Alveolar Macrophage Phagocytosis and Bacteria Clearance in Mice
Summary
Here we report common methods to analyze the phagocytic function of murine alveolar macrophages and bacterial clearance from the lung. These methods study in vitro phagocytosis of fluorescein isothiocyanate beads and in vivo phagocytosis of Pseudomonas aeruginosa Green Fluorescent Protein. We also describe a method for clearing P. aeruginosa in mice.
Abstract
Alveolar macrophages (AMs) guard the alveolar space of the lung. Phagocytosis by AMs plays a critical role in the defense against invading pathogens, the removal of dead cells or foreign particles, and in the resolution of inflammatory responses and tissue remodeling, processes that are mediated by various surface receptors of the AMs. Here, we report methods for the analysis of the phagocytic function of AMs using in vitro and in vivo assays and experimental strategies to differentiate between the pattern recognition receptor-, complement receptor-, and Fc gamma receptor-mediated phagocytosis. Finally, we discuss a method to establish and characterize a P. aeruginosa pneumonia model in mice to assess bacterial clearance in vivo. These assays represent the most common methods to evaluate AM functions and can also be used to study macrophage function and bacterial clearance in other organs.
Introduction
AMs are the major resident phagocytes in the alveoli at the resting stage and one of the major players of innate immune responses through the recognition and internalization of inhaled pathogens and foreign particles1,2. It has been reported that AMs are essential for the rapid clearance of many pulmonary pathogens such as P. aeruginosa and Klebsiella pneumonia3,4, so a deficiency in AM phagocytosis often results in respiratory infections, such as acute pneumonia, which cause higher mortality and morbidity rates.
AMs also initiate innate inflammatory responses in the lung by producing cytokines and chemokines such as TNF-α and IL-1β, which crosstalk with other cells of the alveolar environment to produce chemokines and recruit inflammatory neutrophils, monocytes, and adaptive immune cells in the lung5. For example, IL-1β produced by AMs helps to prime the release of the neutrophil chemokine CXCL8 from epithelial cells6. Moreover, AMs contribute to the phagocytosis of apoptotic polymorphonuclear leukocytes (PMNs), failure of which leads to the sustained leakage of intracellular enzymes from PMNs to the surrounding tissue, resulting in tissue damage and prolonged inflammation7,8,9.
Phagocytosis by the AMs is mediated by a direct recognition of pathogen-associated molecular patterns at the pathogen surface by the pattern recognition receptors (PRRs) of the AMs or by the binding of opsonized pathogens with immune effector receptors of the AMs10. For the latter, AMs can recognize the targets opsonized with immunoglobulin (IgG) through their Fcγ receptors (FcγR) or the pathogens coated with complement fragments, C3b and C3bi, through their complement receptors (CR)11. Among complement receptors, the CR of the immunoglobulin superfamily (CRIg) is selectively expressed in tissue macrophages12, and a recent finding highlighted the role of the CRIg in AM phagocytosis in the context of P. aeruginosa pneumonia13.
Many original studies use methods to evaluate macrophage phagocytosis to describe the molecular mechanisms of macrophage function14,15. However, methods like in vivo phagocytosis require a precise quantification of phagocytosis. Here, we summarize a detailed methodology for both in vitro and in vivo phagocytosis using fluorescein isothiocyanate (FITC)-glass beads and P. aeruginosa green fluorescent protein (GFP), respectively. Further, we explain the method of differentiating among PRR-, CR-, and FcγR-mediated phagocytosis. Finally, we report a method to characterize bacterial clearance in mouse with respect to P. aeruginosa pneumonia.
Protocol
This protocol follows the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Eastern Virginia Medical School.
1. Fluorescent Beads Phagocytosis
- Euthanize the mouse (C57BL/6J, 6 weeks old, female) by CO2 asphyxiation as per IACUC protocols for the ethical euthanasia of animals.
- Lay the mouse belly-up on a dissection board covered with paper towels. Pin its paws down with its limbs spread-eagle and hook a string under its front teeth to pull its head back so that the trachea is positioned straight and level.
- Wet the mouse's throat, chest, and belly with 70% ethanol to disinfect and prevent the fur from sticking to the tools.
- Using regular forceps, pull up the skin at the centerline of the body, and cut with surgical scissors up the centerline to the top of the throat.
- Using the blunt end of standard surgical scissors, carefully move away the muscle and connective tissues on the throat and use spring scissors (microscissors) to expose the trachea.
- Gripping a cartilage ring with the forceps, carefully make a small incision (~1.5 mm), using microscissors, on the ventricle face of the trachea and insert an 18 G cannula into the trachea.
- Gently lavage 3 mL of phosphate-buffered saline (PBS), 1 mL at a time. Each time gently withdraw the fluid into the syringe and reinfuse it back into the lung, 3x in succession. After collecting the BALF, perform cervical dislocation to ensure euthanasia. Transfer the collected PBS (~2.8 mL), which is bronchoalveolar lavage fluid (BALF), to a tube, centrifuge at 1,000 x g for 10 min, and collect the pellet. Add 1 mL of fresh PBS to the tube and centrifuge at 1,000 x g for 10 min to wash the debris and collect the pelleted alveolar macrophages.
- Resuspend the pellet in 2 mL of Dulbecco's modified Eagle's medium (DMEM) with 10% nonheat-inactivated fetal bovine serum (FBS) and culture primary alveolar macrophages in the same media for 2 days on a glass-bottom dish at 37 °C in a humidified atmosphere.
- Aspirate the old media, wash it with 1 mL of PBS, and add 2 mL of fresh media. Add FITC beads (carboxylated latex beads, 2 µm in diameter, 50 beads/cell) and incubate for 1 h at 37 °C in a humidified atmosphere.
- Wash extensively with PBS (1 mL at a time, for a total of five washes) to remove extracellular beads. Image 100 cells randomly and count the cells with intracellular beads (488 nm).
NOTE: Phagocytic indexes are the number of ingested beads divided by the total number of macrophages; the percentage of phagocytic cells is the number of macrophages that ingest at least one bead divided by the total number of macrophages16.- Alternatively, after a 1 h incubation with beads, wash the cells with 3 mL of PBS and process them for flow cytometry for the quantification of phagocytosis. Similarly, process AMs without beads as unstained cells or control cells. Calculate the percentage positivity and mean fluorescence intensity, using flow cytometry software, by selecting those options in the software.
2. FcγR- and CR-mediated Phagocytosis
- For the opsonization, incubate 2 x 108 sheep red blood cells (SRBCs) with 50 µL of rabbit anti-SRBC-IgM or 50 µL of rabbit anti-SRBC-IgG for 30 min at room temperature11.
- Incubate IgM-opsonized SRBCs with 50 µL of C5-deficient (C5D) human serum for 30 min at 37 °C to fix the complement fragments C3b and C3bi on IgM-coated SRBCs.
- Seed murine macrophage cells (MH-S cells) (10,000 cells/well) in a 96-well plate and incubate overnight to get a ~70% confluence. Add 100 µL of 1 x 107/mL opsonized SRBCs to each well of MH-S cells and incubate for 1 h at 37 °C. Wash unbound SRBCs very quickly (~1 min) with 100 µL of ammonium chloride-potassium (ACK) lysis buffer.
- Lyse the cells with 0.1% SDS and add 50 µL of 2,7-diaminofluorene (DAF) containing 3% hydrogen peroxide and 6 M urea. Measure the absorbance of the hemoglobin-catalyzed fluorene blue formation at 620 nm.
- Determine the number of SRBCs by using a standard curve at 620 nm absorbance values with a known number of SRBCs. Similarly, process MH-S cells incubated with nonopsonized SRBCs to use as negative controls.
3. PRR-mediated Phagocytosis
- Follow steps 1.1-1.9 for the isolation and culturing of mouse primary alveolar macrophages.
- After 2 days, remove the media, wash the cells with 1 mL of PBS, and add 500 µL of fresh media containing Alexa Fluor-488-conjugated zymosan-A bioparticles (100 particles/dish).
- Incubate for 1 h at 37 °C. Stop the phagocytosis by adding 500 µL of ice-cold PBS.
- Wash the cells extensively with PBS (1 mL at a time, for a total of five washes). Fix the cells with 4% paraformaldehyde for 10 min at room temperature.
- Wash the cells extensively with PBS (1 mL at a time, for a total of five washes) and keep the cells in 500 µL of PBS.
- Image the cells under differential interference contrast and a fluorescent channel at 488 nm. Count AMs containing zymosan-A bioparticles and determine the percentage of phagocytosis.
4. In Vivo Phagocytosis by Alveolar Macrophages
NOTE: Inoculate P. aeruginosa GFP on a nutrient agar plate and incubate the plate at 37°C overnight. On the next day, inoculate the single colony to 2 mL of nutrient broth and grow the bacteria at 37°C overnight.
- The next day, anesthetize mice with an intraperitoneal administration of 100 mg/kg ketamine and 10 mg/kg xylazine. Confirm proper anesthetization via a lack of response to the toe pinch.
- Lay the mouse on a flat board with a rubber band across the upper incisors and place it in a semirecumbent (45°) position with the ventral surface and rostrum facing upward. Using curved forceps, partially retract the tongue. Using a microsprayer, intratracheally administer 50 µL (5 x 106 colony-forming units [CFU])17 of P. aeruginosa GFP into the lungs of the anesthetized mice.
- After 1 h of infection, follow steps 1.1-1.7.
- Resuspend the cells in PBS and cytocentrifuge them (1,000 x g, 1 min at room temperature) onto a glass slide.
- Differentially stain the cytospin slides for alveolar macrophages, neutrophils, and lymphocytes, according to the manufacturer's instructions.
- Randomly select 100 AMs, count the AMs containing intracellular bacteria, and determine the percentage of phagocytosis.
5. In Vivo Bacteria Clearance Using P. aeruginosa
- Inoculate P. aeruginosa on a P. aeruginosa isolation agar plate and incubate the plate at 37°C overnight. Inoculate the single colony to 2 mL of lysogeny broth (LB) and grow the bacteria at 37°C overnight. Calculate the CFU, using the following formula.
CFU/mL = (number of colonies x dilution factor)/volume of the culture plate.
Dilute the culture with PBS to get the desired CFU/mL. - In trial 1, intratracheally inject a sublethal dose of ~2.5 x 105 CFU/mL P. aeruginosa into anesthetized wild-type (WT) and TRIM72KO mice, as stated in step 4.2. Measure the body weight daily for 6 days.
- In trial 2, inject a second dose of P. aeruginosa (5 x 105 CFU/mL) into the mice that survived in trial 1 and measure the body weight for 4 days.
- In trial 3, using a different set of mice, inject WT and TRIM72KO mice with a lethal dose (3 x 107 CFU/mL) of P. aeruginosa and record the mortality within 2 days of injection. Animals showing signs of severe stress such as significant body weight loss, hunched back, anorexia, or dyspnea will be assessed by the attending vets. Untreatable animals will be sacrificed immediately.
- Either at death or after euthanasia at day 2 after the injection, collect the whole-lung tissue for the quantification of the lung bacterial burden at peak infection.
- To test the lung bacterial burden, add 200 µL of normal saline to the lung tissues and homogenize them, using a previously tested setting on an electronic homogenizer that completely disrupts the lung tissue without breaking bacteria. Adjust the total volume of the lung homogenate to 1 mL and plate 100 µL of lung lysate on Pseudomonas isolation agar plates at 10-fold serial dilutions.
- Incubate the plates at 37 °C for 24 h and count the bacterial colonies to determine the CFU per whole lung.
6. Statistical Analysis
- Use Student's t-test to determine the statistical significance of the difference between the two groups. Consider a difference statistically significant when p < 0.05. All data are presented as means ± standard error of the mean (SEM).
Representative Results
We first performed the experiment to analyze phagocytosis by mouse primary AMs. Throughout all analyses, we compared AMs isolated from WT and TRIM72KO mice. As shown in Figure 1A, fluorescence microscopy revealed that phagocytosis of FITC-glass beads by mouse primary AMs occurs after 1 h of incubation. Figure 1B shows the analysis of phagocytosis by flow cytometry. The quantification of phagocytosis measured by microscopy and flow cytometry is represented in Figure 1C and Figure 1D, respectively. FcγR- and CR-mediated phagocytosis by MH-S cells is represented in Figure 2A, and Figure 2B shows the quantification. These results show that the expression of TRIM72 in MH-S cells resulted in a more than fivefold decrease in complement phagocytosis. Representative images of Alexa Fluor-488-conjugated zymosan-A particle ingestion by primary AMs isolated from WT or TRIM72KO mice are shown in Figure 2C, and the quantification is presented in Figure 2D. In vivo phagocytosis results are represented in Figure 3. Figure 3A shows differential staining identifying the presence of AMs, neutrophils, and lymphocytes, and GFP+ phagocytic cells. The percentage of BALF cells and the quantification of phagocytosis is represented in Figure 3B and Figure 3C, respectively. The percentage of body weight loss in mice after the intratracheal administration of a sublethal dose of P. aeruginosa is shown in Figure 4A, and the percentage of survival of mice at day 2 after a lethal dose of P. aeruginosa is indicated in Figure 4B. Figure 4C shows a scatter plot for the whole-lung bacterial burden at death or at day 2 after the P. aeruginosa infection in mice.
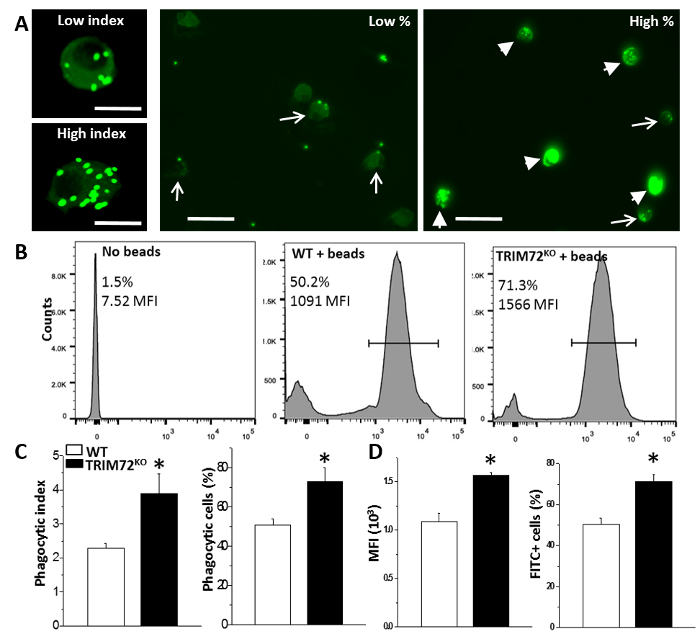
Figure 1: Phagocytosis in mouse primary alveolar macrophages. (A) Representative images of low versus high phagocytic indexes showing primary AMs containing green fluorescent beads (left); representative images showing low and high percentages of phagocytic AMs. Arrows: low phagocytic index AMs; arrowheads: high phagocytic index AMs. The scale bar = 25 µm for the left two images and 50 µm for the right two images. (B) Representative flow cytometry detection of phagocytizing cells in no-beads control, WT + beads, and TRIM72KO + beads AMs. The bars define the bead-containing cell population (in percentage) and the mean fluorescence intensity (MFI). (C) Statistics of the average phagocytic index and the percentage of phagocytic AMs in WT and TRIM72KO AMs; n = 5 for both groups, *p < 0.05. (D) Statistics of flow cytometry MFI and the percentage of FITC+ cells in WT and TRIM72KO AMs; n = 3 for both groups, *p < 0.05. This figure is reprinted with permission of the American Thoracic Society13. Please click here to view a larger version of this figure.
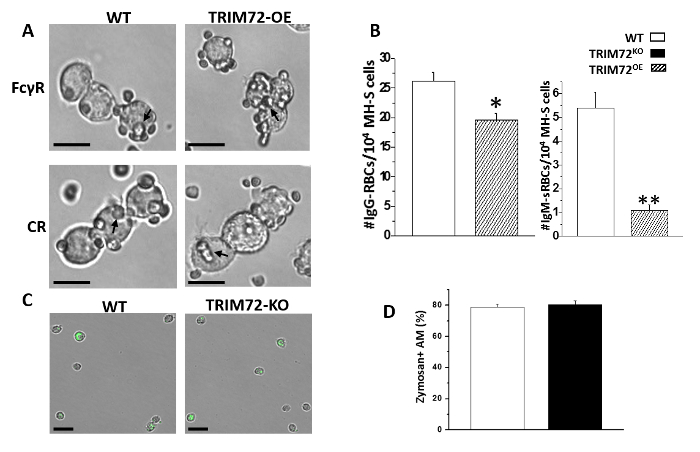
Figure 2: FcγR- and CR-mediated phagocytosis. (A) Representative images of opsonized sheep red blood cells' (SRBCs) phagocytosis by MH-S cells. The arrows point to some SRBCs. The scale bar = 25 µm. (B) Quantification of SRBCs phagocytosis by MH-S cells overexpressing TRIM72 (TRIM72OE) in the presence of IgG (FcγR-mediated phagocytosis) or IgM (CR-mediated phagocytosis); n = 6 for each group, *p < 0.05 and **p < 0.005 compared with WT control. (C) Representative images of Alexa Fluor-488-conjugated zymosan particle ingestion by primary AMs isolated from WT or TRIM72KO mice. The arrows in the panel A and B indicate zymosan + cells. The scale bar = 50 µm. (D) Statistical results of the percentage of zymosan-containing AMs; n = 3 for each group, p > 0.05. This figure is reprinted with permission of the American Thoracic Society13. Please click here to view a larger version of this figure.
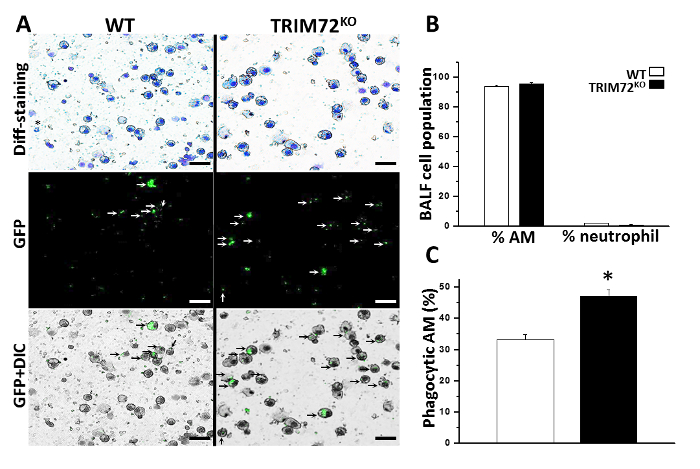
Figure 3: In vivo phagocytosis of P. aeruginosa GFP. (A) Representative images of bronchoalveolar lavage fluid (BALF) cell cytospin slides from WT and TRIM72KO mice 1 h after the injection of P. aeruginosa GFP. Kwik-Diff staining identifies AMs (large, round cells) and neutrophils; GFP identifies phagocytic cells (white arrows) and GFP+ differential interference contrast (DIC) identifies internalized GFP bacteria (black arrows) in AMs. The scale bars = 50 µm. (B) Quantification of the percentage of AMs and neutrophils in BALF of WT and TRIM72KO mice 1 h after the P. aeruginosa injection. (C) Quantification of the percentage of phagocytic AMs in WT and TRIM72KO mice; n = 3 for each group, *p < 0.05. The data are presented as mean (± SEM). This figure is reprinted with permission of the American Thoracic Society13. Please click here to view a larger version of this figure.
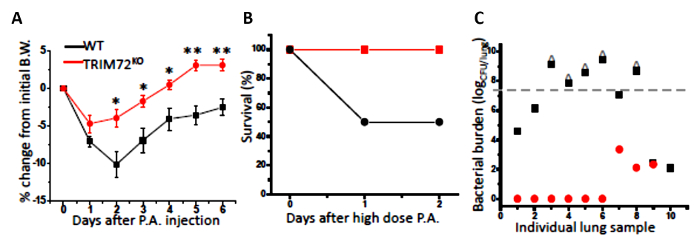
Figure 4: In vivo bacteria clearance using P. aeruginosa (P.A.). (A) The percentage of body weight (B.W.) loss of naive WT and TRIM72KO mice after the first intraperitoneal injection of 2.5 x 105 CFU/mL PAO1 (a clinical isolate of P. aeruginosa); n = 13 for WT (black squares), n = 8 for TRIM72KO (red circles), *p < 0.05, **p < 0.005 compared with WT. (B) The percentage of survival at day 2 after the 3 x 107 CFU/mL P. aeruginosa intraperitoneal injection; n = 10 for WT (solid black circles) and TRIM72KO (solid red squares), *p < 0.05 for WT versus TRIM72KO groups. (C) A scatter plot of the whole-lung bacterial burden at day 2 of the P. aeruginosa injection in WT and TRIM72KO. The gray dashed line designates the injected bacterial dose; ^ designates mice who have died. For WT versus TRIM72KO groups, p < 0.05. This figure is reprinted with permission of the American Thoracic Society13. Please click here to view a larger version of this figure.
Discussion
While performing a gas exchange function, the lung persistently confronts foreign particles, pathogens, and allergens. AMs provide the first line of defense by virtue of their main function, namely phagocytosis. AMs also coordinate with other immune cells in destroying the pathogens and in the resolution of inflammation. Here, we described methods for specifically assessing phagocytosis by AMs isolated from the mouse lung. The protocol presented in this manuscript explains a detailed study of phagocytosis both in vivo and in vitro, which can also be used to study the macrophage function and bacterial clearance in other organs.
The described in vitro phagocytosis relies on the simple idea of incubating serum-treated FITC beads or opsonized SRBCs with cultured AMs to initiate phagocytosis. This method includes steps of extensive washing and the lysis of nonphagocytized RBCs to reduce the background. Care must be taken not to detach the adhered AMs. The wash step involving ACK lysis solution should be done within 1 min, as a longer washing time leads to the lysis of the macrophages.
In addition, in the imaging analysis, we characterized both the percentage of phagocytizing cells and the phagocytic index to gain a comprehensive view of the AM phagocytosis function. We have also explained the method to differentiate among PRR-, FcγR-, and CR-mediated phagocytosis in the murine macrophage MH-S cell line. In conjunction with genetic modulation, this step is useful when trying to gain mechanistic insights on the specific phagocytosis pathway that was affected by the target gene manipulation.
Previous reports documented methods to evaluate the bacterial uptake; the most notable method is the gentamicin protection assay14. In the protocol presented here, we have also shown an imaging method to measure in vivo phagocytosis. There are a few key factors that make this method better in comparison to the previously published methods. The method described here involves an intratracheal injection of P. aeruginosa GFP followed by differential staining of BALF cells. This method highlights the use of fluorescent bacteria, which helps to identify ingested bacteria within the phagocytes. In addition, the differential staining of BALF cells helps to specifically differentiate the phagocytic capacity of AMs from other immune cells in the in vivo environment and to evaluate the relative contribution of different phagocytes to clear the bacterial loads from the lung. A minor limitation of this method is that it requires a careful intratracheal administration to avoid variability between mice.
Further, we explained the method to establish a P. aeruginosa bacteria clearance in mice in the context of pneumonia. To characterize this method, we determined the body weight loss and mortality after the intratracheal administration of P. aeruginosa and used a quantitative assay to determine the lung bacteria burden. Mortality is an important parameter to assess the comprehensive immune response of the body. We used lung samples from this experiment to assess bacterial burden. However, bacterial burden can also be determined from lung samples harvested within 48h following non-lethal dose of bacterial injection. The success of the assay will be determined by a standardization of the timing of the injection, the quality of the pathogen and the dose of injection, and the optimization of a homogenization method that completely disrupts the lung without breaking the bacterial cell membrane.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work is supported by grant R01HL116826 to X. Zhao.
Materials
| 18-G Needle | Nipro Medical | CI+1832-2C | Molecular Biology grade |
| 2,7-diaminofluorene (DAF) | Sigma-Aldrich | D17106 | Molecular Biology grade |
| 70% Ethanol | Decon Labs Inc. | 18C27B | Analytical grade |
| 96-well plate | Corning | 3603 | Cell Biology grade |
| ACK lysis buffer | Life Technologies | A10492 | Molecular Biology grade |
| Alexa fluor-488 Zymosan-A-bioparticle | Thermofisher Scientific | Z23373 | Molecular Biology grade |
| C5 deficient serum | Sigma-Aldrich | C1163 | Biochemical reagent |
| Centrifuge | Labnet International | C0160-R | |
| Cytospin 4 Cytocentrifuge | Thermofisher Scientific | A78300101 Issue 11 | |
| DMEM Cell Culture Media | Gibco | 11995-065 | Cell Biology grade |
| FBS | Atlanta Biologicals | S11550 | Cell Biology grade |
| Flow Cytometer | BD Biosciences | FACSCalibur | |
| Flow Jo Software | FlowJo, LLC | ||
| Forceps | Dumont | 0508-SS/45-PS-1 | Suitable for laboratory animal dissection |
| FITC-carboxylated latex beads | Sigma-Aldrich | L4530 | Cell Biology grade |
| GFP-P. aeruginosa | ATCC | 101045GFP | Suitable for cell infection assays |
| Glass bottom dish | MatTek Corp. | P35G-0.170-14-C | Cell Biology grade |
| High-Pressure Syringe | Penn-Century | FMJ-250 | Suitable for laboratory animal use |
| Homogenizer | Omni International | TH-01 | |
| Hydrogen peroxide | Sigma-Aldrich | H1009 | Analytical grade |
| Inverted Fluorescence Microscope | Olympus | IX73 | |
| Ketamine Hydrochloride | Hospira | CA-2904 | Pharmaceutical grade |
| Shandon Kwik-Diff Stains | Thermofisher Scientific | 9990700 | Cell Biology grade |
| LB Agar | Fisher Scientific | BP1425 | Molecular Biology grade |
| LB Broth | Fisher Scientific | BP1427 | Molecular Biology grade |
| MicroSprayer Aerosolizer | Penn-Century | IA-1C | Suitable for laboratory animal use |
| Paraformaldehyde | Sigma-Aldrich | P6148 | Reagent grade |
| PBS | Gibco | 20012-027 | Cell Biology grade |
| rabbit anti-SRBC-IgG | MP Biomedicals | 55806 | Suitable for immuno-assays |
| rabbit anti-SRBC-IgM | Cedarline Laboratories | CL9000-M | Suitable for immuno-assays |
| Scissors | Miltex | 5-2 | Suitable for laboratory animal dissection |
| Small Animal Laryngoscope | Penn-Century | LS-2 | Suitable for laboratory animal use |
| Sodium Dodecyl Sulfate (SDS) | BioRad | 1610301 | Analytical grade |
| Spring Scissors (Med) | Fine Science Tools | 15012-12 | Suitable for laboratory animal dissection |
| Spring Scissors (Small) | Fine Science Tools | 91500-09 | Suitable for laboratory animal dissection |
| sheep red blood cells (SRBCs) | MP Biomedicals | 55876 | Washed, preserved SRBCs |
| Urea | Sigma-Aldrich | U5378 | Molecular Biology grade |
| Xylazine | Akorn Animal Health | 59399-110-20 | Pharmaceutical grade |
References
- Hussell, T., Bell, T. J. Alveolar macrophages: plasticity in a tissue-specific context. Nature Reviews Immunology. 14, 81-93 (2014).
- Belchamber, K. B. R., Donnelly, L. E. Macrophage Dysfunction in Respiratory Disease. Results and Problems in Cell Differentiation. 62, 299-313 (2017).
- Broug-Holub, E., et al. Alveolar macrophages are required for protective pulmonary defenses in murine Klebsiella pneumonia: elimination of alveolar macrophages increases neutrophil recruitment but decreases bacterial clearance and survival. Infection and Immunity. 65, 1139-1146 (1997).
- Knapp, S., et al. Alveolar macrophages have a protective antiinflammatory role during murine pneumococcal pneumonia. American Journal of Respiratory and Critical Care Medicine. 167, 171-179 (2003).
- Bhatia, M., Zemans, R. L., Jeyaseelan, S. Role of chemokines in the pathogenesis of acute lung injury. American Journal of Respiratory Cell and Molecular Biology. 46, 566-572 (2012).
- Marriott, H. M., et al. Interleukin-1beta regulates CXCL8 release and influences disease outcome in response to Streptococcus pneumoniae, defining intercellular cooperation between pulmonary epithelial cells and macrophages. Infection and Immunity. 80, 1140-1149 (2012).
- Greenlee-Wacker, M. C. Clearance of apoptotic neutrophils and resolution of inflammation. Immunological Reviews. 273, 357-370 (2016).
- Haslett, C. Granulocyte apoptosis and its role in the resolution and control of lung inflammation. American Journal of Respiratory and Critical Care Medicine. 160, 5-11 (1999).
- Cox, G., Crossley, J., Xing, Z. Macrophage engulfment of apoptotic neutrophils contributes to the resolution of acute pulmonary inflammation in vivo. American Journal of Respiratory Cell and Molecular Biology. 12, 232-237 (1995).
- Groves, E., Dart, A. E., Covarelli, V., Caron, E. Molecular mechanisms of phagocytic uptake in mammalian cells. Cellular and Molecular Life Sciences. 65, 1957-1976 (2008).
- Mosser, D. M., Zhang, X. Measuring Opsonic Phagocytosis via Fcγ Receptors and complement receptors on macrophages. Current Protocols in Immunology. , (2011).
- He, J. Q., Wiesmann, C., van Lookeren Campagne, M. A role of macrophage complement receptor CRIg in immune clearance and inflammation. Molecular Immunology. 45, 4041-4047 (2008).
- Nagre, N., et al. Inhibition of Macrophage Complement Receptor CRIg by TRIM72 Polarizes Innate Immunity of the Lung. American Journal of Respiratory Cell and Molecular Biology. 58 (6), 756-766 (2018).
- Miksa, M., Komura, H., Wu, R., Shah, K. G., Wang, P. A Novel Method to Determine the Engulfment of Apoptotic Cells by Macrophages using pHrodo Succinimidyl Ester. Journal of Immunological Methods. 342 (1-2), 71-77 (2009).
- Su, H., Chen, H., Jen, C. J. Severe exercise enhances phagocytosis by murine bronchoalveolar macrophages. Journal of Leukocyte Biology. 69, 75-80 (2001).
- Amiel, E., Lovewell, R. R., O’Toole, G. A., Hogan, D. A., Berwin, B. Pseudomonas aeruginosa. evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infection and Immunity. 78, 2937-2945 (2010).
- Giannoni, E., Sawa, T., Allen, L., Wiener-Kronish, J., Hawgood, S. Surfactant Proteins A and D Enhance Pulmonary Clearance of Pseudomonas aeruginosa. American Journal of Respiratory Cell and Molecular Biology. 34, 704-710 (2006).

