Force-Clamp Rheometry for Characterizing Protein-based Hydrogels
Summary
A new force-clamp rheometry technique is used to investigate the mechanical properties of low-volume protein-based hydrogel samples tethered between a voice-coil motor and a force sensor. An analog proportional-integral-derivative (PID) system allows for the 'clamping' of the force experienced to the desired protocol.
Abstract
Here, we describe a force-clamp rheometry method to characterize the biomechanical properties of protein-based hydrogels. This method uses an analog proportional-integral-derivative (PID) system to apply controlled-force protocols on cylindrical protein-based hydrogel samples, which are tethered between a linear voice-coil motor and a force transducer. During operation, the PID system adjusts the extension of the hydrogel sample to follow a predefined force protocol by minimizing the difference between the measured and set-point forces. This unique approach to protein-based hydrogels enables the tethering of extremely low-volume hydrogel samples (< 5 µL) with different protein concentrations. Under force-ramp protocols, where the applied stress increases and decreases linearly with time, the system enables the study of the elasticity and hysteresis behaviors associated with the (un)folding of proteins and the measurement of standard elastic and viscoelastic parameters. Under constant-force, where the force pulse has a step-like shape, the elastic response, due to the change in force, is decoupled from the viscoelastic response, which comes from protein domain unfolding and refolding. Due to its low-volume sample and versatility in applying various mechanical perturbations, force-clamp rheometry is optimized to investigate the mechanical response of proteins under force using a bulk approach.
Introduction
Apart from having unique physical properties, protein-based hydrogels hold the promise of revolutionizing force spectroscopy by enabling the measurement of several billion molecules in one 'pull', thus enabling the study of proteins in crowded environments, similar to those encountered in skin and other tissues. Protein domains remain folded inside hydrogels, allowing the study of their biomechanical response to force, binding partners, and chemical conditions. Additionally, the biomechanical response of protein domains inside hydrogels resembles the response seen with single-molecule force spectroscopy techniques. For example, chemical denaturants and oxidizing agents decrease the stability of the folded state, both at the single protein domain level1,2,3 and at the macroscopic level4,5,6,7. Similarly, osmolytes increase the stability of single proteins8,9, leading to a decrease in the viscoelastic response of hydrogels, for the same force conditions7,10.
Several approaches have been implemented to synthesize protein-based hydrogels, by either using physical interactions11,12 or covalent cross-linking4,13. Covalent reactions allow for fixed cross-linking locations and these hydrogels can recover the initial state upon a removal of the mechanical or chemical perturbations. A successful approach for covalent cross-linking relies on forming covalent carbon-carbon bonds between exposed tyrosine amino acids using ammonium persulfate (APS) as an oxidant and a ruthenium (II) salt as an initiator (Figure 1)14. Upon exposure to white light, a solution of concentrated proteins can be turned into a hydrogel. By controlling when the reaction starts, the protein-APS mix can be injected into any casting form, such as polytetrafluoroethylene (PFTE) tubes (Figure 1B and 1C), allowing the use of an extremely small solution volume15. Furthermore, the use of white light to trigger the cross-linking reaction results in a limited bleaching of fluorescent proteins and allows the formulation of composite hydrogels with fluorescent markers (Figure 1). Other protein-based hydrogel formation methods use cross-linking based on the SpyTag-SpyCatcher covalent interaction16, amine cross-linking via glutaraldehyde13, or biotin-streptavidin interactions17.
Dynamic mechanical analysis (DMA) is currently a technique extensively used to study polymer-based hydrogels13,18. While DMA can apply constant force protocols to biomaterials, it requires Young's moduli over 10 kPa, and large sample volumes of more than 200 µL19. Due to these limitations, protein hydrogels are generally too soft to be investigated by this technique. As engineered polyproteins are harder to synthesize than polymers, since they require a living system to produce, such high volumes are inefficient, at best4,15. Furthermore, most biological tissues are softer than 10 kPa. Several approaches were developed for biological samples, especially in the study of muscle elasticity20,21. These techniques can also operate under feedback to apply constant force but are optimized for samples with small diameters (in the micron range) exposed to force for very short times (typically less than 1 s).
Protein-based hydrogels were successfully studied with modified rheometry techniques. For example, casting the hydrogel in a ring shape allows the use of extensional rheometry to measure the change in the experienced force as a function of extension4,22. Other approaches for studying the rheological properties of protein-based hydrogels use controlled shear-stress rheometry. These techniques can also achieve low sample volume and tolerate soft materials. However, these methods lack the ability to mimic the pulling forces that cause protein unfolding in vivo, and Young's modulus is calculated based on complex theories that require various assumptions and corrections23.
We have recently reported a new approach that utilizes a small volume of proteins, polymerized inside tubes with diameters < 1 mm. Our first implementation of this technique was operating in length-clamp mode, where the gel was extended following the desired protocol15. In this method, the proteins experience a continuous change in both extension and force while the domains unfold, making the data interpretation cumbersome. Recently, we have reported a new force-clamp rheometry technique, where a feedback loop can expose low-volume protein hydrogels to a predefined force protocol7 (Figure 2). An analog PID system compares the force measured by the force sensor with the set point sent from the computer and adjusts the gel extension by moving the voice coil to minimize the difference between the two inputs. This 'clamping' of the force now allows for new types of experiments to measure the biomechanics of protein hydrogels.
In the force-ramp mode, a tethered protein hydrogel experiences a constant increase and decrease of force with time. The PID compensates for any viscoelastic deformation by changing the extension in a non-linear way, depending on the type of protein and hydrogel formulation. The main advantage of force ramp is that it allows the quantification of standard parameters, such as Young's modulus and energy dissipation, due to an unfolding and refolding of protein domains.
In constant-force mode, the applied force changes in a step-like fashion. In this mode, the gel extends and contracts elastically when the force is increased or decreased, respectively, followed by a time-dependent deformation. This viscoelastic deformation, taking place while the gel experiences a constant force, is directly related to domain unfolding/refolding. In a simplified way, this extension can be seen as the equivalent of several billion single molecule traces averaged together and measured all at once. Constant-force protocols can be used to study the creep and relaxation of protein hydrogels as a function of force and time. As a function of force, for BSA-based protein hydrogels, we have recently shown that there is a linear dependency between the elastic and viscoelastic extension and recoil with the applied strain7.
Here we detail the operation of a force-clamp rheometer using composite gels made from a mixture of protein L (8 domains24, depicted as L8) and a protein L-eGFP construct (L-eGFP), which makes the overall hydrogel fluorescent and easy to demonstrate.
Protocol
1. Reagents Solution Preparation
- Prepare a starting protein solution by dissolving/diluting the protein of interest to the desired concentration, using a Tris buffer [20 mM tris(hydroxymethyl)aminomethane and 150 mM NaCl, pH 7.4].
NOTE: The smallest protein concentration for which cross-linking leads to hydrogels depends on the protein used and is typically > 1 mM. - Prepare stocks of ammonium persulfate (APS) (1 M) and tris(bipyridine)ruthenium(II) chloride ([Ru(bpy)3]2+) (6.67 mM) solutions by dissolving APS and [Ru(bpy)3]2+ powders in the Tris buffer.
2. Protein-based Hydrogel Synthesis
- Fix a 23 G needle on a 1 mL syringe with a pressed plunger.
- Cut a 10 cm polytetrafluoroethylene (PTFE) tube (with an inner diameter of 0.022 in and an outer diameter of 0.044 in) using a razor blade. Attach the needle and syringe to one end of the PTFE tube.
- Insert the second end of the tube into a silane solution and fill the tube by retracting the syringe plunger. Leave the tube for ~30 min.
- Remove the silane solution and dry the tube with compressed air.
NOTE: Make sure that all of the silane solution is dried and that no residue is left in the tube. - Mix the protein solution with APS and [Ru(bpy)3]2+ in a 1.5 mL tube using a constant volume ratio [e.g., 15:1:1 or 15:0.5:0.5 (v:v:v)].
- Vortex the photoactive solution until it is mixed completely.
- Centrifuge the mix at maximum speed (e.g., 14,000 x g) to remove any bubbles from the solution.
- Insert the open end of the treated PTFE tube into the photoactive mixture and draw the solution into the tube by retracting the syringe plunger.
- Place the loaded tube ~10 cm away from a 100 W mercury lamp to prevent heating it and keep it there for up to 30 min at room temperature (Figure 1B).
NOTE: In some cases, the exposure time to light can be as low as 30 s. Shorter gelation times are used here for fluorescent gels, to limit photobleaching. - Remove the tube from the needle and cut the edges of the tube near the hydrogel ends with a razor blade.
- Use a blunted 24 G needle to extrude the hydrogel into the Tris solution (Figure 1C).
NOTE: Blunted needles are used to avoid any notches or damage to the hydrogel sample. - Visually inspect the gels for any defects that might form during the extrusion or due to bubbles and discard the gels with defects.
3. Protein-based Hydrogel Attachment and Force-Clamp Rheometer Set-up
- Start the instrument control program. Turn on the voice-coil motor. Set the coil position to a value toward the end of the range (e.g., 7.5 mm).
NOTE: The voice-coil position is recommended to be toward the end of the maximum movement range, to maximize the possible extension of the hydrogel. - Displace the hooks in the z-direction and align them at the bend in the x-direction (which is the pulling coordinate; see Figure 2B). Record the values of the micrometer screws for the x-direction.
- Cut 2 sterile sutures into strands of equal length (2 – 3 cm; see Figure 3A and B).
- Tie a loose double overhand knot into each of the strands and place the 2 loops on the hook connected to the force sensor (Figure 3C and 3D).
- Fill the experimental chamber with Tris buffer and transfer the hydrogel sample into the filled chamber using medical tweezers.
- Place the voice coil and force sensor hooks close to the solution surface and align the hooks in all directions using the x/y/z-positioning manipulators.
- Using medical tweezers, hang both sides of the protein hydrogel sample on the hooks connected to the voice coil and force sensor (Figure 3C).
- Tighten 1 suture loop around the hydrogel sample on the voice coil hook by holding both ends of the suture loop with medical tweezers and pulling them simultaneously (Figure 3D).
- Repeat step 3.8 for the loop connected to the force sensor (Figure 3D).
NOTE: Avoid an extreme tightening of the sutures to prevent any structural damage and transversal cutting of the hydrogel sample. - Tighten the suture loops on the bends of each hook to prevent any slippage; use these bends as reference points to find the zero separation between the hooks in step 3.2. Cut the excess lengths of the sutures using medical scissors (Figure 3D).
- Move the attached hydrogel using z-manipulators along the z-axis toward the experimental chamber to immerse the hydrogel in the experimental solution.
- Align the hydrogel sample in y–z using the manipulators such that the gel is not under any stress.
- Zero the force sensor and separate the two hooks using the x-micrometer stages until the gel starts to experience force. Once this happens, slightly turn back the micrometer screw in the x-direction.
- Record the position of both manipulators for the voice coil motor and the sensor and use the difference between these values and the ones measured in step 3.2 to calculate the exact separation between the tethering hooks at the start of the experiment.
- Set the range for the slack curve to ~1.5 – 2 mm and measure the gel slack (Figure 4A).
NOTE: For each slack measurement, try to keep the start of the slack regime near the initial voice coil position, allowing for an optimal number of data points to fit the 2 regimes (Figure 4A). The gel length can be determined with a micron resolution using the separation between the hooks and the intersection between the 2 regimes in the slack curve (see also step 5.1). As the force sensor might drift with time due to variations in the experimental conditions, the part of the slack curve where the gel is not under force reports on this possible drift. The program controlling the instrument compensates automatically for this difference when sending the set-point command to the PID loop (Figure 4A inset).
4. Protein-based Hydrogel Characterization using Controlled Force-Ramp and Constant-Force Measurements
- Force-ramp experiments
- To perform a force-ramp cycle by increasing the force at the desired loading rate (e.g., 0.01 mN/s), input the starting and final forces and the duration of the protocol as a flipped "V". Then, hold the gel at 0 mN (or low force) for > 200 s to allow the protein domains to refold and the gel elasticity to recover.
- Save the trace.
- Constant-force experiments
- Perform a constant-force protocol by applying a low force (e.g., 0.1 mN) for 30 s and then increase the force to a constant force (e.g., 1 mN) for a defined amount of time (e.g., 120 s), followed by quenching the force back to the same low value (e.g., 0.1 mN) for > 300 s to allow the protein domains to refold and the gel elasticity to recover.
- Following the first pulse, adjust the PID settings to maximize the response time of the feedback loop (see Figure 2D).
NOTE: For stiff gels and for small changes in force, the response time of the loop is limited by the electronics of the force sensor and the response time of the coil, and can be as low as 5 ms7. For softer gels and large changes in force, the response time is dictated by the elasticity of the hydrogels (Figure 2D). - Save the trace.
5. Data Analysis
- Utilizing the measured separation between the hooks and the calculated coil position, when the gel starts to experience force (Δx in the Figure 4A insert), compute the gel length L using the equation:
L = L0 + ∆x
Here, L0 is the separation between the hooks, measured from the position of the micrometer screws before the experiment (step 3.14).
NOTE: For gels with low protein concentrations that do not result in a complete cross-linking, the measured length will change from trace-to-trace. Also, over long periods of time, proteins inside hydrogels might experience aging effects25, which result in an overall lengthening of the gel. - Normalize the measured extension to the gel length to obtain the strain.
- Normalize the measured force to the transversal surface area by using the inner diameter of the tube used for the polymerization.
Representative Results
Figure 1A shows the scheme of the photoactive reaction used to synthesize the L-EGP/L8 hydrogel. Figure 1B shows the hydrogel mixture in the PTFE tube before and after the photoactivation. Figure 1C presents the extruded L-eGFP-L8 hydrogel inside a Tris solution. The hydrogel sample has no structural defects such as notches. Hydrogels with clearly visible damage should be discarded.
The rendering of the assembled and exploded views of the force-clamp rheometer are presented in Figure 2A and 2B. Figure 2C shows the force-clamp rheometer scheme, where the hydrogel sample is tethered between the hooks connected to the linear voice-coil and the force sensor and immersed in a buffer solution. The analog PID system adjusts the hydrogel extension by controlling the linear-voice coil position to follow the force set point. Figure 2D shows the tuning of the PID using various increments for the integral gain.
Figure 3 shows a typical attachment process of a hydrogel sample. After tethering the hydrogel between the aligned hooks, the suture loops are tightened around the hydrogel near the bends to prevent the sample from slipping and to allow for the precise determination of the hydrogel length.
Force-ramp Measurement and Analysis of Protein-based Hydrogels:
Representative measurements of the force-ramp protocol are shown in Figure 4A – 4C. Each new pull starts with a slack measurement, as shown in Figure 4A. Then, the force curve is obtained by applying an inverted "V" protocol as the load increases and decreases linearly with time. Afterward, the hydrogel is held at a 0 mN force for 200 s, to allow the protein domains inside the hydrogel sample to refold (Figure 4B). During stress, the PID system modifies the hydrogel extension represented by the coil position to follow the predefined force set point. For each slack curve, we fit 2 lines (Figure 4C). The blue line is used to fit the first regime when the hydrogel recoils and the orange line is used to fit the regime when the hydrogel becomes slack. The intersection between the two lines is used to calculate the true gel length with a micrometer resolution (Figure 4A). Afterward, the extension of the hydrogel sample is calculated by subtracting the initial coil position from the coil position trace (Figure 4D). Figure 4F presents the stress-strain curve. The stress is calculated by dividing the applied force by the cross-sectional area of the hydrogel sample and the strain is calculated by dividing the extension (Figure 4E) by the true gel length calculated from the slack curve as presented in Figure 4A.
Constant-force Measurement and Analysis of Protein-based Hydrogels:
Representative measurements of a constant-force protocol are shown in Figure 5A – 5D. A constant force of 0.1 mN is applied to the hydrogel sample for 30 s, the force is then changed to 1 mN for 120 s, and finally, the force is quenched back to 0.1 mN for 300 s to allow the protein domains to refold (Figure 5A). During the first 30 s at low force, there is no notable change in the gel extension. When increasing the force to 1 mN, the hydrogel shows a fast elastic extension. Following this initial extension, the hydrogel keeps extending over time, while keeping the force constant (1 mN). Afterward, the force is quenched back to the initial low value (0.1 mN) and the hydrogel recovers to its initial length (Figure 5B). The extension of the hydrogel sample (Figure 5C) and the force are used to calculate the strain (top) and stress (bottom) in a similar fashion as in the force ramp measurements (Figure 5D).
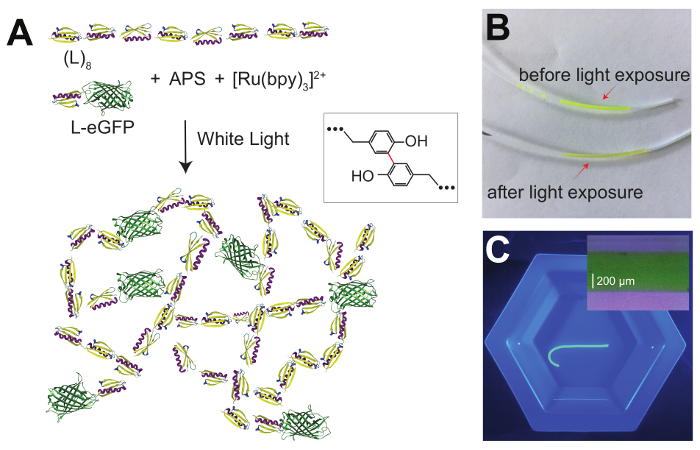
Figure 1: L-eGFP/(L)8-based hydrogel synthesis. (A) This panel shows the schematics of an L-eGFP/(L)8 protein hydrogel synthesis using a photoactivated reaction. The protein is mixed with APS and [Ru(bpy)3]2+ and exposed to white light, which promotes the formation of covalent bonds between adjacent tyrosine amino acids (inset). (B) This panel shows an L-eGFP/(L)8-, [Ru(bpy)3]2+-, and APS-mixture loaded into a PTFE tube using a 23 G needle before exposure to white light (top), and after (bottom). (C) This panel shows an extruded L-eGFP/(L)8-based hydrogel into a Tris solution. The inset shows a magnified image of an L-eGFP/(L)8-based hydrogel. The diameter distribution is 552 ± 8 μm, in agreement with the interior diameter of the PFTE tube used during the polymerization (558 μm). Please click here to view a larger version of this figure.
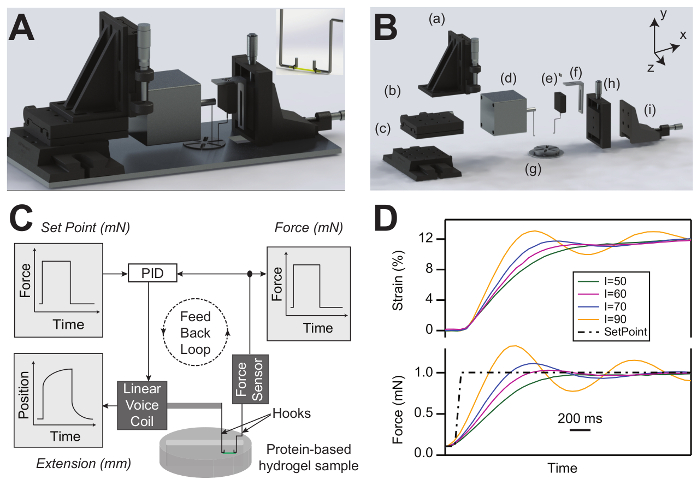
Figure 2: Force-clamp rheometer design and set-up. (A) Rendering of the assembled force-clamp hydrogel rheometer. The inset shows a protein-based hydrogel sample attached to the voice coil and force sensor hooks inside the solution chamber. (B) Rendering of the exploded view of the force-clamp hydrogel rheometer: (a – c) the x–y–z manipulators for adjusting the voice-coil hook position, (d) the linear voice coil motor, (e) the force transducer, (f) the force transducer holder, (g) the solution chamber, and (h – i) the x–y manipulators for adjusting the force transducer position. (C) Scheme of the force-clamp hydrogel rheometer set-up. The scheme shows a protein-based hydrogel sample attached to a force sensor and voice coil hooks using medical sutures. The analog PID system changes the hydrogel length by adjusting the voice-coil position to follow the force set point. (D) PID system response using different integral-gain values (I) to reach the force set point (dashed line). The colored traces represent the measured force (bottom) and strain (top) derived from the PID system response. Please click here to view a larger version of this figure.
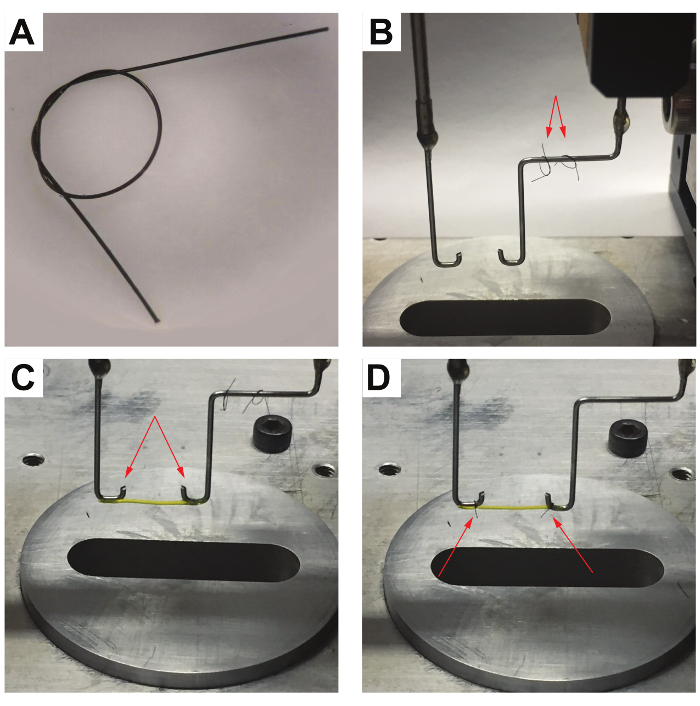
Figure 3: L-eGFP/(L)8-based hydrogel attachment process. (A) Close-up view of a suture loop, tied with a loose double overhand knot used to attach the hydrogel sample to the hooks. (B) The two suture loops are placed on the force sensor hook used in protein-based hydrogel attachment. (C) An L-eGFP/(L)8-based hydrogel sample is hung between the hooks (indicated by the arrows). (D) The suture loops on the side of the voice coil (left) and the force sensor hook (right) are tightened around the hydrogel sample at the bend of each hook to prevent the sample from slipping during the measurements. Afterward, the excess sutures are trimmed using medical scissors (indicated by the red arrows). Please click here to view a larger version of this figure.
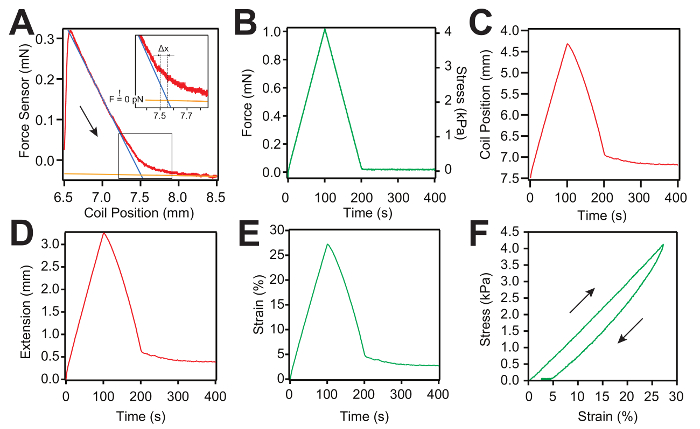
Figure 4: Representative force-ramp measurement and data analysis curves for an L-eGFP/(L)8-based hydrogel sample. (A) Typical slack measurement curve (red) used to determine the zero force of the force sensor and the true length of the hydrogel. Two linear curves (the blue and the orange lines) are used to fit both regimes: first, when the gel is under force (blue line), and second, when the gel becomes slack (plateau – orange line). The intersection between the two lines is used to calculate the true hydrogel length at zero force. The arrow shows the direction of the motion. The inset shows the location of the zero force and the correction of the hydrogel length. (B) Representative force-ramp curve applied to the hydrogel sample. (C) Trace representing the coil position movement as a function of time. The coil starts from the initial position defined in the protocol at step 3.1 (7.5 mm). (D) Representative curve of the extension of the hydrogel sample as a function of time. The extension is calculated as the displacement between the measured coil position and its initial position. (E) Representative strain-versus-time curve. The strain is calculated by dividing the extension by the true gel length calculated from the slack measurement. (F) Representative stress-strain curve of a hydrogel sample. Please click here to view a larger version of this figure.
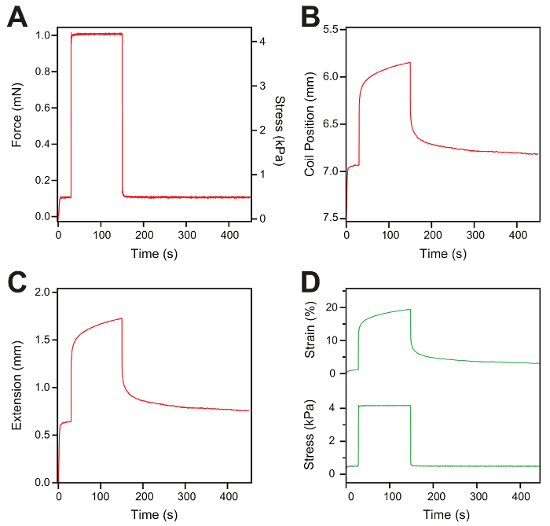
Figure 5: Constant-force measurement and data analysis. (A) Representative trace of the constant-force protocol applied to a hydrogel. The hydrogel is exposed to 0.1 mN for 30 s, then the force is increased to 1 mN for 120 s, and finally, the force is quenched back to 0.1 mN for 300 s. (B) This panel shows a coil position trace versus the time representing the change in the length of the hydrogel sample following the force protocol. (C) This panel shows the gel extension measured from the displacement of the voice-coil. (D) Representative figure of stress (bottom) and strain traces (top) after the data analysis. Please click here to view a larger version of this figure.
Discussion
Herein, we describe a force-clamp rheometry technique to investigate the biomechanical response of low-volume protein-based hydrogels. Additionally, a protocol is provided to synthesize a uniform cylindrical low-volume protein hydrogel sample. A protocol is also presented which describes how to tie different types of protein-based hydrogels with various elasticities without causing any mechanical deformation or damage to the protein-based hydrogel samples or slippage of the gel on the hooks. The analog PID system, together with the linear voice coil and the force sensor, enable the application of controlled-force protocols such as force ramp and constant force. Recently, this technique has been used to study the biomechanical response of different cross-linked concentrations of BSA-based hydrogels in different experimental solutions7.
An important aspect when formulating and working with protein hydrogels is the reproducibility of the measurements. If gels are formulated with a too low protein concentration or incomplete cross-linking, permanent plastic deformations will appear during extension7. These plastic deformations would drastically limit the data interpretation, as the viscoelastic effects would come from both the domain unfolding and the molecular rearrangement inside the gel. An easy test to see if there complete cross-linking is to immerse the hydrogel in chemical denaturants, such as 6 M guanidinium chloride1. In this case, there should not be any viscoelastic effects in the stress-versus-strain curves, as all domains are unfolded chemically, and the molecules are now behaving as simple polymers4,7,26. Furthermore, the gel should recover its initial elasticity when immersed back in the initial buffer7.
If there are variations in the measured response between traces obtained with different gels, several aspects should be considered for troubleshooting: the protein aggregation in solution, a non-homogeneous mixing of the protein with the cross-linking chemicals, the presence of bubbles, the binding of the protein-based hydrogel to the PFTE tube due to incorrect silanization. Residues of silane on the tube walls may contaminate the hydrogel and lead to structural defects. To avoid this error, more compressed air is needed to ensure a total removal of the silane from the tube. Additionally, bubbles can form during the suction of the hydrogel photoactive mix into the PTFE tube. These bubbles can lead to sample damage and affect the biomechanical response of the hydrogel. To prevent any bubble formation, the end of the PTFE tube must be inside solution mix during the loading process and the syringe plunger must be retracted slowly. Another typical error is an over-tightening of the suture loops around the hydrogel samples during the attachment process, which may lead to a notch formation and the cutting of the hydrogel. The moving range of the voice coil limits the maximum extension of the attached hydrogel sample. This limitation has to be taken into account when measuring gels that extend several hundred percent of their initial length. For example, to extend a hydrogel for more than 200%, an initial length of less than 4 mm is required.
Protein-based hydrogels are a unique class of biomaterials due to their biocompatibility and high stretchability derived from proteins, their main building units, and the inherent folding transition that is characteristic for proteins. Additionally, these hydrogels have an excellent potential for tissue engineering, drug delivery systems, and biological ink (bioink) for 3D printing27. The force-clamp hydrogel rheometer can be used to investigate a large variety of proteins. Furthermore, the force-clamp rheometer enables the application of constant-force protocols on low-volume hydrogel samples. These experiments allow the decoupling of the elastic and viscoelastic behaviors and study (un)folding mechanics in a bulk approach.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We acknowledge financial support from Research Growth Initiative (Award No. 101X340), National Science Foundation, Major Research Instrumentation Program (Grant No. PHY-1626450), Greater Milwaukee Foundation (Shaw Award) and University of Wisconsin System (Applied Research Grant).
Materials
| SI-KG4A force transducer | World Precision Instruments (WPI) | SI-KG4A | |
| Linear Voice Coil Motor | Equipement Solutions | LFA2010 | |
| Bovine serum albumin | Rocky Mountain Biologicals (RMBIO) | BSA-AAF-1XG / 100 G | |
| Trizma | Sigma-Aldrich | T1503-1KG | |
| Sodium chloride | Sigma-Aldrich | S7653-1KG | |
| Ammonium persulfate | Sigma-Aldrich | 248614-100G | |
| Tris(bipyridine)ruthenium(II) chloride | Sigma-Aldrich | 544981-1G | |
| EXPRESS MEDICAL SUPPLIES 6-0 NYLON SUTURE 12/PK | Fisher Scientific | NC0395626 | |
| 1mL Syringe Only, Luer-Lok Tip | BD | 309628 | |
| Silane, Sigmacote | Sigma-Aldrich | SL2-25ML | |
| Microbore PTFE Tubing, 0.022"ID x 0.042"OD, 100 ft/roll | Cole-Parmer | EW-06417-21 | |
| Hypodermic Needle, 23 Gauge | Healthcare Supply Pros | 305194 | |
| Jensen Global JG24-1.5X Red IT Dispensing Tips – 24 gauge | KIMCO | JG24-1.5X | |
| USH-103D USHIO 100W Short Arc Mercury Lamp | ALB | USH-103D USHIO | |
| Medical Tweezers | |||
| Medical scissors | |||
| Olympus | |||
| The computer code and CAD design of the custom parts can be made available on request to the corresponding author. |
References
- Cao, Y., Li, H. How do chemical denaturants affect the mechanical folding and unfolding of proteins?. Journal of Molecular Biology. 375 (1), 316-324 (2008).
- Carrion-Vazquez, M., et al. Mechanical and chemical unfolding of a single protein: a comparison. Proceedings of the National Academy of Sciences of the United States of America. 96 (7), 3694-3699 (1999).
- Wiita, A. P., et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 450 (7166), 124-127 (2007).
- Lv, S., et al. Designed biomaterials to mimic the mechanical properties of muscles. Nature. 465 (7294), 69-73 (2010).
- Plumere, N., et al. A redox hydrogel protects hydrogenase from high-potential deactivation and oxygen damage. Nature Chemistry. 6 (9), 822-827 (2014).
- Kong, N., Peng, Q., Li, H. B. Rationally Designed Dynamic Protein Hydrogels with Reversibly Tunable Mechanical Properties. Advanced Functional Materials. 24 (46), 7310-7317 (2014).
- Khoury, L. R., Nowitzke, J., Shmilovich, K., Popa, I. Study of Biomechanical Properties of Protein-Based Hydrogels Using Force-Clamp Rheometry. Macromolecules. 51 (4), 1441-1452 (2018).
- Auton, M., Rosgen, J., Sinev, M., Holthauzen, L. M. F., Bolen, D. W. Osmolyte effects on protein stability and solubility: A balancing act between backbone and side-chains. Biophysical Chemistry. 159 (1), 90-99 (2011).
- Popa, I., Kosuri, P., Alegre-Cebollada, J., Garcia-Manyes, S., Fernandez, J. M. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nature Protocols. 8 (7), 1261-1276 (2013).
- Aioanei, D., Brucale, M., Tessari, I., Bubacco, L., Samori, B. Worm-Like Ising Model for Protein Mechanical Unfolding under the Effect of Osmolytes. Biophysical Journal. 102 (2), 342-350 (2012).
- Wheeldon, I. R., Gallaway, J. W., Barton, S. C., Banta, S. Bioelectrocatalytic hydrogels from electron-conducting metallopolypeptides coassembled with bifunctional enzymatic building blocks. Proceedings of the National Academy of Sciences of the United States of America. 105 (40), 15275-15280 (2008).
- Sathaye, S., et al. Rheology of peptide- and protein-based physical hydrogels: are everyday measurements just scratching the surface?. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 7 (1), 34-68 (2015).
- Ma, X., et al. A Biocompatible and Biodegradable Protein Hydrogel with Green and Red Autofluorescence: Preparation, Characterization and In Vivo Biodegradation Tracking and Modeling. Scientific Reports. 6, 19370 (2016).
- Fancy, D. A., Kodadek, T. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proceedings of the National Academy of Sciences of the United States of America. 96 (11), 6020-6024 (1999).
- Saqlain, F., Popa, I., Fernandez, J. M., Alegre-Cebollada, J. A Novel Strategy for Utilizing Voice Coil Servoactuators in Tensile Tests of Low Volume Protein Hydrogels. Macromolecular Materials and Engineering. 300 (3), 369-376 (2015).
- Sun, F., Zhang, W. B., Mahdavi, A., Arnold, F. H., Tirrell, D. A. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proceedings of the National Academy of Sciences of the United States of America. 111 (31), 11269-11274 (2014).
- Thompson, M. S., et al. Self-assembling hydrogels crosslinked solely by receptor-ligand interactions: tunability, rationalization of physical properties, and 3D cell culture. Chimie. 21 (8), 3178-3182 (2015).
- Kocen, R., Gasik, M., Gantar, A., Novak, S. Viscoelastic behaviour of hydrogel-based composites for tissue engineering under mechanical load. Biomedical Materials. 12 (2), (2017).
- Desai, M. S., et al. Elastin-Based Rubber-Like Hydrogels. Biomacromolecules. 17 (7), 2409-2416 (2016).
- Fusi, L., Brunello, E., Yan, Z., Irving, M. Thick filament mechano-sensing is a calcium-independent regulatory mechanism in skeletal muscle. Nature Communications. 7, (2016).
- McDonald, K. S. Ca2+ dependence of loaded shortening in rat skinned cardiac myocytes and skeletal muscle fibres. Journal of Physiology-London. 525 (1), 169-181 (2000).
- Wu, J. H., et al. Rationally designed synthetic protein hydrogels with predictable mechanical properties. Nature Communications. 9, (2018).
- Bharadwaj, N. A., Ewoldt, R. H. Single-point parallel disk correction for asymptotically nonlinear oscillatory shear. Rheologica Acta. 54 (3), 223-233 (2015).
- Valle-Orero, J., Rivas-Pardo, J. A., Popa, I. Multidomain proteins under force. Nanotechnology. 28 (17), 174003 (2017).
- Valle-Orero, J., et al. Mechanical Deformation Accelerates Protein Ageing. Angewandte Chemie International Edition. 56 (33), 9741-9746 (2017).
- Fang, J., et al. Forced protein unfolding leads to highly elastic and tough protein hydrogels. Nature Communications. 4, (2013).
- Gungor-Ozkerim, P. S., Inci, I., Zhang, Y. S., Khademhosseini, A., Dokmeci, M. R. Bioinks for 3D bioprinting: an overview. Biomaterial Science. , (2018).

