使用 CRISPR 介质基础编辑器对 BRCA1 变体进行功能评估
Summary
BRCA1突变患者患癌症的风险较高,这值得准确评估BRCA1变异的功能。在此,我们描述了使用CRISPR介质细胞素基础编辑器对BRCA1变种进行功能评估的协议,该基编辑器支持活细胞中目标C:G到T:A的转换。
Abstract
最近的研究已经调查了与 BRCA1 基因突变相关的风险,使用各种功能评估方法,如荧光记者检测,胚胎干细胞可行性测定,和治疗药物的灵敏度检测。虽然它们澄清了许多 BRCA1 变种,但这些涉及使用外源表达 BRCA1 变种的检测与过度表达问题有关,不能应用于转录后调节。为了解决这些限制,我们之前报告了通过CRISPR介导细胞素基础编辑器对 BRCA1 变异进行功能分析的方法,该方法可诱导活细胞中有针对性的核苷酸替代。使用此方法,我们识别出其功能仍然模糊不清的变体, 包括 c.-97C=T、c.154C=T、c.3847C=T、c.5056C=T 和 c.4986+5G=A,并确认CRISPR介质基础编辑器是重新分类 BRCA1中不确定意义变体的有用工具。在这里,我们描述了使用基于CRISPR的细胞素基础编辑器对 BRCA1 变种进行功能分析的协议。该协议为选择目标地点、功能分析和评估 BRCA1 变种提供了准则。
Introduction
乳腺癌1型易感基因(BRCA1)是一种广为人知的肿瘤抑制基因。由于BRCA1基因与DNA损伤的修复有关,该基因的突变将导致单个基因患癌症的风险更大。乳腺癌、卵巢癌、前列腺癌和胰腺癌与BRCA1基因2的遗传性功能丧失(LOF)突变有关。BRCA1变种的功能评估和鉴定可能有助于预防和诊断各种疾病。为了解决BRCA1变种的功能问题,已经开发并广泛用于研究BRCA1变种的致病性,如胚胎干细胞活性测定、荧光记者检测和治疗性药物灵敏度检测3、4、5、6。虽然这些方法评估了很多BRCA1变种的功能,但涉及外源表达BRCA1变种的方法在过度表达方面存在局限性,可能会影响下游调控、基因剂量和蛋白质折叠7。此外,这些检测不能利用后脚本法规,如mRNA拼接,成绩单稳定性,以及未翻译区域8,9的影响。
CRISPR-Cas9系统使活细胞和生物体10具有针对性的基因组编辑。通过单导RNA,Cas9可以在特定基因组位点诱导染色体DNA中的双链断裂(DSBs),以激活两个DNA修复途径:易出错的非同源端连接(NHEJ)通路和无差错同源修复(HDR)通路11。HDR 是一种精确的修复机制:然而,Cas9 为 HDR 诱导的 DSB 通常会导致不需要的插入和删除 (indel) 突变。此外,它需要同源捐赠者DNA模板来修复DNA损伤,效率相对较低。最近,Cas9 nickase (nCas9) 已与细胞丁去氨酶域融合,用于定位 C:G 到 T:A 替换,无需均匀的 DNA 模板和 DNA 双链断裂 12、13、14、15。利用细胞素基础编辑器,我们开发出一种对BRCA1变种16进行功能分析的新方法。
在这项研究中,我们使用CRISPR介导细胞素基础编辑器,BE314,它诱导有效的C:G到T:A点突变,用于实施BRCA1变异的功能评估,并成功识别了几个BRCA1变种的功能(图1)。
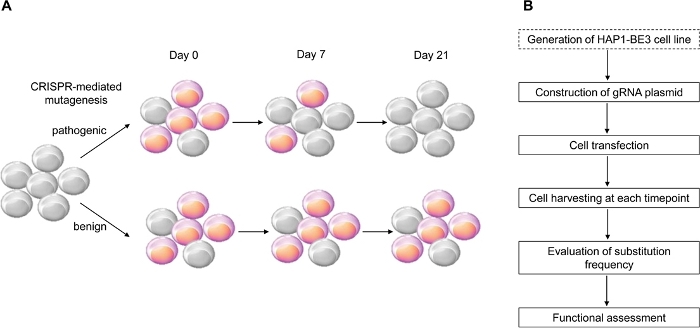
图1:功能评估工作流程概述。(A) 显示 BRCA1功能评估的示意图。因为 BRCA1 的LOF影响细胞的生存能力,当 BRCA1 突变是致病的,细胞死亡的通道数增加。(B) BRCA1功能评估的阶段。点缀框是可选的。它可以被 gRNA 表达和 BE3 表达质粒 DNA 的共变性所取代。 请点击这里查看此数字的较大版本。
Protocol
Representative Results
Discussion
此协议描述了使用CRISPR冥想细胞素基础编辑器对 BRCA1 变种进行功能评估的简单方法。该协议描述了在目标位置设计 gRNA 的方法,以及从中表达的质粒 DNA 的构建方法。细胞素基础编辑器在活动窗口中诱导核苷酸转换(如果 BE3,核苷酸 4-8 在 GRNA 目标序列的 PAM-分端)。研究人员应该仔细选择目标序列,因为活动窗口中的所有细胞氨酸都可以代替胸腺素。此外,正如第 5 步所述,应仔细分析?…
Divulgations
The authors have nothing to disclose.
Acknowledgements
这项工作得到了韩国国家研究基金会的支持(赠款2017M3A9B4062419,2019R1F1A1057637和2018R1A5A2020732到Y.K.)。
Materials
| BamHI | NEB | R3136 | Restriction enzyme |
| Blasticidin | Thermo Fisher Scientific | A1113903 | Drug for selecting transduced cells |
| BsaI | NEB | R0535 | Restriction enzyme |
| DNeasy Blood & Tissue Kit | Qiagen | 69504 | Genomic DNA prep. kit |
| Dulbecco’s modified Eagle’s medium | Gibco | 11965092 | Medium for HEK293T/17 cells |
| Fetal bovine serum | Gibco | 16000036 | Supplemetal for cell culture |
| FuGENE HD Transfection Reagent | Promega | E2311 | Transfection reagent |
| Gibson Assembly Master Mix | NEB | E2611L | Gibson assembly kit |
| Iscove’s modified Dulbecco’s medium | Gibco | 12440046 | Medium for HAP1 cells |
| lentiCas9-Blast | Addgene | 52962 | Plasmids DNA for lentiBE3 cloning |
| Lipofectamine 2000 | Thermo Fisher Scientific | 11668027 | Transfection reagent |
| Opti-MEM | Gibco | 31985070 | Transfection materials |
| pCMV-BE3 | Addgene | 73021 | Plasmids DNA for lentiBE3 cloning |
| Penicillin-Streptomycin | Gibco | 15140 | Supplemetal for cell culture |
| Phusion High-Fidelity DNA Polymerase | NEB | M0530SQ | High-fidelity polymerase |
| pMD2.G | Addgene | 12259 | Plasmids DNA for virus prep. |
| pRG2 | Addgene | 104174 | gRNA cloning vector |
| psPAX2 | Addgene | 12260 | Plasmids DNA for virus prep. |
| QIAprep Spin Miniprep kit | Qiagen | 27106 | Plasmid DNA prep. Kit |
| QIAquick Gel extraction Kit | Qiagen | 28704 | Gel extraction kit |
| QIAquick PCR Purification Kit | Qiagen | 28104 | PCR product prep. kit |
| Quick Ligation Kit | NEB | M2200 | Ligase for gRNA cloning |
| T7 Endonuclease I | NEB | M0302 | Materials for T7E1 assay |
| XbaI | NEB | R0145 | Restriction enzyme |
References
- Roy, R., Chun, J., Powell, S. N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nature Reviews Cancer. 12 (1), 68-78 (2011).
- Kuchenbaecker, K. B., et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. Journal of the American Medical Association. 317 (23), 2402-2416 (2017).
- Millot, G. A., et al. A guide for functional analysis of BRCA1 variants of uncertain significance. Human Mutation. 33 (11), 1526-1537 (2012).
- Santos, C., et al. Pathogenicity evaluation of BRCA1 and BRCA2 unclassified variants identified in Portuguese breast/ovarian cancer families. Journal of Molecular Diagnostics. 16 (3), 324-334 (2014).
- Starita, L. M., et al. A Multiplex Homology-Directed DNA Repair Assay Reveals the Impact of More Than 1,000 BRCA1 Missense Substitution Variants on Protein Function. American Journal of Human Genetics. 103 (4), 498-508 (2018).
- Anantha, R. W., et al. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. Elife. 6, (2017).
- Gibson, T. J., Seiler, M., Veitia, R. A. The transience of transient overexpression. Nature Methods. 10 (8), 715-721 (2013).
- Quann, K., Jing, Y., Rigoutsos, I. Post-transcriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Frontiers in Genetics. 6, 242 (2015).
- Saunus, J. M., et al. Posttranscriptional regulation of the breast cancer susceptibility gene BRCA1 by the RNA binding protein HuR. Recherche en cancérologie. 68 (22), 9469-9478 (2008).
- Knott1, G. J., Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science. 361, 866-869 (2018).
- Sander, J. D., Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology. 32 (4), 347-355 (2014).
- Hess, G. T., et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nature Methods. 13 (12), 1036-1042 (2016).
- Kim, K., et al. Highly efficient RNA-guided base editing in mouse embryos. Nature Biotechnology. 35 (5), 435-437 (2017).
- Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533 (7603), 420-424 (2016).
- Park, D. S., et al. Targeted Base Editing via RNA-Guided Cytidine Deaminases in Xenopus laevis Embryos. Molecules and Cells. 40 (11), 823-827 (2017).
- Kweon, J., et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene. 39 (1), 30-35 (2020).
- Gibson, D. G. Enzymatic assembly of overlapping DNA fragments. Methods in Enzymology. 498, 349-361 (2011).
- Nageshwaran, S., et al. CRISPR Guide RNA Cloning for Mammalian Systems. Journal of Visualized Experiments. (140), (2018).
- Findlay, G. M., et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 562 (7726), 217-222 (2018).
- Kweon, J., Kim, D. E., Jang, A. H., Kim, Y. CRISPR/Cas-based customization of pooled CRISPR libraries. PLoS One. 13 (6), 0199473 (2018).
- Kim, Y., et al. A library of TAL effector nucleases spanning the human genome. Nature Biotechnology. 31 (3), 251-258 (2013).
- Sayers, E. W., et al. GenBank. Nucleic Acids Research. 47 (1), 94-99 (2019).
- Hwang, G. H., et al. Web-based design and analysis tools for CRISPR base editing. BMC Bioinformatics. 19 (1), 542 (2018).
- Kim, D., Kim, D. E., Lee, G., Cho, S. I., Kim, J. S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nature Biotechnology. 37 (4), 430-435 (2019).
- Clement, K., et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nature Biotechnology. 37 (3), 224-226 (2019).
- Hu, J. H., et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 556 (7699), 57-63 (2018).
- Nishimasu, H., et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 361 (6408), 1259-1262 (2018).
- Walton, R. T., Christie, K. A., Whittaker, M. N., Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. , (2020).
- Kim, D., et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nature Biotechnology. 35 (5), 475-480 (2017).
- Zuo, E., et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 364 (6437), 289-292 (2019).
- Jin, S., et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 364 (6437), 292-295 (2019).
- Grunewald, J., et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 569 (7756), 433-437 (2019).
- Doman, J. L., Raguram, A., Newby, G. A., Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nature Biotechnology. 38 (5), 620-628 (2020).
- Gaudelli, N. M., et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 551 (7681), 464-471 (2017).
- Anzalone, A. V., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 576 (7785), 149-157 (2019).

