Studying Organelle Dynamics in B Cells During Immune Synapse Formation
Summary
Herein we describe two approaches to characterize cell polarization events in B lymphocytes during the formation of an IS. The first, involves quantification of organelle recruitment and cytoskeleton rearrangements at the synaptic membrane. The second is a biochemical approach, to characterize changes in composition of the centrosome, which undergoes polarization to the immune synapse.
Abstract
Recognition of surface-tethered antigens by the B cell receptor (BCR) triggers the formation of an immune synapse (IS), where both signaling and antigen uptake are coordinated. IS formation involves dynamic actin remodeling accompanied by the polarized recruitment to the synaptic membrane of the centrosome and associated intracellular organelles such as lysosomes and the Golgi apparatus. Initial stages of actin remodeling allow B cells to increase their cell surface and maximize the quantity of antigen-BCR complexes gathered at the synapse. Under certain conditions, when B cells recognize antigens associated to rigid surfaces, this process is coupled to the local recruitment and secretion of lysosomes, which can facilitate antigen extraction. Uptaken antigens are internalized into specialized endo-lysosome compartments for processing into peptides, which are loaded onto major histocompatibility complex II (MHC-II) molecules for further presentation to T helper cells. Therefore, studying organelle dynamics associated with the formation of an IS is crucial to understanding how B cells are activated. In the present article we will discuss both imaging and a biochemical technique used to study changes in intracellular organelle positioning and cytoskeleton rearrangements that are associated with the formation of an IS in B cells.
Introduction
B lymphocytes are an essential part of the adaptive immune system responsible for producing antibodies against different threats and invading pathogens. The efficiency of antibody production is determined by the ability of B cells to acquire, process and present antigens encountered either in a soluble or surface-tethered form1,2. Recognition of antigens attached to the surface of a presenting cell, by the BCR, leads to the formation of a close intercellular contact termed IS3,4. Within this dynamic platform both BCR-dependent downstream signaling and internalization of antigens into endo-lysosome compartments takes place. Uptaken antigens are processed and assembled onto MHC-II molecules and subsequently presented to T lymphocytes. Productive B-T interactions, termed B-T cell cooperation, allow B lymphocytes to receive the appropriate signals, which promote their differentiation into antibody-producing plasma cells or memory cells8.
Two mechanisms have been involved in antigen extraction by B cells. The first one relies on the secretion of proteases originating from lysosomes which undergo recruitment and fusion at the synaptic cleft5,6. The second one, depends on Myosin IIA-mediated pulling forces that triggers the invagination of antigen containing membranes which are internalized into clathrin-coated pits7. The mode of antigen extraction relies on the physical properties of the membrane in which antigens are found. Nevertheless, in both cases, B cells undergo two major remodeling events: actin cytoskeleton reorganization and polarization of organelles to the IS. Actin cytoskeleton remodeling involves an initial spreading stage, where actin-dependent protrusions at the synaptic membrane increase the surface in contact with the antigen. This is followed by a contraction phase, where BCRs coupled with antigens are concentrated at the center of the IS by the concerted action of molecular motors and actin cytoskeleton remodeling8,9,10,11. Polarization of organelles also relies on the remodeling of actin cytoskeleton. For instance, the centrosome becomes uncoupled from the nucleus, by local depolymerization of associated actin, which allows the repositioning of this organelle to the IS5,12. In B cells, repositioning of the centrosome to one cell pole (IS) guides lysosome recruitment to the synaptic membrane, which upon secretion can facilitate the extraction and/or processing of surface-tethered antigens6. Lysosomes recruited at the IS are enriched with MHC-II, which favors the formation of peptide-MHC-II complexes in endosomal compartments to be presented to T cells13. Additionally, the Golgi apparatus has also been observed to be closely recruited to the IS14, suggesting that Golgi-derived vesicles from the secretory pathway could be involved in antigen extraction and/or processing.
Altogether, intracellular organelle and cytoskeleton rearrangements in B cells during synapse formation are the key steps that allow efficient antigen acquisition and processing required for their further activation. In this work we introduce detailed protocols on how to perform imaging and biochemical analysis in B cells to study the intracellular remodeling of organelles associated with the formation of an IS. These techniques include: (i) Immunofluorescence and image analysis of B cells activated with antigen-coated beads and on antigen-coated coverslips, which allows visualization and quantification of intracellular components that are mobilized to the IS and (ii) isolation of centrosome-enriched fractions in B cells by ultracentrifugation on sucrose gradients, which allows the identification of proteins associated with the centrosome, potentially involved in regulating cell polarity.
Protocol
NOTE: The following steps were performed using IIA1.6 B cells.
1. B cell activation with antigen-coated beads
- Preparation of antigen-coated beads
- To activate B cells, use NH2-beads covalently coated with antigen (Ag-coated beads), which are prepared using 50 µL (~20 x 106 beads) of 3 µm NH2-beads with activating (BCR-ligand+) or non-activating (BCR-ligand-) antigens.
- For IIA1.6 B cells use anti-IgG-F(ab')2 fragment as BCR-ligand+ and anti-IgM-F(ab')2 or bovine serum albumin (BSA) as BCR-ligand-. To activate primary B cells or IgM+ B cell line use anti-IgM-F(ab')2 as BCR-ligand+ and BSA as BCR-ligand-.
NOTE: To avoid the binding of ligands to the Fc receptor, use F(ab') or F(ab')2 antibodies fragments instead of full-length antibodies. - To proceed with the preparation of Ag-coated beads, place the beads in low protein binding microcentrifuge tubes to maximize the beads recovery during the sample manipulation. Add 1 mL of 1x phosphate-buffered saline (PBS) to wash beads and centrifuge at 16,000 x g for 5 min. Aspirate the supernatant.
- Resuspend the beads with 500 µL of 8% glutaraldehyde to activate the NH2 groups and rotate for 4 h at room temperature (RT).
CAUTION: The glutaraldehyde stock solution should only be used in a chemical fume hood. Follow the instructions on the material safety data sheet (MSDS). - Centrifuge beads at 16,000 x g for 5 min, remove the glutaraldehyde and wash the beads three more times with 1 mL of 1x PBS.
CAUTION: The glutaraldehyde solution should be discarded as a hazardous chemical waste. - Resuspend the activated beads in 100 µL of 1x PBS. The sample can be divided into two low protein binding microcentrifuge tubes: 50 µL for the BCR-ligand+ and 50 µL for BCR-ligand-.
- To prepare the antigen solution use two 2 mL low protein binding microcentrifuge tubes containing 100 µg/mL of antigen solution in 150 µL of PBS: One tube with BCR-ligand+ and other with BCR-ligand-.
- Add 50 µL of activated beads solution to each tube containing 150 µL of antigen solution, vortex and rotate overnight at 4 °C.
- Add 500 µL of 10 mg/mL BSA to block the remaining reactive NH2 groups on the beads and rotate for 1 h at 4 °C.
- Centrifuge the beads at 16,000 x g for 5 min at 4 °C and remove the supernatant. Wash the beads with cold 1x PBS three more times.
- Resuspend the activated beads in 70 µL of 1x PBS.
- To determine the final concentration of Ag-coated beads, dilute a small volume of beads in PBS (1:200) and count using a hemocytometer. Then store at 4 °C until use.
NOTE: Ag-coated beads should not be stored for more than 1 month.
- Preparation of poly-L-lysine coverslips
- Before the B cell activation assay with Ag-coated beads, prepare poly-L-lysine-coated coverslips (PLL-coverslips). Use a 50 mL tube containing 40 mL of 0.01% w/v of PLL solution and immerse the 12 mm-diameter coverslips into the solution. Rotate overnight at RT.
- Wash the coverslips with 1x PBS and leave to dry on a 24-well plate lid covered with paraffin film. Proceed with B cell activation.
- B cell activation
- To start B cell activation with beads, first dilute the IIA1.6 B cell line to 1.5 x 106 cells/mL in CLICK medium (RPMI-1640 supplemented with 2 mM L-Alanine-L-Glutamine + 55 µM beta-mercaptoethanol + 1 mM pyruvate + 100 U/mL Penicillin + 100 µg/mL streptomycin) + 5% heat-inactivated fetal bovine serum [FBS]).
- Combine 100 µL (150,000 cells) of IIA1.6 B cells with Ag-coated beads at 1:1 ratio in 0.6 mL tubes. Mix gently using a vortex and seed onto PLL-coverslips. Incubate for different time points in a cell culture incubator (37 °C / 5% CO2). The typical activating time points are 0, 30, 60 and 120 min. For time 0, place the PLL-coverslips into the 24-well plate lid on ice. Add the cells-Ag-coated bead mixture and incubate for 5 min on ice.
NOTE: It is important to mix by vortex instead of pipetting up and down, because this could reduce the number of beads in the sample, due to their accumulation in the plastic tip of the pipette. Use beads coated with BCR-ligand- as a negative control for each assay. We recommend activating the longest incubation time first and then the following time points. Calculate intervals of activation for samples such that they are ready for fixation at the same time. - Add 100 µL of cold 1x PBS to each coverslip to stop the activation and continue with the immunofluorescence protocol. See protocol section 3.
2. B cell activation on antigen-coated coverslips
- Preparation of antigen-coated coverslips
- Before activation, prepare the antigen solution (1x PBS containing 10 µg/mL BCR-ligand+ and 0.5 µg/mL rat anti-mouse CD45R/B220).
NOTE: Consider preparing the coverslips the day before the assay. B220 improves B cell adhesion, however, the BCR-Ligand+ is sufficient to generate the spreading response. - Place the 12 mm coverslip onto a 24-well plate lid covered with paraffin film, add 40 µL of antigen solution onto each coverslip and incubate at 4 °C overnight. Seal the plate to avoid evaporation of antigen solution.
- Wash the coverslips with 1x PBS and air dry.
- Before activation, prepare the antigen solution (1x PBS containing 10 µg/mL BCR-ligand+ and 0.5 µg/mL rat anti-mouse CD45R/B220).
- B cell activation
- To start the activation, dilute IIA1.6 B cells to 1.5 x 106 cells/mL in CLICK medium + 5% FBS.
- Add 100 µL of cells onto an antigen-coated coverslip (Ag-coverslip) and activate for different time points in a cell incubator at 37 °C / 5% CO2. The typical activating time points are 0, 30 and 60 min.
NOTE: As in section 1, we recommend starting with the longest activating time point in order to fix all the samples at the same time. - For time 0, place the 24-well plate lid containing the Ag-coverslip on ice and add the cells. Incubate for 5 min on ice.
- Carefully aspirate the media on each Ag-coverslip and then add 100 µL of cold 1x PBS to stop the activation. Continue with the immunofluorescence protocol. See section 3.
3. Immunofluorescence
NOTE: To avoid cross-reactivity of the secondary antibody with the BCR of IIA 1.6 B cells, do not use mouse-derived primary antibodies.
- Remove the 1x PBS and proceed with the fixation of each coverslip.
NOTE: To decide which fixation medium to use, check the antibody/dye data sheet. For example, for actin labeling by phalloidin use paraformaldehyde (PFA) fixation medium, and for the centrosome labeling by pericentrin use methanol fixation.- Add 50 µL of 1x PBS supplemented with 4% PFA and incubate for 10 min at RT.
CAUTION: Formaldehyde is toxic. Please read the MSDS before working with this chemical. PFA solutions should be only made under a chemical fume hood wearing gloves and safety glasses. PFA solution should be discarded as a hazardous chemical waste. - Alternatively, add 50 µL of cold methanol and incubate for 20 min.
- Add 50 µL of 1x PBS supplemented with 4% PFA and incubate for 10 min at RT.
- Wash three times with 1x PBS.
NOTE: Coverslips can be stored at 4 °C at this point for maximum three days in 1x PBS. - Remove 1x PBS and add 50 µL of blocking buffer (2% BSA + 0.3 M glycine in 1x PBS) onto each coverslip. Incubate at RT for 10 min.
- Aspirate gently and add 50 µL of permeabilization buffer (PB) (0.2% BSA + 0.05% saponin in 1x PBS). Incubate at RT for 20 min.
- Prepare the antibodies or dyes in PB (use 30 µL per coverslip) and incubate at RT for 1 h or overnight at 4 °C. Refer to the Table of Materials for additional details.
- To label the microtubule organizing center (MTOC) or centrosome use the following antibodies: anti-γ-Tubulin (1:500), anti-Cep55 (1:500), anti-α-tubulin (1:500), anti-pericentrin (1:1000).
NOTE: For centrosome labeling B cells can be transfected with a centrin-GFP expression plasmid. - For labeling Golgi apparatus use anti-Rab6a (1:500).
NOTE: Other antibodies or dyes can also be used. - For labeling lysosomes, use anti-Lamp1 (1:200).
NOTE: Anti-H2-DM and anti-MHC-II also can be used to label antigen processing compartments15,16. - For labeling endoplasmic reticulum use anti-Sec61a (1:500).
- For actin cytoskeleton consider the following: polymerized actin can be visualized by Phalloidin conjugated to fluorescent dyes.
NOTE: B cells transfected with a LifeAct-GFP/RFP expression plasmid can also be used to label actin.
- To label the microtubule organizing center (MTOC) or centrosome use the following antibodies: anti-γ-Tubulin (1:500), anti-Cep55 (1:500), anti-α-tubulin (1:500), anti-pericentrin (1:1000).
- Wash the coverslips three times with PBS.
- Dilute the secondary antibody or dyes in PBS (use 30 µL per coverslip) and incubate 1 h at RT.
NOTE: Avoid exposing the samples to direct light to preserve the quality of the fluorescence signal. - Wash the coverslips three times with PBS.
- Remove the PBS solution from coverslips.
- Add 4 µL of mounting reagent to a microscope slide. Mount the coverslip onto the slide with the cell side facing down. Allow the slides to dry for 30 min at 37 °C or at RT overnight.
NOTE: Consider using an "anti-fade" mounting reagent (see the Table of Materials). - Acquire fluorescence images on a confocal or epifluorescence microscope with a 60x or 100x oil immersion objective. For each acquisition consider the transmitted light or bright field to easily identify B cells interacting with beads.
NOTE: Take three-dimensional (3D) images, covering the entire cell using z-stacks. We recommend taking 0.5 µm thick stacks, however this depends on the microscope.
4. Image analysis
NOTE: The following algorithms are described for ImageJ software. However, this can be performed using an equivalent software. Also, consider that for all fluorescence intensity measurements we use the integrated fluorescence density ("RawIntDen" in ImageJ), because this parameter considers the total amount of fluorescence in each pixel of the image, taking the area into account.
- Analysis of the distribution of organelles in B cells activated with Ag-coated beads
NOTE: To quantify the polarization of cell components to the IS, we define an arbitrary value as a measure of proximity to the IS. The index ranges between -1 (anti-polarized) and 1 (Fully polarized, object on the bead), as was previously presented by Reversat et al.12.- Estimate the polarity index for the centrosome and Golgi apparatus (Figure 1B).
NOTE: This algorithm can be used for organelles that are confined to a one point.- First define the bead and cell areas to analyze using the circle tool selection to delimit the boundaries of both, and then save them as regions of interest (ROI). See Figure 1, Figure 2, Figure 3, and Figure 4.
- Determine the cell center (CC) and the bead center (BC) by running Analyze | Measure on cell and bead areas respectively. X and Y values obtained from the Résultats window determine the center coordinates.
- Manually determine the center of the centrosome or Golgi apparatus (Organelle) using the point tool selection in ImageJ and run Analyze | Measure. X and Y values obtained from the Résultats window determine the coordinates.
- Then, draw an angle from CC to Organelle (a) and CC to BC (b) using the angle tool selection and run Analyze | Measure. The angle value in the results window shows the angle (α) between both vectors (a and b).
- Calculate the polarity index using the following formula:
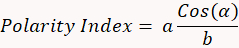
- Estimate the polarity index for lysosomes (Figure 1F).
NOTE: We use this algorithm to analyze the polarity of organelles that display a more dispersed distribution, such as lysosomes.- Define the bead and cell areas to analyze, using the circle tool selection to delimit the boundaries of both, and save them as ROI. Once the bead and cell areas have been determind, set the fluorescence channel and project the image into one z-stack (Image | Stacks | Z-Project [Sum slices]), then run Analyze | Measure and extract the mass center (MC) coordinates (MX and MY) from the results windows.
- Apply the same algorithm mentioned before changing Organelle for MC. Thus, the angle (α) is defined by CC-MC (a) and CC-BC (b).
- Calculate the polarity using the following formula:
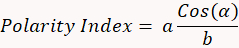
- Estimate lysosome recruitment to the synaptic area (Figure 2B).
NOTE: This algorithm is used to quantify an organelle adjacent to the IS.- Once the bead and cell areas have been determined, determine the angle of the vector between the CC and BC and then rotate the image to achieve a 0° angle.
- Set the fluorescent channel and project the image into one z-stack (Image | Stacks | Z-Project [Sum slices]), eliminate all the fluorescence that falls outside the cell and bead area together (run edit | clear outside in ImageJ).
- Using the rectangle tool selection, draw a rectangle adjacent to the bead and measure the synaptic area fluorescence (SAF). This rectangle is a quarter of the cell width.
- Select the entire image and run Analyze | Measure to obtain the whole cell fluorescence (WCF).
- Calculate the organelle fluorescence percentage adjacent to the IS using the following formula:

- Quantify recruitment of cellular components to the centrosome.
NOTE: This algorithm, adapted from Obino et al.5, is used to quantify the enrichment of organelles at the centrosome area. Briefly, we consider the centrosome area as the domain in which the centrosome-associated organelle fluorescence remains constant or above 70% in the fluorescence/radius plot. It is essential to set this parameter at resting conditions because this radius could change upon activation.- Once bead and cell areas have been determined, define the localization of the centrosome using the point tool selection (Figure 3B).
- First, determine the maximum area around the centrosome that is possible to quantify, by drawing a 3 µm-radius circle surrounding the centrosome.
- Use the ImageJ plugin Radial Profile, which measures the fluorescence in concentric circles and displays a fluorescence/radius plot.
- Identify the maximum radius within which at least 70% of the fluorescence intensity is maintained.
- Calculate the fluorescence density ratio using the following formula:
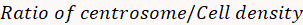 =
= 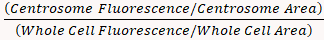
NOTE: The fluorescence density ratio indicates the concentration of an organelle at the centrosome compared to its distribution in the whole cell.
- Estimate the polarity index for the centrosome and Golgi apparatus (Figure 1B).
- Analysis of cell spreading and distribution of organelles in B cells activated on Ag-coverslips
- Estimate organelle distribution at the IS (Figure 4B).
NOTE: This algorithm allows the quantification of the XY distribution of organelles and their concentration at the center of the IS. We define a ratio of fluorescence density between the central and the total synaptic area. The values obtained can vary from negative to positive, indicating a peripheral or a central distribution of the organelle, respectively.- Determine the slice in which the cell is in contact with the cover.
NOTE: To delimit the cell boundaries one can label the plasma membrane or actin which is enriched in the periphery of the IS. - Change the type of the slice to 8-bit, then binarize (Process | Binary | Make Binary) and connect the nearest outsider points Process | Binary | Outline. Delimit the boundaries of the cell area (CA) using the polygon tool selection.
NOTE: This step is useful to increase the contrast of the cell boundaries and make it easier to identify. Also, at this point, it is possible to apply the Analyze particle plugin of ImageJ to determine the CA automatically. However, other cells in the same field can interfere with the results. - Take CA parameters (height and width) and delimit a central rounded area, which is separated from the boundaries by a quarter of the height and width values, consider this area as the Cell Center Area ( CCA).
- Calculate the fluorescence density distribution at the center of the IS using the following formula:

- Determine the slice in which the cell is in contact with the cover.
- Measure organelle distribution in Z planes (Figure 4C).
NOTE: This analysis determines the general distribution of organelle fluorescence across Z planes of B cells activated onto Ag-coverslips, showing the percentage of fluorescence per Z fraction.- To quantify the fluorescence distribution in Z, first determine the plane of the IS where the cell is in contact with the Ag-coverslip and then the plane corresponding to the upper limit of the cell.
- Draw a line across the cell center.
- Reslice the image in Z (Image | Stacks | Reslice), to obtain an XZ image.
- Measure the height and divide the XZ image into 10 consecutive rectangles of the same height (Z fraction) from the bottom (IS interface) to the top (upper side of the cell) and quantify the fluorescence signal in each one.
- Normalize the fluorescence intensity of each Z fraction by the sum of the total fluorescence of the 10 fractions.
- Plot the percentage of fluorescence intensity per Z fraction of the cell.
- Estimate organelle distribution at the IS (Figure 4B).
5. Isolation of centrosome-enriched fraction from resting and activated B cells
NOTE: Keep all solutions at 4 °C during the experiment to avoid protein degradation. This protocol was adapted from previous work17,18.
- Activate 2 x 107 B cells with Ag-coated beads in 2 mL of CLICK medium + 2% heat-inactivated FBS (ratio 1:1). Consider non-activated B cells as resting B cells.
- Add cytochalasin D (2 µM) and nocodazole (0.2 µM) and incubate for 1 h at 37 °C.
NOTE: These drugs are used to gently detach the centrosome from the nucleus, by depolymerizing actin cytoskeleton and microtubules, respectively, to avoid nuclear contamination. - Wash each sample with 5 mL of cold 1x TBS (50 mM Tris-HCl, pH 7.6, 150 mM NaCl) and then with 1 mL of 0.1x TBS supplemented with 8% sucrose.
- Resuspend the cells with 150 µL of centrosome lysis buffer (1 mM HEPES, pH 7.2, 0.5% NP-40, 0.5 mM MgCl2, 0.1% beta-Mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF) or protease inhibitor cocktail) and pipette up and down until viscosity decreases, which indicates cell lysis is identified in the sample. Put the sample in a 1.5 mL tube.
- Centrifuge at 10,000 x g for 10 min at 4 °C, to separate the organelles from the nucleus.
- Carefully recover the supernatant and place it on top of a 1.5 mL tube with 300 µL of gradient buffer (GB) (10 mM PIPES pH 7.2, 0.1% Triton X-100, 0.1% beta-mercaptoethanol) containing 60% sucrose.
- Centrifuge at 10,000 x g for 30 min at 4 °C to concentrate the centrosomes in the 60% sucrose fraction.
- Meanwhile, prepare a discontinuous gradient in 2 mL ultracentrifuge tubes by overlaying 450 µL of GB + 70% sucrose with 270 µL of GB + 50% sucrose and then 270 µL of GB + 40% sucrose.
- After the first centrifugation (centrosome-concentrated fraction), discard the upper fraction (less dense portion) until reaching the interface and vortex the remaining sample in the tube. Then, overlay on top of the discontinuous gradient previously prepared with the centrosome enriched sample.
NOTE: Be careful not to disrupt the gradient, all pipetting procedures must be carefully performed. - Centrifuge at 40,000 x g for 1 h at 4 °C with minimal acceleration and with the centrifuge brake set to off, to avoid disrupting the gradient.
- Collect 12 fractions of 100 µL into separate tubes beginning from the top.
- Identify the centrosome enriched fractions by immunoblot using γ-tubulin as the centrosome marker.
NOTE: We usually find centrosome-enriched extracts between fractions 6 and 8.
Representative Results
The present article shows how B cells can be activated using immobilized antigen on beads or coverslips to induce the formation of an IS. We provide information on how to identify and quantify the polarization of different organelles by immunofluorescence and how to characterize proteins that undergo dynamic changes in their association to the centrosome, which polarizes to the IS, using a biochemical approach.
The imaging of B cells by immunofluorescence allows us to follow the dynamics of organelles such as the centrosome, Golgi apparatus and lysosomes, which are recruited to the IS upon B cell activation. One can obtain quantitative parameters to measure the polarization of these organelles to the IS and compare them under different conditions. As we show in Figure 1A, the Centrosome and Golgi Apparatus are recruited to the IS upon B cell activation. Recruitment was not observed in B cells stimulated with BCR-ligand- indicating that BCR engagement is required to mobilize the centrosome and Golgi apparatus to the IS (Figure 1A). To obtain a polarity index for each organelle, as a measurement of its proximity to the IS, we considered three features: the distance between the organelle and the center of the cell, the distance between the bead center and the cell center, and the angle between these two vectors (Figure 1B). This index ranges between -1 (anti-polarized) and 1 (fully polarized, object on the bead). Figure 1C and 1D show graphs where polarity indexes of each organelle are plotted versus time of activation. In agreement with immunofluorescence staining, both the centrosome and Golgi apparatus, display more positive polarity indexes upon longer time points of activation, reflecting how they are progressively recruited to the IS. This algorithm is applicable for organelles confined to one point.
Additionally, we quantified the polarization of organelles that display a dispersed distribution, such as lysosomes (Figure 1E), by applying the same algorithm mentioned before, but changing the point coordinate by the mass center coordinates of lysosomes (Figure 1F). We can observe that the polarity indexes of lysosome pools reach more positive values upon activation, which indicates that the lysosomes are being mobilized towards the synapse upon B cell activation (Figure 1G).
We also defined two algorithms to determine organelles that are in contact with the synaptic interface (bead) and the amount of organelles at the proximity of the synapse, but not necessarily in contact. As we show in Figure 2, we quantified lysosomes contacting the synaptic interface (Figure 2A), by dividing the LAMP-1 signal at the bead area by the total fluorescence in the cell plus the bead (Figure 2B). It is important to consider that the diameter of the bead area determines the ranges of the percentages obtained. For example, in the quantification displayed (Figure 2B), we draw a circle with a 3 µm diameter to measure the fluorescence at the bead, which is a restrictive parameter considering the bead size (3 µm). Using this parameter, the results show that lysosomes are progressively recruited to the synaptic interface upon B cell activation, reaching 10-15% of the total mass (Figure 2C). Another approach shown is the quantification of lysosomes to the synaptic area. Figure 2D shows that lysosomes progressively accumulate up to 25% of their lysosomes at the IS. Overall, by performing both types of analysis one can evaluate the polarization and docking of organelles at the IS.
During B cell activation, changes in the pool of centrosome-associated proteins have been documented5. For instance, one of these proteins that changes their association at the Centrosome during B cell activation is actin. In this case actin becomes depleted within this region, which allows the centrosome to become detached from the nucleus, promoting its polarization to the IS5. Here we present the quantification of actin at the Centrosome and how it is depleted upon B cell activation (Figure 3). To define the centrosome area used to quantify associated actin, we measured the fluorescence intensity of this label in concentric circles surrounding the centrosome. The radius was defined based on the point where at least 70% of the fluorescence intensity of the label is maintained (Figure 3B). Using this approach one can quantify the decrease in actin density at the centrosome upon B cell activation, as shown in Figure 3C.
To obtain greater resolution in the distribution of organelles at the IS interface, we activated B cells on antigen-coated coverslips, labeling actin, the Golgi apparatus and endoplasmic reticulum (Figure 4A). The distribution of organelles at the IS can be performed by diving the synaptic area in a central and peripheral area, applying a criteria shown in Figure 4B. This was based on previous work showing that BCRs gather within this central area, which most likely corresponds to the central supramolecular activation complex (cSMAC)10,19. Figure 4A shows that organelles recruited to the IS, such as the Golgi apparatus and the endoplasmic reticulum display opposite distributions. Indeed, their distribution indexes correlate with their immunofluorescence staining shown in Figure 4C. The Golgi apparatus shows a positive value, which means that it is concentrated at the center of the IS while the endoplasmic reticulum shows negative values, which means it is mainly localized at the peripheral region of the IS. Additionally, it is possible to take the values of the synaptic area previously determined, to measure the spreading area during B cell activation, as it is shown in Figures 4D and 4E. These results show an increase in the spreading area upon B cell activation, as previously described10,20.
As in the bead assay, we can determine the distribution of organelles toward the immune synapse by taking the XZ images of B cells activated on antigen-coated coverslips and measuring the organelle fluorescence across the Z dimension (Figure 4F). The fluorescence intensity of actin in the Z plane was measured, as it is represented in the XZ plane in Figure 4F, where a progressive enrichment of actin at the synaptic interface (fractions 1 and 2) can be observed (Figure 4G).
In addition to the B cell imaging analysis, we performed the isolation of centrosome-enriched fractions to quantify centrosome-associated proteins (Figure 5A). This approach can be important to complement the study of IS formation in B cells, given that the centrosome polarizes to the synaptic membrane and has been shown to coordinate lysosome trafficking involved in antigen extraction and presentation21. This method was previously described using large amounts of cells (1 x 109 cells)17,18 but we have standardized a simplified version, which uses a reduced amount of cells and can be performed using lower-volume ultracentrifuge-tubes (2-3 mL). As we show in Figure 5B, we analyzed different cell fractions by immunoblotting and determined which ones correspond to centrosome-rich fractions, by gamma tubulin staining. Here we show an example of proteins that undergo changes in their accumulation at the centrosome, such as Actin and Arp2, which have been previously reported5.
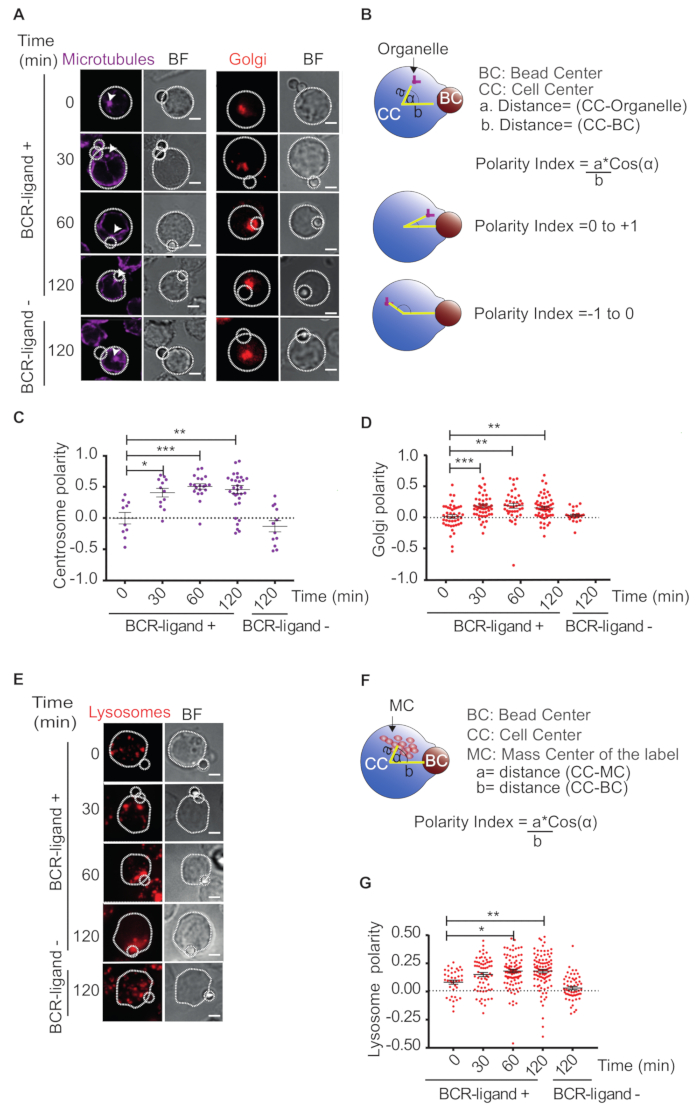
Figure 1: Polarization of organelles towards the IS. (A) Immunofluorescence staining of the Golgi apparatus (Rab6a) and microtubules (α-tubulin) in IIA1.6 B cells incubated with BCR-ligand+ (0, 30, 60 and 120 min) or BCR-ligand- beads (120 min). Arrowheads indicate the centrosome. BF = Bright field. Scale bar = 3 µm. (B) Scheme depicting how to calculate the polarity index of organelles (centrosome or Golgi apparatus) toward the IS. a = Distance between the center of the cell (CC) to the organelle. b = Distance from the CC and to the bead center (BC). (C, D) Representative dot plots showing polarity indexes of organelles at different time points of activation. Each dot represents one cell. (E) Immunofluorescence staining of Lysosomes (Lamp1) in B cells incubated with BCR-ligand+ (0, 30, 60 and 120 min) or BCR-ligand- beads (120 min). BF = Bright field. Scale bar = 3 µm. (F)Scheme representing how to calculate the polarity index of Lysosomes. a = Distance between CC to the mass center (MC). b = Distance from CC to BC. (G)Representative dot plots showing polarity indexes of lysosomes at different time points of activation. Each dot represents one cell. 2-way ANOVA with Sidak's post-test was performed. Means with SEM are shown. Please click here to view a larger version of this figure.
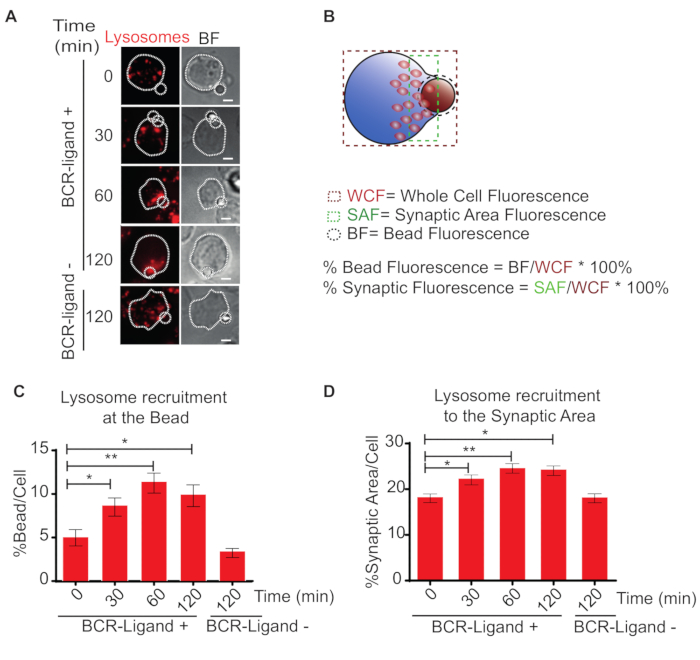
Figure 2: Accumulation of lysosomes at the IS. (A) Immunofluorescence staining of lysosomes (Lamp1) in B cells incubated with BCR-ligand+ (0, 30, 60 and 120 min) or BCR-ligand-beads (120 min). BF = bright field. Scale bar = 3 µm. (B) Scheme depicting how to calculate the accumulation of organelles such as lysosomes at the bead and the synaptic area. Fluorescence of the whole cell (WCF), bead fluorescence (BF) and the synaptic area fluorescence (SAF) are indicated. (C, D) Representative bar graphs for the accumulation of lysosomes at the bead and the synaptic area. N = 1. 20 > Cells. 2-way ANOVA with Sidak's post-test was performed. Means with SEM are shown. Please click here to view a larger version of this figure.
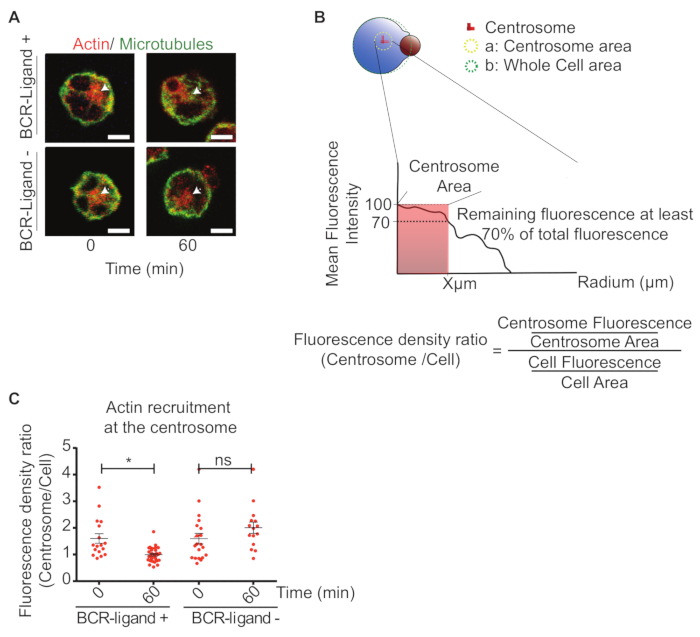
Figure 3: Quantification of actin at the centrosome during IS formation. (A) Immunofluorescence staining of actin (Phalloidin) and microtubules (α-Tubulin) in B cells incubated with BCR-ligand+ (0 and 60 min) or BCR-ligand- beads (0 and 60 min). Arrow heads indicate the centrosome. Scale bar = 3 µm. (B) Scheme depicting how to calculate the recruitment or depletion of Centrosome associated components. The centrosome area is calculated by using a radius (X µm) that maintain at least 70% of the maximum fluorescence. (C) Representative dot plots showing actin at the centrosome under activating (BCR-ligand+) and non-activating (BCR-ligand-) conditions. N = 1. 15 > Cells. 2-way ANOVA with Sidak's post-test was performed. Means with SEM are shown. Please click here to view a larger version of this figure.
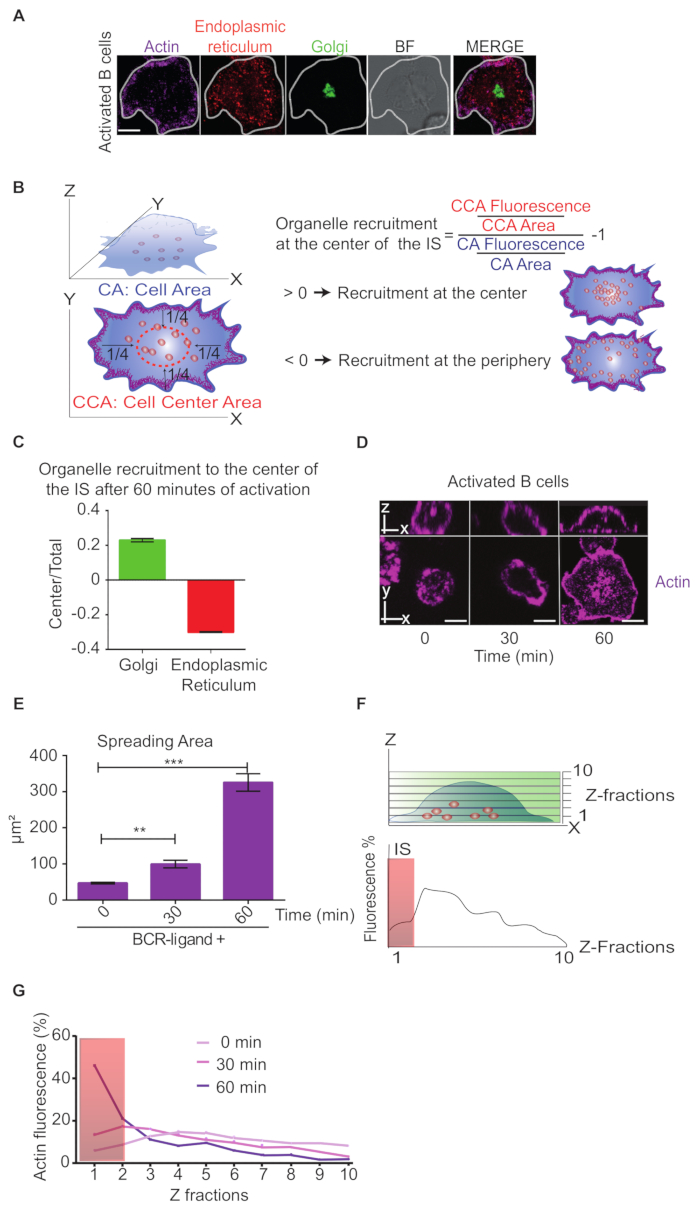
Figure 4: Distribution of cellular components at the synaptic interface. (A) Immunofluorescence staining of actin (Phalloidin), Endoplasmic Reticulum (Sec61a), Golgi apparatus (transfected with a KDELR-DN-GFP a Negative Dominant of KDEL Receptor, which localizes in the Golgi). B cells were seeded onto coverslips coated with a BCR-ligand+ for 60 min. BF = bright field. Scale bar = 5 µm. (B) Scheme depicting how to determine the spreading area of the cell and the recruitment of organelles at the center of the IS. In the scheme, CA indicates the cell area delimited by cortical actin and CCA indicates the center cell area.(C) Golgi apparatus and endoplasmic reticulum distribution at the IS, represented by a bar graph where a value >0 indicates an enrichment in the center of the IS and < 0 indicates a peripheral distribution. N = 1. 20 > Cells. Means with SEM are shown. (D) Representative images of B cells labelled for actin (Phalloidin) activated onto Ag-coated coverslips at different time points (0, 30 and 60 min), showing the XZ and XY plane. (E) Graph showing the spreading area of B cells in E. (F) Scheme depicting how to determine the distribution of the signal of interest in the Z plane, and the plot per Z fraction. The rectangle indicates the IS interface. (G) Distribution of actin across the Z plane represented by a line plot of the percentage of fluorescence distribution versus each Z fraction. The rectangle represents the IS interface. N = 1. 25 > Cells. Means with SEM are shown. Please click here to view a larger version of this figure.
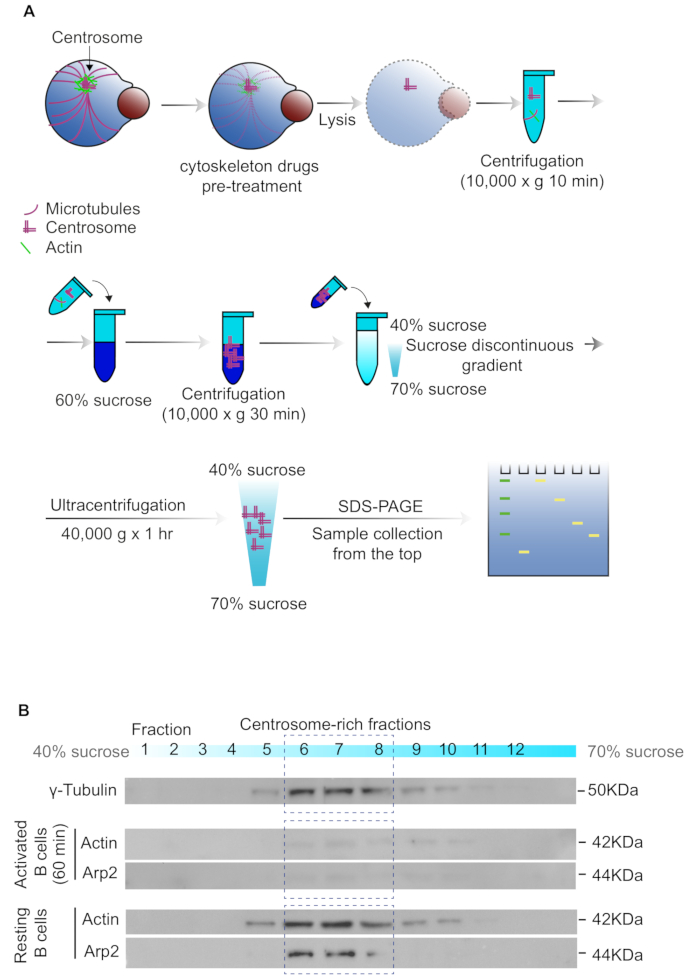
Figure 5: Centrosome enriched fractions of B cells. (A) Workflow of how to obtain centrosome-enriched fractions by sucrose gradient ultracentrifugation. (B) Immunoblot of fractions obtained from resting and activated B cells (60 min). Centrosome-rich fractions are detected by labeling with γ-tubulin and are indicated within the dashed rectangle (fraction 6 to 8). Centrosome associated proteins (Actin and Arp2) are shown in centrosome-rich fractions in activating and resting conditions. Please click here to view a larger version of this figure.
Discussion
We describe a comprehensive method to study how B lymphocytes re-organize their intracellular architecture to promote the formation of an IS. This study includes the use of imaging techniques to quantify the intracellular distribution of organelles, such as centrosome, Golgi apparatus and lysosomes during B cell activation, and how they polarize to the IS. Additionally, we describe a biochemical approach to study changes in the centrosome composition upon B cell activation.
To promote the formation of an IS in B cells, we used immobilized antigens (antigen-coated beads) instead of soluble immune-complexes because the former triggers the establishment of a discrete contact site where cytoskeleton rearrangements and organelle polarization can easily be studied. A critical aspect in imaging B cell activation is the preparation of the samples. All image acquisitions should ideally obtain a 1:1 cell : bead ratio, to quantify the polarization of organelles to a unique site of antigen contact. However, in some cases B cells can capture two or more beads, thereby making it difficult to choose a single site of antigen encounter. To avoid the formation of bead aggregates, it is important to vortex antigen-conjugated beads thoroughly before adding them to the cells. Additionally, for immunofluorescence experiments, we recommend excluding from quantification cells that capture more than one bead, in order to avoid misinterpretations during image analysis. Another critical step in the preparation of samples is the fixation/permeabilization condition. For instance, we recommend fixing cells with methanol to optimize microtubule staining and 4% PFA for phalloidin staining. When simultaneously labeling actin cytoskeleton and microtubules, an alternative can be to label the actin pool by transfecting cells with a LifeAct-GFP/RFP expression plasmid in combination with microtubule staining. Image acquisition used to quantify the polarization of the Centrosome and other intracellular organelles can be performed with an epifluorescence microscope. However, to obtain an accurate quantification at different z-planes, for instance in the spreading assay, confocal microscopy is required.
For the centrosome isolation protocol described here, a critical step is obtaining an appropriate amount of samples to be able to quantify proteins by immunoblot. Given that B cells have a large nucleus and their cytoplasm is less than 30% of their total cell volume, obtaining higher yields of cytoplasmic proteins can be challenging. For this reason, we recommend using approximately 20 x 106 B cells for each activation time point for centrosome isolation.
A significant limitation of these methods is that activation with antigen-conjugated beads does not include the complexity of the environment in which surface-tethered antigens are presented to B cells in vivo. This limitation can be addressed by supplementing the culture or beads with other co-stimulatory factors, such as cytokines and components of the extracellular matrix. However, it is crucial to use the appropriate controls in these cases in order to correctly interpret the results.
The significance of the methods presented in this work relies on the integrated approach used to study the B cell immune synapse: (1) the polarization of organelles and their distribution and (2) the changes in protein composition at the centrosome. Due to the large number of cells required to perform a significative analysis and the large amount of data generated in this process, the different quantification algorithms presented in this work facilitate the analysis of cell imaging. Moreover, the algorithms used for cell polarization are easy to automatize by MACROS functions in ImageJ, which is a useful tool to decrease repetitive tasks and save time.
These experimental procedures can be extrapolated to other types of cells that establish polarized phenotypes in order to accomplish a certain cellular function. Additionally, these protocols can be optimized by including other assays, for instance, the activity of the associated proteins such as kinases or proteases in Centrosome-enriched fractions. Overall, these studies can provide mechanistic insight into cell signaling and molecular pathways that regulate the establishment of cell polarity.
Divulgations
The authors have nothing to disclose.
Acknowledgements
M.-I.Y. is supported by a research grant from FONDECYT #1180900. J.I., D.F. and J.L. were supported by fellowships from the Comisión Nacional de Ciencia y Tecnología. We thank to David Osorio from Pontificia Universidad Católica de Chile for the video recording and editing.
Materials
| IIA1.6 (A20 variant) mouse B-lymphoma cells | ATCC | TIB-208 | Murine B-cell lymphoma of Balb/c origin that expresses an IgG-containing BCR on its surface without FcγIIR |
| 100% methanol | Fisher Scientific | A412-4 | |
| 10-mm diameter cover glasses thickness No. 1 circular | Marienfield-Superior | 111500 | |
| 2-mercaptoethanol | Thermo Fisher Scientific | 21985023 | |
| Alexa 488 fluor- donkey ant-rabbit IgG | LifeTech | A21206 | 1:500 dilution recomended but should be optimized |
| Alexa Fluor 546 goat anti-Rabbit IgG | Thermo Fisher Scientific | A-11071 | 1:500 dilution recomended but should be optimized |
| Alexa Fluor 647-conjugated phalloidin | Thermo Fisher Scientific | A21238 | 1:500 dilution recomended but should be optimized |
| Amaxa Nucleofection kit V | Lonza | VCA-1003 | Follow the manufacturer's directions for mixing the transfection reagents with the DNA |
| Amaxa Nucleofector model 2b | Lonza | AAB-1001 | Program L-013 used |
| Amino- Dynabeads | ThermoFisher | 14307D | |
| Anti-pericentrin | Abcam | ab4448 | 1:200 dilution recomended but should be optimized |
| Anti-rab6 | Abcam | ab95954 | 1:200 dilution recomended but should be optimized |
| Anti-sec61 | Abcam | ab15575 | 1:200 dilution recomended but should be optimized |
| BSA | Winkler | BM-0150 | |
| CaCl2 | Winkler | CA-0520 | |
| Culture plate T25 | BD | 353014 | |
| Fiji Software | Fiji col. | ||
| Fluoromount G | Electron Microscopy Science | 17984-25 | |
| Glutamine | Thermo Fisher Scientific | 35050061 | |
| Glutaraldehyde | Sigma | G7651 | |
| Glycine | Winkler | BM-0820 | |
| Goat-anti-mouse IgG antibody | Jackson ImmunoResearch | 315-005-003 | IIA1.6 positive ligand |
| Goat-anti-mouse IgM antibody | Thermo Fisher Scientific | 31186 | IIA1.6 negative ligand |
| HyClone Fetal bovine serum | Thermo Fisher Scientific | SH30071.03 | Heat inactivate at 56 oC for 30 min |
| KCl | Winkler | PO-1260 | |
| Leica SP8 TCS microscope | Leica | ||
| NaCl | Winkler | SO-1455 | |
| Nikon Eclipse Ti-E epifluorescence microscope | Nikon | ||
| Parafilm | M | P1150-2 | |
| Paraformaldehyde | Merck | 30525-89-4 | Dilute to 4% with PBS in a safety cabinet, use at the moment |
| Penicillin-Streptomycin | Thermo Fisher Scientific | 15140122 | Liquid |
| Polybead Amino Microspheres 3.00μm | Polyscience | 17145-5 | |
| Poly-L-Lysine | Sigma | P8920 | Dilute with sterile water |
| Rabbit anti- alpha tubulin antibody | Abcam | ab6160 | 1:1000 dilution recommended but should be optimized |
| Rabbit anti mouse lamp1 antibody | Cell signaling | 3243 | 1:200 dilution recomended but should be optimized |
| Rabbit anti-cep55 | Abcam | ab170414 | 1:500 dilution recomended but should be optimized |
| Rabbit Anti-gamma Tubulin antibody | Abcam | ab16504 | 1:1000 for Western Blot |
| RPMI-1640 | Biological Industries | 01-104-1A | |
| Saponin | Merck | 558255 | |
| Sodium pyruvate | Thermo Fisher Scientific | 11360070 | |
| Sucrose | Winkler | SA-1390 | |
| Triton X-100 | Merck | 9036-19-5 | |
| Tube 50 ml | Corning | 353043 |
References
- Carrasco, Y. R., Batista, F. D. B. Cells Acquire Particulate Antigen in a Macrophage-Rich Area at the Boundary between the Follicle and the Subcapsular Sinus of the Lymph Node. Immunity. 27 (1), 160-171 (2007).
- Batista, F. D., Harwood, N. E. The who, how and where of antigen presentation to B cells. Nature Reviews Immunology. 9 (1), 15-27 (2009).
- Grakoui, A., et al. The immunological synapse: a molecular machine controlling T cell activation. Science (New York, N.Y.). 285 (5425), 221-227 (1999).
- Batista, F. D., Iber, D., Neuberger, M. S. B cells acquire antigen from target cells after synapse formation. Nature. 411 (6836), 489-494 (2001).
- Obino, D., et al. Actin nucleation at the centrosome controls lymphocyte polarity. Nature Communications. 7, (2016).
- Yuseff, M. I., et al. Polarized Secretion of Lysosomes at the B Cell Synapse Couples Antigen Extraction to Processing and Presentation. Immunity. 35 (3), 361-374 (2011).
- Spillane, K. M., Tolar, P. Mechanics of antigen extraction in the B cell synapse. Molecular Immunology. 101, 319-328 (2018).
- Schnyder, T., et al. B Cell Receptor-Mediated Antigen Gathering Requires Ubiquitin Ligase Cbl and Adaptors Grb2 and Dok-3 to Recruit Dynein to the Signaling Microcluster. Immunity. , (2011).
- Wang, J. C., et al. The Rap1 – cofilin-1 pathway coordinates actin reorganization and MTOC polarization at the B cell immune synapse. Journal of Cell Science. , 1094-1109 (2017).
- Fleire, S. J., Goldman, J. P., Carrasco, Y. R., Weber, M., Bray, D., Batista, F. D. B cell ligand discrimination through a spreading and contraction response. Science. 312 (5774), 738-741 (2006).
- Vascotto, F., et al. The actin-based motor protein myosin II regulates MHC class II trafficking and BCR-driven antigen presentation. Journal of Cell Biology. 176 (7), 1007-1019 (2007).
- Reversat, A., et al. Polarity protein Par3 controls B-cell receptor dynamics and antigen extraction at the immune synapse. Molecular biology of the cell. 26 (7), 1273-1285 (2015).
- Lankar, D., et al. Dynamics of Major Histocompatibility Complex Class II Compartments during B Cell Receptor–mediated Cell Activation. The Journal of Experimental Medicine. 195 (4), 461-472 (2002).
- Duchez, S., et al. Reciprocal Polarization of T and B Cells at the Immunological Synapse. The Journal of Immunology. 187 (9), 4571-4580 (2011).
- Pérez-Montesinos, G., et al. Dynamic changes in the intracellular association of selected rab small GTPases with MHC class II and DM during dendritic cell maturation. Frontiers in Immunology. 8 (MAR), 1-15 (2017).
- Le Roux, D., Lankar, D., Yuseff, M. I., Vascotto, F., Yokozeki, T., Faure-Andre´, G., Mougneau, E., Glaichenhaus, N., Manoury, B., Bonnerot, C., Lennon-Dume´nil, A. M. Syk-dependent Actin Dynamics Regulate Endocytic Trafficking and Processing of Antigens Internalized through the B-Cell Receptor. Molecular biology of the cell. 18 (September), 3451-3462 (2007).
- Reber, S. Chapter 8 Isolation of Centrosomes from Cultured Cells. Methods in Molecular Biology. 777 (2), 107-116 (2011).
- Gogendeau, D., Guichard, P., Tassin, A. M. Purification of centrosomes from mammalian cell lines. Methods in Cell Biology. 129, (2015).
- Obino, D., et al. Vamp-7–dependent secretion at the immune synapse regulates antigen extraction and presentation in B-lymphocytes. Molecular Biology of the Cell. 28 (7), 890-897 (2017).
- Harwood, N. E., Batista, F. D. Early Events in B Cell Activation BCR: B cell receptor. Annual Review of Immunology. , (2010).
- Yuseff, M. I., Lennon-Duménil, A. M. B cells use conserved polarity cues to regulate their antigen processing and presentation functions. Frontiers in Immunology. 6, 1-7 (2015).

