oligodendrocytes) are found in layer 2. The pellet at the bottom mainly contains microglial cells. Please click here to view a larger version of this figure.
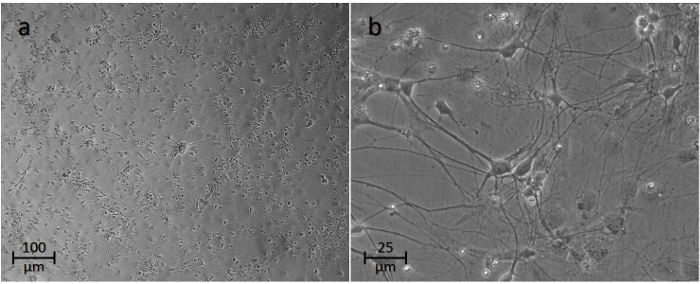
Figure 3: Light Microscopy of Neuronal Cell Cultures after 7 days of Isolation. At 10X magnification (a), axonal connections among various conglomerates of neurons can be seen. At 40X magnification (b), the neurons can be visualized more closely with their axonal projections. Please click here to view a larger version of this figure.
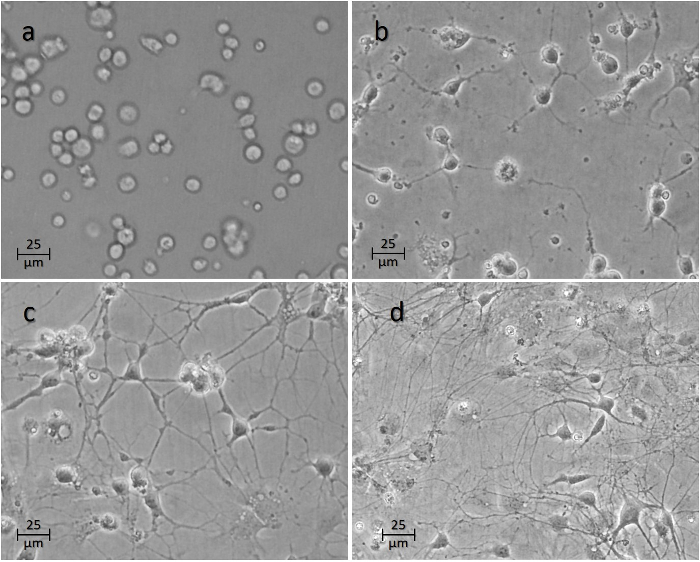
Figure 4: Time Course of the Neurons after Seeding onto the Wells. Image (a) shows the cells 1 h, (b) 24 h, (c) 48 h, and (d) 1 week after seeding. Please click here to view a larger version of this figure.
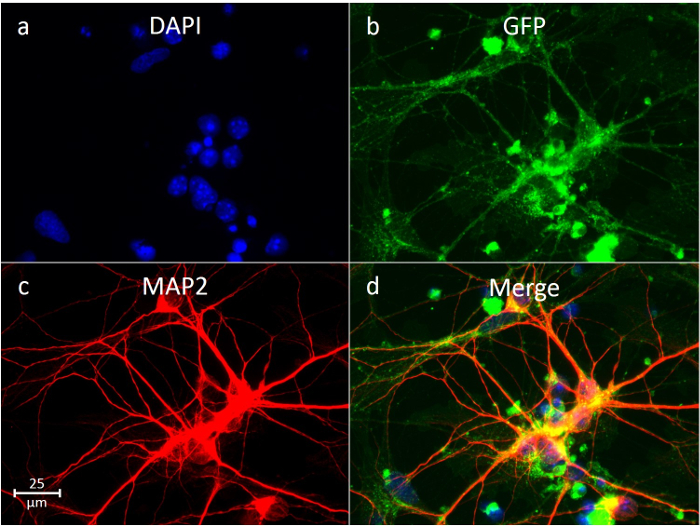
Figure 5: Immunofluorescent Stain of Neurons using Neuronal Cytoskeletal Marker MAP2. Nuclear staining (DAPI) is shown in blue (a), with cytoplasmic staining (GFP) in green (b) and neuron cytoskeletal protein (MAP2) in red (c). The merged images (d) are combined, showing the relative abundance of neurons. Please click here to view a larger version of this figure.
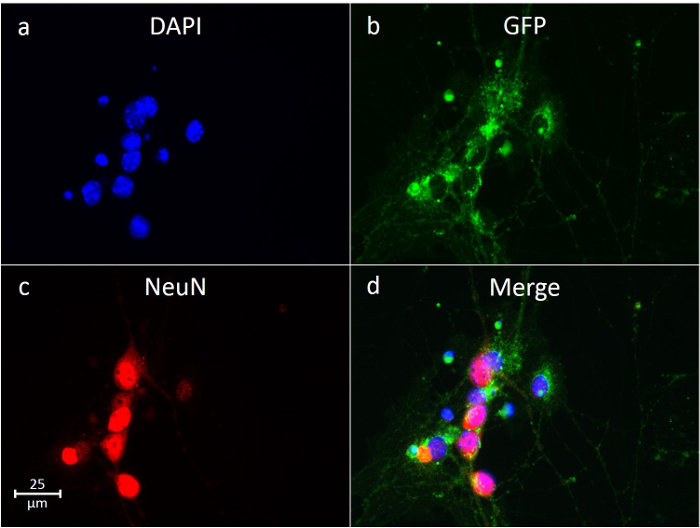
Figure 6: Immunofluorescent Stain of Neurons using Neuronal Nuclear Marker NeuN. Nuclear staining (DAPI) is shown in blue (a), with cytoplasmic staining (GFP) in green (b) and neuronal nuclei (NeuN) in red (c). The merged images (d) are combined, showing the relative abundance of neurons. Please click here to view a larger version of this figure.
| Media | Storage | Ingredients | Preparation | ||
| HABG | 4 °C/24h | 100 mL | – Thaw B27 in 4 °C fridge O/N – Filter sterilize (0.22 µm) |
||
| Hibernate A | 97.8 mL | ||||
| B27 [2%] | 2 mL | ||||
| GlutaMAX [0.5 mM] | 0.25 mL | ||||
| Neurobasal Media | 4 °C/1 week | 100 mL | – Thaw B27 in 4 °C fridge O/N – Filter sterilize (0.22 µm) - Aliquot 50 mL portions into 50 mL tubes (store at 4 °C) |
||
| Neurobasal A | 97 mL | ||||
| B27 [2%] | 2 mL | ||||
| GlutaMAX [0.5 mM] | 0.25 mL | ||||
| Pen/Strep [1%] | 1 mL | ||||
| Digestion Media | Day of Isolation | 10 mL | – Shake vigorously – Let it sit in 37°C bath for 30 min (ideally prepared 30 min prior to use) – Filter sterilize (0.22 µm) |
||
| HA-Ca | 10 mL | ||||
| Papain | 25 mg | ||||
| GlutaMAX [0.5 mM] | 0.025 mL | ||||
| Density Gradient | Day of Isolation | 1 Gradient | |||
| Layer | OptiPrep | HABG | |||
| (0.13 mL) | (5.29 mL) | ||||
| 1 Bottom | 0.26 mL | 1.24 mL | |||
| 2 | 0.19 mL | 1.31 mL | |||
| 3 | 0.15 mL | 1.35 mL | |||
| 4 Top | 0.11 mL | 1.39 mL | |||
Table 1: Preparation of the Media and Solutions used in this Protocol.
| Coating Substrate | Storage | Ingredients | Preparation | |
| PDL | -20 °C Freeze-thaw once |
Poly-D-Lysine Hydrobromide | 5 mg | – Aliquot 4 mL into 15 mL tubes and store immediately in -20 °C – 0.5 mL/well (24-well plate) |
| Sterile Water | 50 mL | |||
| Laminin | Day of coating | Laminin (1 mg/mL) | 80 µL | – Enough for 8 wells in a 24-well plate – 0.35 mL/well (24-well plate) |
| Neurobasal Medium | 2.72 mL | |||
Table 2: Preparation of the Coating Substrates used to Coat the Wells upon Which the Neurons are Cultured.
Discussion
This technique allows for the reliable culture of spinal cord neurons. Once proficiency in the technique is achieved, it takes approximately 3.5 h to complete. We have been able to carry out the isolation of neurons from 2 separate litters (16 mice total) in approximately 4 h. The key step in feasibility is being able to proficiently extract the spinal cords from the mice. The yield allows for plating several wells and for the ability to test the neurons under various conditions. We have been able to treat the neurons after maturity and to reliably evaluate protein expression using Western blot analysis. Furthermore, using glass slides inside the wells for neuron attachments allows for the further staining analysis of the neurons.
This procedure is advantageous in that it does not require embryonic tissue and the associated labor intensity. Furthermore, it is not necessary to use adult spinal cords to collect enough tissue for adequate neuron isolation. This prevents the factor of age compounding behavior in neuron physiology, as seen with adult mice. However, this technique is not without limitations. As mentioned earlier, aged neurons are excluded by using neonatal mice, and this can be a limitation if age-related behavior and biology is the area of investigation. There is a learning curve to the procedure, which is necessary not only to optimize the yield, but to ensure that the isolated neurons are healthy. The spinal cord extraction technique can take some time to master, especially with newly born mice. We have noticed that prolonged time associated with the extraction of the spinal cords leads to neurons that can be cultured for a significantly shorter period. This is somewhat alleviated by the placement of the neurons on ice; however, time still plays a key role.
There are key steps in the procedure that help to ensure the optimal yield of neurons. If the neuron yield is not as expected, a few steps should be evaluated. First, the number of spinal cords should ideally be more than 4. When 3 spinal cords or less are used, the yield is usually less than 1 million neurons. Adequate preparation of the digestion medium must be ensured (step 4.5). Leaving the tissue for too long in the digestion medium will cause excessive breakdown of cells, while using digestion medium that has not been activated at 37 °C will result in fewer isolated neurons. Finally, the trituration technique (step 4.7) must be carried out with precision. Low yields usually result from inadequate trituration. A good indicator is seeing whether the solution becomes cloudy during the trituration – if this does not happen, it is likely that the trituration is too gentle.
If the yield is as expected but the neurons do not appear to regenerate processes within the first 1-2 days, the problem is usually a failure in proper coating, with either inadequate washing or drying (step 2.2). If the cells initially project processes but subsequently become apoptotic before the 5-7 day mark, there could be several issues. First, ensure that the HABG solution is prepared within 24 h of isolation and that fresh B27 supplement that has been thawed out on the same day of HABG preparation is used. The antioxidants in this supplement need to be fresh and not expired. Next, ensure that no bubbles are introduced during the trituration process (step 4.7). The introduction of air bubbles contributes to this issue and may be related to the drying of the neurons. Also, ensure that the culture is not overwhelmed with contamination, which can be attributed to failures in sterility.
Here, we describe the use of 24-well plates upon which to seed the neurons. Sometimes, it is necessary to carry out time and/or dosage trials, such as when using pharmacological agents. The technique can be modified and the neurons can be seeded on to 48-well plates instead. Since the surface area of a well in a 48-well plate is half of that in a 24-well plate, the volumes of coating solutions used should be cut in half. The cells should also be coated at half the volume (i.e. 0.5 mL instead of 1.0 mL). However, their concentration should remain the same (i.e. 1-5 x 105 cells/mL). While this study has been carried out using C57BL/6 mice, the technique should be applicable across other genetically manipulated strains.
Spinal cord ischemia and protection is an area of growing interest12,13,14. Clinically, thoracoabdominal aortic surgery can result in devastating paralysis related to spinal cord ischemia and reperfusion, the pathophysiology of which remains to be completely understood15,16. Understanding how neurons respond to the stress of ischemia is feasible using this model. Our group has previously published on spinal cord neuronal injury related to ischemic insult through oxygen glucose deprivation using this technique17. The model has also been extended to include the culture of astrocytes, separate from neurons18.
In conclusion, this technique is valuable for the study of spinal cord pathophysiology and neuronal microbiology. The serum-free culture allows for precise environmental control. It enables direct access for the application of pharmacological agents. Combined with additional techniques, such as oxygen glucose deprivation, this technique can be useful to expand our understanding of the pathophysiology of determinantal spinal cord ischemia.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors have no acknowledgements.
Materials
| Hibernate A Medium – 500 mL | Thermo-Fisher | A1247501 | https://www.thermofisher.com/order/catalog/product/A1247501 |
| Hibernate A Minus Calcium – 500 mL | Brainbits | HA-Ca | http://www.brainbitsllc.com/hibernate-a-minus-calcium/ |
| Glutamax 100X – 100 mL | Thermo-Fisher | 35050061 | https://www.thermofisher.com/order/catalog/product/35050079 |
| B27 Supplement 50X – 10 mL | Thermo-Fisher | 17504044 | https://www.thermofisher.com/order/catalog/product/17504044 |
| Papain, Lyophilized – 100 mg | Worthington | LS003119 | http://www.worthington-biochem.com/pap/cat.html |
| Neurobasal A Medium – 500 mL | Thermo-Fisher | 10888022 | https://www.thermofisher.com/order/catalog/product/10888022 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo-Fisher | 15140122 | https://www.thermofisher.com/order/catalog/product/15140122 |
| Poly-D-lysine hydrobromide – 5 mg | Sigma-Aldrich | P6407-5MG | http://www.sigmaaldrich.com/catalog/product/sigma/p6407?lang=en®ion=US |
| Mouse Laminin – 1 mg | Thermo-Fisher | 23017015 | https://www.thermofisher.com/order/catalog/product/23017015 |
| Trypan Blue – 20 mL | Sigma-Aldrich | T8154-20ML | http://www.sigmaaldrich.com/catalog/product/sigma/t8154?lang=en®ion=US |
| OptiPrep Density Gradient Medium – 250 mL | Sigma-Aldrich | D1556-250ML | http://www.sigmaaldrich.com/catalog/product/sigma/d1556?lang=en®ion=US |
| Dichlorodimethylsilane (DMDCS, Sigma Silicoat) | Sigma-Aldrich | 440272-100ML | http://www.sigmaaldrich.com/catalog/product/aldrich/440272?lang=en®ion=US |
| Chloroform | Sigma-Aldrich | 288306-1L | http://www.sigmaaldrich.com/catalog/product/sial/288306?lang=en®ion=US |
| Glass Pippette – 9" | Sigma-Aldrich | 13-678-20C | http://www.sigmaaldrich.com/catalog/product/sigma/cls7095d9?lang=en®ion=US |
| Pipette bulb – 5 mL | Sigma-Aldrich | Z186678-3EA | http://www.sigmaaldrich.com/catalog/product/aldrich/z186678?lang=en®ion=US&cm_sp=Insite-_-prodRecCold_xviews-_-prodRecCold10-1 |
| BRAND® Petri dish, glass – 60×15 mm | Sigma-Aldrich | BR455717-10EA | http://www.sigmaaldrich.com/catalog/product/aldrich/br455717?lang=en®ion=US |
| Sterile 24 Well Cell Culture Plate | Sigma-Aldrich | M8812-100EA | http://www.sigmaaldrich.com/catalog/product/sigma/m8812?lang=en®ion=US |
| Hausser Hemacytometer (glass counting chamber) | Fischer Scientific | 02-671-6 | https://www.fishersci.com/shop/products/hausser-bright-line-phase-hemacytometer-hemacytometer/026716 |
| Glass Slides – 12 mm sterile cover glass – uncoated | Neuvitro | GG-12-1.5-Pre | http://www.neuvitro.com/german-coverslip-12mm-diameter.htm |
| NeuN Rabbit Monoclonal Antibody – 100 µL | Abcam | ab177487 | After fixing in paraformaldehyde and blocking with 5% BSA, cells on a 12 mm coverslip were incubated in the antibody diluded to 1:200 for 18 hours in 4 °C |
| MAP-2 Mouse Monoclonal Antibody – 50 µL | Abcam | ab11267 | After fixing in paraformaldehyde and blocking with 5% BSA, cells on a 12 mm coverslip were incubated in the antibody diluded to 1:500 for 18 hours in 4 °C |
References
- Qayumi, A. K., et al. Animal model for investigation of spinal cord injury caused by aortic cross-clamping. J Invest Surg. 10 (1-2), 47-52 (1997).
- Fang, B., et al. Ischemic preconditioning protects against spinal cord ischemia-reperfusion injury in rabbits by attenuating blood spinal cord barrier disruption. Int J Mol Sci. 14 (5), 10343-10354 (2013).
- Taira, Y., Marsala, M. Effect of proximal arterial perfusion pressure on function, spinal cord blood flow, and histopathologic changes after increasing intervals of aortic occlusion in the rat. Stroke. 27 (10), 1850-1858 (1996).
- Stoppini, L., Buchs, P. -. A., Muller, D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 37 (2), 173-182 (1991).
- Ahlemeyer, B., Baumgart-Vogt, E. Optimized protocols for the simultaneous preparation of primary neuronal cultures of the neocortex, hippocampus and cerebellum from individual newborn (P0. 5) C57Bl/6J mice. J Neurosci Methods. 149 (2), 110-120 (2005).
- Brewer, G. J., Torricelli, J. R. Isolation and culture of adult neurons and neurospheres. Nat Protoc. 2 (6), 1490-1498 (2007).
- Banker, G. A., Cowan, W. M. Rat hippocampal neurons in dispersed cell culture. Brain Res. 126 (3), 397-425 (1977).
- Graber, D. J., Harris, B. T. Purification and culture of spinal motor neurons from rat embryos. Cold Spring Harb Protoc. 2013 (4), 319-326 (2013).
- Anderson, K. N., Potter, A. C., Piccenna, L. G., Quah, A. K., Davies, K. E., Cheema, S. S. Isolation and culture of motor neurons from the newborn mouse spinal cord. Brain Res Prot. 12 (3), 132-136 (2004).
- Gingras, M., Gagnon, V., Minotti, S., Durham, H. D., Berthod, F. Optimized protocols for isolation of primary motor neurons, astrocytes and microglia from embryonic mouse spinal cord. J Neurosci Methods. 163 (1), 111-118 (2007).
- Brewer, G. J., et al. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 35 (5), 567-576 (1993).
- Fang, B., et al. Ischemic preconditioning protects against spinal cord ischemia-reperfusion injury in rabbits by attenuating blood spinal cord barrier disruption. Int J Mol Sci. 14 (5), 10343-10354 (2013).
- Su, M., Zhong, W., Ren, S. Dose-dependent protection of reseveratrol against spinal cord ischemic-reperfusion injury in rats. Trop J Pharm Res. 15 (6), 1225-1233 (2016).
- Haapanen, H., et al. Remote ischemic preconditioning protects the spinal cord against ischemic insult: An experimental study in a porcine model. J Thorac Cardiovasc Surg. 151 (3), 777-785 (2016).
- Conrad, M. F., Ye, J. Y., Chung, T. K., Davison, J. K., Cambria, R. P. Spinal cord complications after thoracic aortic surgery: long-term survival and functional status varies with deficit severity. J Vasc Surg. 48 (1), 47-53 (2008).
- Wong, D. R., et al. Delayed spinal cord deficits after thoracoabdominal aortic aneurysm repair. Ann Thorac Surg. 83 (4), 1345-1355 (2007).
- Freeman, K. A., et al. Alpha-2 agonist attenuates ischemic injury in spinal cord neurons. J Surg Res. 195 (1), 21-28 (2015).
- Freeman, K. A., et al. Spinal cord protection via alpha-2 agonist-mediated increase in glial cell-line-derived neurotrophic factor. J Thorac Cardiovasc Surg. 149 (2), 578-586 (2015).

