High-Throughput In Vitro Assay using Patient-Derived Tumor Organoids
Summary
A highly accurate in vitro high-throughput assay system was developed to evaluate anticancer drugs using patient-derived tumor organoids (PDOs), similar to cancer tissues but are unsuitable for in vitro high-throughput assay systems with 96-well and 384-well plates.
Abstract
Patient-derived tumor organoids (PDOs) are expected to be a preclinical cancer model with better reproducibility of disease than traditional cell culture models. PDOs have been successfully generated from a variety of human tumors to recapitulate the architecture and function of tumor tissue accurately and efficiently. However, PDOs are unsuitable for an in vitro high-throughput assay system (HTS) or cell analysis using 96-well or 384-well plates when evaluating anticancer drugs because they are heterogeneous in size and form large clusters in culture. These cultures and assays use extracellular matrices, such as Matrigel, to create tumor tissue scaffolds. Therefore, PDOs have a low throughput and high cost, and it has been difficult to develop a suitable assay system. To address this issue, a simpler and more accurate HTS was established using PDOs to evaluate the potency of anticancer drugs and immunotherapy. An in vitro HTS was created that uses PDOs established from solid tumors cultured in 384-well plates. An HTS was also developed for assessment of antibody-dependent cellular cytotoxicity activity to represent the immune response using PDOs cultured in 96-well plates.
Introduction
Human cancer cell lines are widely accepted to study the biology of cancer and evaluate anticancer agents. However, these cell lines do not necessarily preserve the original characteristics of their source tissue because their morphology, gene mutation, and gene expression profile can change during culture over long periods. Furthermore, most of these cell lines are cultured in a monolayer or used as murine xenografts, neither of which physically represent tumor tissue1,2. Thus, the clinical efficacy of anticancer agents may not be the same as that observed in cancer cell lines. Therefore, in vitro systems, such as ex vivo assays using patient-derived tumor xenografts or patient-derived tumor organoids (PDOs) and tumor spheroid models that accurately reproduce the structure and function of tumor tissues, have been developed. Increasing evidence suggests that these models predict patients' response to anticancer agents by being directly comparable to the corresponding cancer tissue. These in vitro systems have been established for different tumor tissue types, and associated high-throughput assay systems (HTS) for drug screening have also been developed3,4,5,6,7. Heterogeneous ex vivo organoid cultures of primary tumors obtained from patients or patient-derived tumor xenografts have gained considerable traction in recent years because of their ease of culture and ability to maintain the complexity of cells in the stromal tissue8,9,10. These models are expected to enhance understanding of the biology of cancer and facilitate the evaluation of drug efficacy in vitro.
A series of novel PDOs were recently created from different types of tumor tissue, designated as F-PDO, under the Fukushima Translational Research Project. The PDOs form large cell clusters with a morphology similar to that of the source tumor and can be cultured for more than six months11. The comparative histology and comprehensive gene expression analyses showed that the features of the PDOs are close to those of their source tumor tissues, even after prolonged growth under culture conditions. Furthermore, a suitable HTS was established for each type of PDO in 96-well and 384-well plates. These assays were used to evaluate several molecular targeted agents and antibodies. Here, standard chemotherapeutics (paclitaxel and carboplatin) used for endometrial cancer were evaluated using F-PDOs derived from a patient who did not respond to paclitaxel and carboplatin. Accordingly, the cell growth inhibitory activity of paclitaxel and carboplatin against this PDO was weak (IC50: >10 µM). In addition, previous research has reported that the sensitivity of some F-PDOs to chemotherapeutic agents and molecular targeted agents is consistent with the clinical efficacy11,12,13. Finally, changes in the higher-order structure of the PDOs caused by anticancer agents were analyzed using a three-dimensional cell analysis system12,13. The results of the evaluation of anticancer agents using a PDO-based HTS are comparable with the clinical results obtained for these agents. Here, a protocol is presented for a simpler and more accurate HTS that can be used to evaluate the potency of anticancer agents and immunotherapy using the PDO models.
Protocol
All experiments involving human-derived materials were performed under the Declaration of Helsinki and approved in advance by the ethics committee of Fukushima Medical University (approval numbers 1953 and 2192; approval dates March 18, 2020, and May 26, 2016, respectively). Written informed consent was obtained from all patients who provided the clinical specimens used in this study.
1. Culture of PDOs
NOTE: F-PDOs form cell clusters that exhibit a variety of heterogeneous morphologies and grow in suspension culture (Figure 1). Furthermore, F-PDOs can be cultured for more than 6 months and can be cryopreserved for future use.
- Thawing of stored PDOs (day 0)
- Thaw and seed PDOs (e.g., RLUN007, lung adenocarcinoma) on day 0 (Figure 1). First, add 15 mL of medium for PDOs (see Table of Materials) containing 1% of B-27 supplement and 30 ng/mL of epidermal growth factor to a sterile 50 mL centrifuge tube.
- After removing the frozen vial from liquid nitrogen storage, gently agitate the PDOs in a 37 °C water bath for 2 min. Next, remove the vial from the water bath, wipe the vial with 70% ethanol, and then move the vial into a biological safety cabinet.
- Transfer the contents of the vial to the tube containing 15 mL of medium for PDOs. Mix the PDOs and medium by gently pipetting up and down five times with a 3 mL transfer pipette. Centrifuge the tube at 200 x g for 3 min at ~25 °C and discard the supernatant. Resuspend the PDO pellet in 5 mL of fresh medium and transfer to a 25 cm2 flask (see Table of Materials) with gentle pipetting. Finally, culture the PDOs in an incubator at 37 °C in 5% CO2.
- Change the medium twice per week (days 3-7). Centrifuge the PDO suspension to precipitate the cell clusters, and replace 4 mL of the medium (80% volume). Resuspend the cells in the fresh medium.
NOTE: The majority of RLUN007 appear as cell clusters that are 100-500 µm in diameter. When the color of phenol red in the medium changes to yellow as shown in Figure 1A, replace the medium more frequently. If the medium turns yellow on the day following replacement, and each cell cluster merges to form larger clusters that are >500 µm in diameter, passage at a split ratio of 1:2. - Subculture (days 8-28) of PDOs
NOTE: Given the difficulty of actually measuring the number of single cells, the timing of passage is determined based on an appropriate cell cluster density and the size of the PDO pellet after centrifugation (Figure 1B). In the case of RLUN007, the volume of the PDO pellet reaches 30 µL (saturated density in 25 cm2 flasks) approximately 2 weeks after thawing.- For the transfer from one 25 cm2 flask to two 25 cm2 flasks (P1), transfer the PDO suspension into a centrifuge tube and centrifuge at 200 x g for 2 min at approximately 25 °C. Estimate the volume of the PDO pellet, and discard the supernatant. Resuspend the PDO pellet using a 5 mL pipette in 5 mL of fresh medium. Pipette gently up and down five times at low speed. Then, transfer half of the volume of the PDO suspension (2.5 mL) into two flasks, and add 2.5 mL of fresh medium to each flask. Culture the cells at 37 °C in 5% CO2.
- For the transfer from two 25 cm2 flasks to one 75 cm2 flask (P2), transfer the PDO suspension from two flasks into two centrifuge tubes and centrifuge the PDO at 200 x g for 2 min at approximately 25 °C. Next, estimate the volume of the PDO pellet and resuspend the pellet in 2.5 mL of fresh medium (per tube). Thereafter, combine the PDO suspension in one tube with that in the other and transfer it to a 75 cm2 flask containing 10 mL of fresh medium. Culture the cells at 37 °C in 5% CO2. Transfer the sub cultured PDOs from two 25 cm2 flasks to one 75 cm2 flask approximately 1 week after the first passage (Figure 1C).
2. Growth inhibition HTS
NOTE: The growth inhibitory activity of anticancer agents against PDOs is evaluated by measuring the intracellular ATP content, as shown in Figure 2. This step is performed using a commercially available cell viability assay kit (see Table of Materials).
- On day 0, culture the PDOs (e.g., RLUN007) in flasks until adequate numbers of cell clusters are available for the assay. One day before seeding, transfer the PDO suspension from a 75 cm2 flask to a 15 mL tube and centrifuge at 200 x g for 2 min to measure the PDO pellet volume. Then, resuspend the PDO pellet in 15 mL of fresh medium and transfer it back to a 75 cm2 flask. Culture the cells at 37 °C in 5% CO2.
NOTE: The required PDO pellet volume depends on each PDO's dilution rate and the number of 384-well plates used for the assay. For RLUN007, a 200 µL of cell pellet volume is necessary for seeding into ten 384-well plates. - On day 1 (24 h after changing the medium), mince the PDOs using cell fragmentation and dispersion equipment (see Table of Materials) with a filter holder containing a 70 µm mesh filter. Then, dilute 15 mL of the PDO suspension by 10x. Seed 40 µL of the PDO suspension into 384-well ultra-low attachment spheroid (round-bottom) microplates (see Table of Materials) using a cell suspension dispenser (see Table of Materials).
NOTE: For mincing cell clusters, it is recommended to use the commercially available cell fragmentation and dispersion equipment (see Table of Materials). - At 24 h after seeding (day 2), treat the PDOs with 0.04 µL of test agent solutions at final concentration ranges of 20 µM to 1.0 nM (10 serial dilutions) using a liquid handler (see Table of Materials).
- On day 8 (144 h after test substance treatment), add intracellular ATP measuring reagent to the test wells. Mix the plates using a mixer and incubate for 10 min at 25 °C. Measure the intracellular ATP content as luminescence using a plate reader (see Table of Materials).
- To calculate the cell viability, divide the quantity of ATP in the test wells by that in the control wells containing the vehicle, with the background subtracted. To calculate the growth rate over 6 days, divide the quantity of ATP in the vehicle control wells by that in the vehicle control wells 24 h after seeding.
- Calculate the 50% inhibitory concentration (IC50) and area under the curve (AUC) values from the dose-response curves using biological data analysis software (see Table of Materials). The Z factor is a dimensionless parameter that ranges from 1 (infinite separation) to <0, defined as Z = 1 – (3σc+ + 3σc-) / |µc+ – µc-|, where σc+, σc-, µc+, and µc- are the standard deviations (σ) and averages (µ) of the high (c+) and low (c−) controls14, respectively.
3. HTS with a cell picking and imaging system for growth inhibition
NOTE: If there is a large deviation (when the coefficient of variation [CV] at assay is more than 20%) in the data using protocol 2, PDOs of a selected size may be seeded into 96-well or 384-well plates using a cell picking and imaging system (Figure 2). The protocol is the same as that described in steps 2.1 and 2.2 in the previous section. This step is performed using a commercially available cell picking and imaging system (see Table of Materials).
- On day 1, set a 384-well ultra-low attachment spheroid microplate with 40 µL of medium/well as a destination plate on the cell picking and imaging system.
- Fill the picking chamber (see Table of Materials) with 6 mL of culture medium and centrifuge at 1,500 x g for 2 min to remove air bubbles. Add the PDOs suspended in the medium (PDO pellet volume, 4 µL) to the picking chamber and set on the system.
- Stand the chamber for at least 1 minute, followed by dispersion, so that the cell clusters settle at the bottom of the chamber; then, perform scanning of the chamber.
- Set the picking size to 140-160 µm (area 15,386-20,000 µm2) on the system for automatic selection of cell clusters. Next, check the quality of the selected cell clusters on the scanned images and transfer ten cell clusters per well using picking tips (see Table of Materials) into the destination plate.
- Perform the protocol steps from step 2.3 onward.
4. HTS for antibody-dependent cellular cytotoxicity
NOTE: This step is performed using a commercially available system (see Table of Materials), which is an electrical impedance measuring instrument. It is used to evaluate cytolysis of PDOs by antibody-dependent cellular cytotoxicity (ADCC) with monoclonal antibodies and natural killer (NK) cells (Figure 3). NK cells are produced from peripheral blood mononuclear cells using the NK cell production kit (see Table of Materials), following the manufacturer's instructions.
- Measurement of ADCC activity
- On day 0, coat a 96-well plate (see Table of Materials) with 50 µL of 10 µg/mL fibronectin solution (0.5 µg/well) at 4 °C overnight.
- On day 1, after removing the fibronectin solution, add 50 µL of the culture medium to each well to measure the background impedance.
- Prior to seeding, add 5 mL of cell culture dissociation reagent (see Table of Materials) to the PDOs (RLUN007: 100 µL of the PDO pellet in a 75 cm2 flask) and incubate in a CO2 incubator at 37 °C for 20 min to disperse the PDOs. To stop trypsinization, add 5 mL of medium, centrifuge and remove the dissociation reagent. Rinse the PDOs with fresh medium and filter through a 40 µm cell strainer (see Table of Materials).
- Count the number of cells using a cell viability analyzer (see Table of Materials).
- Transfer the PDO suspension to a reservoir and mix using a multi-channel pipettor. Add the PDO suspension into wells at 5 x 104 cells/well in a 96-well plate. Each well contains a final volume of 100 µL. Then, place the plate in a biological safety cabinet at approximately 25 °C for 30 min.
- Transfer the plate to the instrument in a CO2 incubator at 37 °C. Record changes in the impedance signals every 15 min as the cell index.
- Thaw the NK cells in a 37 °C water bath on the same day as the PDO seeding. Transfer the cells from the vial to a 15 mL tube containing 10 mL of medium for NK cells (see Table of Materials) and centrifuge at 300 x g for 5 min at approximately 25 °C. Discard the supernatant and resuspend the cell pellet in 10 mL of fresh medium. After cell counting, adjust the cell density to 1 x 106 cells/mL and transfer to a 75 cm2 flask. Culture the NK cells in a 5% CO2 incubator at 37 °C.
- On day 2, prepare the antibody solutions (phosphate-buffered saline) at 10 times the final concentration in sterile V-bottom 96-well plates.
- Remove 60 µL of medium from each well, and add 10 µL of trastuzumab or cetuximab solution (10 µg/mL, 1 µg/mL, and 0.1 µg/mL) to the PDOs. Transfer the plates back to the instrument in an incubator and record the cell index for 1 h.
- Transfer the NK cell suspension into a 50 mL tube and count the cell number. Centrifuge the cells at 300 x g for 5 min and adjust the NK cell density to 1 x 106 cells/mL and 2 x 106 cells/mL with the medium for the PDOs.
- Add 50 µL of NK cell suspension to the effector (NK) cells at a target (RLUN007) cell ratio of 1:1 or 2:1. The final concentrations of the antibodies are 1 µg/mL, 0.1 µg/mL, and 0.01 µg/mL. Keep the plate at approximately 25 °C for 15 min, and return the plates to the instrument.
- Data collection and analysis
- Convert the cell index to percent cytolysis values using analysis software (see Table of Materials).
- The percent cytolysis refers to the percentage of target cells killed by NK cells versus target cells (PDOs) alone as a control. Subtract the cell indices of the wells containing only NK cells from the index of the sample wells at each time point. Normalize each value to the cell index immediately before the addition of antibodies. Convert the normalized cell index to percent cytolysis using the following equation: % cytolysis = (1 – normalized cell index [sample wells]) / normalized cell index (target alone wells) x 100.
Representative Results
A highly accurate HTS was developed using PDOs and 384-well microplates to evaluate anticancer agents, and the development of an HTS for each PDO was previously reported10,11,12,13. The performance of the HTS was evaluated by calculating the CVs and the Z'-factor. The Z'-factor is a widely accepted method for validation of assay quality and performance, and the assay is suitable for HTS if this value is >0.514. The control datum points in the 384-well plate assay using RLUN007 showed little variability, with CV values of 5.8% and calculated Z'-factors of 0.83, as shown in Figure 4. These results indicate that this assay has high performance for HTS. To investigate the sensitivity of PDOs to anticancer agents using HTS, growth inhibition was assessed using RLUN007 treated with eight anticancer agents, specifically, epidermal growth factor receptor (EGFR) inhibitors (afatinib, erlotinib, gefitinib, lapatinib, osimertinib, and rociletinib) and paclitaxel, which are standard clinical treatments for non-small cell lung cancer, and mitomycin C as a positive control. The IC50 and AUC values of the anticancer agents for each PDO are shown in Figure 4. The RLUN007 showed high sensitivity (IC50 < 2 µM, AUC < 282) for all EGFR inhibitors and other anticancer agents. Sigmoid curves calculated for all the data indicated that the growth inhibitory activity of the anticancer agents could be accurately measured.
The cell picking and imaging system is used when the data vary significantly using the above methodology. The cell picking and imaging system, which picks cell clusters accurately without damaging them, allows for accurate HTS assays by aligning the cell cluster size to exclude cell debris from the assay system. When the system was not used, the CV value was 26.0% and the Z'-factor value was 0.23 (data not shown). However, the CV and Z'-factor values were improved at 6.4% and 0.81, respectively, using the system.
To investigate cytolysis of PDOs with ADCC activity using the electrical impedance measuring instrument, which monitors the number, morphology, and attachment of cells for a long duration, changes in impedance signals were assessed using RLUN007 treated with the antibodies (trastuzumab and cetuximab) and NK cells as effector cells in a 96-well plate. Compared with the control consisting of only target cells, the percent cytolysis increased with time. It reached 45% or 75% after 6 h at an E:T ratio of 1:1 (Figure 5A,C) or 2:1 (Figure 5B,D) without the antibodies. NK cell-mediated cytolysis using trastuzumab was approximately 60% and 90% at a ratio of 1:1 (Figure 5A, 1 µg/mL) and 2:1 (Figure 5B, 1 µg/mL), respectively, at 6 h. In contrast, cetuximab had a dose-dependent impact on NK cell-mediated cytolysis (Figure 5C,D). At the highest concentration of cetuximab, RLUN007 were destroyed at 90% and 100% at a ratio of 1:1 and 2:1, respectively (Figure 5C,D). The effect of trastuzumab was weaker than that of cetuximab, with only 60% cytotoxicity. These results indicate that the PDO assay system can evaluate ADCC activity using real-time impedance-based technology.
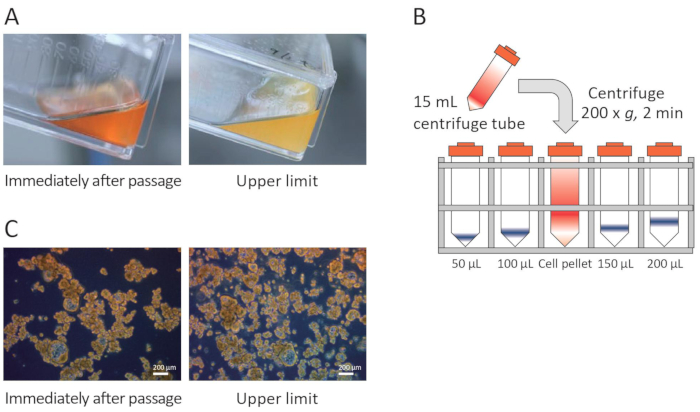
Figure 1: Critical points for PDO culture. (A) Color change in the medium. (B) Measurement of the quantity of PDOs from the pellet size by lining a centrifuge tube containing the PDOs with tubes marked at levels for 50-200 µL. (C) PDO density. Please click here to view a larger version of this figure.
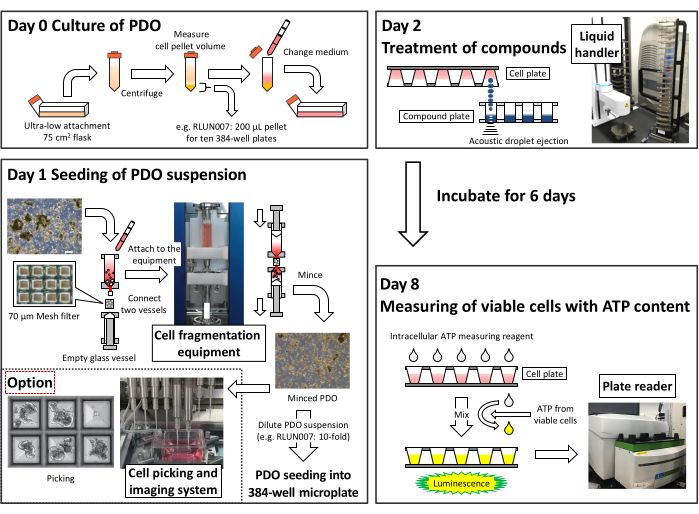
Figure 2: Summary of the protocol used to create a high-throughput assay system using 384-well microplates. Please click here to view a larger version of this figure.
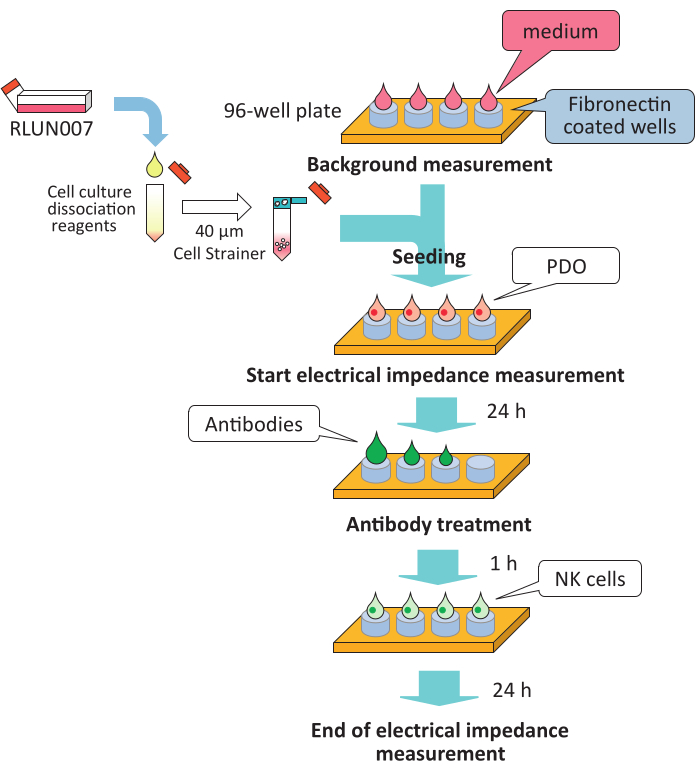
Figure 3: Summary of the protocol for high-throughput assay of ADCC activity. ADCC, antibody-dependent cellular cytotoxicity; NK, natural killer. Please click here to view a larger version of this figure.
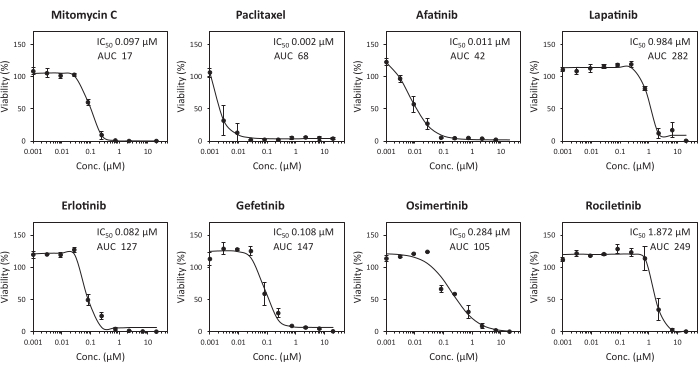
Figure 4: High-throughput assay system for growth inhibition with anticancer agents. Dose-response curve of RLUN007 to anticancer agents. The minced PDOs were seeded in 384-well plates. These were treated for 6 days with ten different concentrations of anticancer agents (between 10 µM and 1.5 nM). The data represent the mean ± standard deviation of triplicate experiments. Please click here to view a larger version of this figure.
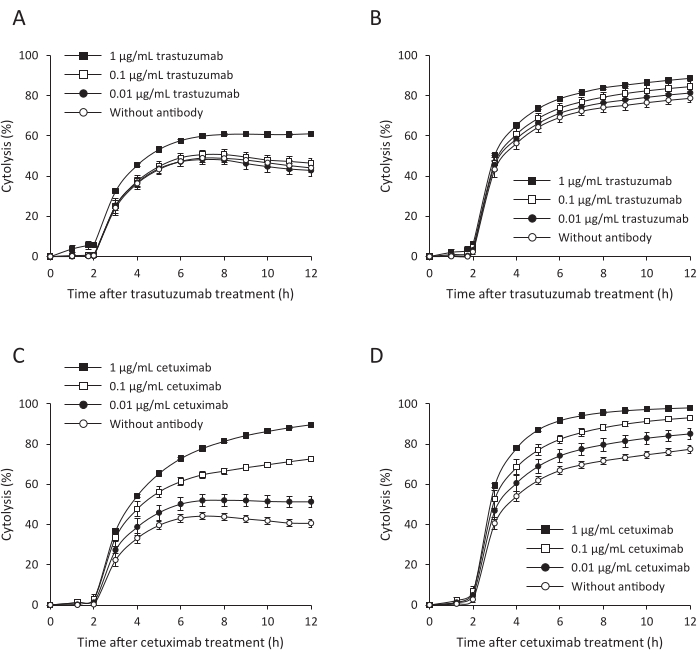
Figure 5: High-throughput assay for ADCC activity. (A,B) Trastuzumab. (C,D) Cetuximab. (A,C) A 1:1 ratio of RLUN007 to effector cells. (B,D) Cytolysis with a ratio of RLUN007: effector cells of 1:2. The activity was measured 12 h after the addition of the effector cells. The data are presented as the mean ± standard deviation of three replicate samples. ADCC, antibody-dependent cellular cytotoxicity. Please click here to view a larger version of this figure.
Discussion
The unique characteristic of PDOs is that they are not enzymatically separated into single cells during culture or assay and maintain cell clusters in culture. Therefore, the number of cells cannot be counted accurately under a microscope. To solve this problem, the number of cells is determined visually by lining a centrifuge tube containing the cells with tubes marked with levels for 50-200 µL (Figure 1B). Furthermore, because it is difficult to measure the pellet volume of cell clusters cultured in a 25 cm2 flask visually, the time of passage was determined using the medium color change from red to yellow and the noticeable increase in single cells or debriscompared with the time of passaging as indicators (Figure 1A,C). This is the point of passaging for PDOs. The quantity of PDO pellets is measured visually after centrifugation at each medium change. When the pellet volume stops increasing, and the medium turns yellow on the day after medium replacement, the medium is considered to be saturated with density, and passaging is performed. The pellet volume is defined for each PDO. If the PDOs do not proliferate, the quantity of medium is changed from 80% to 50% at the time of medium exchange, and the density of PDOs is increased in the culture.
An HTS suitable for PDOs was developed. Its throughput is at least ten to twenty 384-well plates performed using one 75 cm2 flask of PDO, and the number of plates processed per day is at least 50. In addition, the results of the evaluation of various anticancer drugs by HTS using PDOs have already been reported.
When performing HTS, clogging of the mesh filter caused by mincing of the F-PDOs using the cell fragmentation and dispersion equipment is addressed initially by changing the mesh size of the filter to 100 µm. The next step is to reduce the volume of the PDO suspension applied to the glass vessel. When preparing test substance solutions for HTS, low-molecular-weight compounds are usually dissolved in dimethyl sulfoxide, and the antibodies are dissolved in phosphate-buffered saline. The appropriate solvent is used as the test substance, and the control data are obtained from the solvent used.
The following is a description of how to deal with variability in the assay data. If there is large variation in the data in the test using 384-well plates, the assay plate should be changed to a 96-well plate format. The PDO dilution factor (number of seeded cell clusters) is also examined after seeding the plate. Finally, the cell picking and imaging system can be used to select the size of PDOs for the assay. Before adding PDOs to the chamber, single cells and small cell clusters should be removed by low-speed centrifugation to be able to correctly recognize PDOs. If single cells or small cell clusters are visible after adding PDOs to the chamber, multiple dispersions can be performed to remove the single cells. Next, although the cell picking and imaging system has a function that allows the plate to be kept warm, this function is not used because of the evaporation of the culture medium when the system is working for a long period of time. Lastly, the volume of cell clusters is unknown because it is recognized by a planar image. Furthermore, if two or more PDOs overlap, a single PDO cannot be recognized correctly.However, it is possible to remove unwanted PDOs using the remove function by checking them on the scanned image after the move.
The electrical impedance measuring instrument is generally used for adherent target cancer cells to monitor changes in impedance during cell proliferation. Therefore, a change in the cell index of non-adherent PDOs is not detected. In an attempt to solve this problem, it is necessary to investigate the seeding conditions such as PDO density and enzymatic treatment (cell dissociation enzyme and treatment time) depending on the type of PDO. The wells in the plate must also be coated with an appropriate extracellular matrix for seeding PDOs. PDOs are seeded without enzymatic treatment, depending on the type of PDO. RLUN007 was used to measure the impedance by seeding the PDOs on a 96-well plate after dispersing them by enzymatic treatment. RLUN007 was treated with the cell culture dissociation reagent for 20 min at 37 °C to disperse the cells and attach them to the wells of a 96-well plate. Given that dissociated RLUN007 cells immediately form aggregates, it is desirable to seed on the plates just after filtration using a strainer. After transferring the cell suspension from the tube to a reservoir, the reservoir was gently moved from right to left two to three times and pipetted up and down five times before seeding onto the plate. The suspension was also mixed with each addition to the well. The plate was then placed in a biological safety cabinet for 30 min (for PDOs) or 15 min (for NK cells) to allow the cells to distribute evenly in the well. The second important point is that treatment with antibodies and NK cells should be timed to occur before the cell index reaches a plateau and the value is not less than 0.5. In the case of RLUN007, the optimal time to start the assay is 20-22 h after plating, and the cell number for seeding is 5 x 104 cells/well.
In general, the culture and assays for tumor organoids use extracellular matrices such as Matrigel to create tumor tissue scaffolds or enzymes such as trypsin and collagenase to disrupt the organoids3,4,5,6,7. The advantage of this method is that no extracellular matrix or enzymatic treatment is required during culture and assay (except for assays using the electrical impedance measuring instrument), which significantly reduces labor requirements and costs. Furthermore, this method is relatively easy to adapt to HTS assay systems and various measurement systems. However, the use of an extracellular matrix is desirable for some research purposes because it can act as a scaffold for cells and affect morphogenesis, differentiation, and homeostasis in tissues.
In this study, PDOs (RLUN007) with an EGFR mutation (L858R) that is clinically sensitive to EGFR inhibitors and high expression of the EGFR gene (data not shown) were used to evaluate EGFR inhibitors. It was demonstrated that the sensitivity of RLUN007 to EGFR inhibitors was higher than those of other lung cancer-derived F-PDOs13 (Figure 4). Thus, an HTS using PDOs, which retain the characteristics of tumor tissue, is superior for the evaluation of potential anticancer agents and presents opportunities for drug assessment and advances in personalized medicine. Although HTS is suitable for the initial screening of agents, it does not reproduce the tumor microenvironment and thus cannot evaluate the efficacy of drugs in vivo. Therefore, an in vitro system that can mimic human tumor tissue in vivo by co-culture with vascular endothelial cells and other stromal cells or organ-on-a-chip technology in the absence of animal models is now under development.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank the patients who provided the clinical specimens used in this research. This research is supported by grants from the Translational Research Program of Fukushima Prefecture.
Materials
| 384-well Ultra-Low Attachment Spheroid Microplate | Corning | 4516 | Plates for HTS |
| 40-µm Cell Strainer | Corning | 352340 | |
| AdoptCell-NK kit | Kohjin Bio | 16030400 | Kit for NK cell production |
| Cancer Cell Expansion Media plus | Fujifilm Wako Pure Chemical | 032-25745 | Medium for F-PDO |
| ALyS505N-175 | Cell Science & Technology institute | 10217P10 | Medium for NK cells |
| CELL HANDLER | Yamaha Motor | – | Cell picking and imaging system |
| CellPet FT | JTEC | – | Cell fragmentation and dispersion equipment |
| CellTiter-Glo 3D Cell Viability Assay | Promega | G9683 | Cell viability luminescent assay, intracellular ATP measuring reagent |
| Echo 555 | Labcyte | – | Liquid handler |
| EnSpire | PerkinElmer | – | Plate reader |
| E-plate VIEW 96 | Agilent | 300601020 | Plates are specifically designed to perform cell-based assays with the xCELLigence RTCA System |
| Fibronectin Solution | Fujifilm Wako Pure Chemical | 063-05591 | Plate coating for xCELLigence RTCA System |
| F-PDO | Fujifilm Wako Pure Chemical or Summit Pharmaceuticals International | – | The F-PDO can be purchased from Fujifilm Wako Pure Chemicals or Summit Pharmaceuticals International |
| Morphit software, version 6.0 | The Edge Software Consultancy | Biological data analysis software | |
| Multidrop Combi | ThermoFisher Scientific | 5840300 | Cell suspension dispenser |
| Precision Chamber | Yamaha Motor | JLE9M65W230 | Chamber for picking cell clusters using CELL HANDLER |
| Precision Tip | Yamaha Motor | JLE9M65W300 | Micro tip for picking cell clusters using CELL HANDLER |
| RLUN007 | Fujifilm Wako Pure Chemical or Summit Pharmaceuticals International | Lung tumor derived F-PDO | |
| TrypLE Express | ThermoFisher Scientific | 12604021 | Cell culture dissociation reagent |
| Ultra-Low Attachment 25 cm² Flask | Corning | 4616 | Culture flask for PDO |
| Ultra-Low Attachment 75 cm² Flask | Corning | 3814 | Culture flask for PDO |
| Vi-CELL XR Cell Viability Analyzer System | Beckman coulter | – | Cell viability analyzer |
| xCELLigence immunotherapy software, version 2.3 | ACEA Bioscience | – | Analysis software for xCELLigence RTCA System |
| xCELLigence RTCA System | ACEA Bioscience | – | Electrical impedance measuring instrument for cytolysis |
References
- Sharma, S. V., Haber, D. A., Settleman, J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nature Reviews Cancer. 10 (4), 241-253 (2010).
- Shamir, E. R., Ewald, A. J. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nature Reviews Molecular Cell Biology. 15 (10), 647-664 (2014).
- Xu, H., et al. Organoid technology and applications in cancer research. Journal of Hematology & Oncology. 11 (1), 116 (2018).
- Palechor-Ceron, N., et al. Conditional reprogramming for patient-derived cancer models and next-generation living biobanks. Cells. 8 (11), 1327 (2019).
- Broutier, L., et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nature Medicine. 23 (12), 1424-1435 (2017).
- Du, Y., et al. Development of a miniaturized 3D organoid culture platform for ultra-high-throughput screening. Nature Reviews Molecular Cell Biology. 12 (8), 630-643 (2020).
- van de Wetering, M., et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161 (4), 933-945 (2015).
- Meijer, T. G., Naipal, K. A., Jager, A., van Gent, D. C. Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Science OA. 3 (2), (2017).
- Inoue, A., et al. Current and future horizons of patient-derived xenograft models in colorectal cancer translational research. Cancers (Basel). 11 (9), 1321 (2019).
- Hum, N. R., et al. Comparative molecular analysis of cancer behavior cultured in vitro, in vivo, and ex vivo. Cancers (Basel). 12 (3), 690 (2020).
- Tamura, H., et al. Evaluation of anticancer agents using patient-derived tumor organoids characteristically similar to source tissues. Oncology Reports. 40 (2), 635-646 (2018).
- Takahashi, N., et al. An in vitro system for evaluating molecular targeted drugs using lung patient-derived tumor organoids. Cells. 8 (5), 481 (2019).
- Takahashi, N., et al. Construction of in vitro patient-derived tumor models to evaluate anticancer agents and cancer immunotherapy. Oncology Letters. 21 (5), 406 (2021).
- Zhang, J. H., Chung, T. D., Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of Biomolecular Screening. 4 (2), 67-73 (1999).

