Evaluation of Substrate Ubiquitylation by E3 Ubiquitin-ligase in Mammalian Cell Lysates
Summary
We provide a detailed protocol for a ubiquitylation assay of a specific substrate and an E3 ubiquitin-ligase in mammalian cells. HEK293T cell lines were used for protein overexpression, the polyubiquitylated substrate was purified from cell lysates by immunoprecipitation, and resolved in SDS-PAGE. Immunoblotting was used to visualize this post-translational modification.
Abstract
Ubiquitylation is a post-translational modification which occurs in eukaryotic cells that is critical for several biological pathways’ regulation, including cell survival, proliferation, and differentiation. It is a reversible process that consists of a covalent attachment of ubiquitin to the substrate through a cascade reaction of at least three different enzymes, composed of E1 (Ubiquitin-activation enzyme), E2 (Ubiquitin-conjugating enzyme), and E3 (Ubiquitin-ligase enzyme). The E3 complex plays an important role in substrate recognition and ubiquitylation. Here, a protocol is described to evaluate substrate ubiquitylation in mammalian cells using transient co-transfection of a plasmid encoding the selected substrate, an E3 ubiquitin ligase, and a tagged ubiquitin. Before lysis, the transfected cells are treated with the proteasome inhibitor MG132 (carbobenzoxy-leu-leu-leucinal) to avoid substrate proteasomal degradation. Furthermore, the cell extract is submitted to small-scale immunoprecipitation (IP) to purify the polyubiquitylated substrate for subsequent detection by western blotting (WB) using specific antibodies for ubiquitin tag. Hence, a consistent and uncomplicated protocol for ubiquitylation assay in mammalian cells is described to assist scientists in addressing ubiquitylation of specific substrates and E3 ubiquitin ligases.
Introduction
Post-translational modifications (PTMs) are an important mechanism regarding protein regulation, which is essential for cell homeostasis. Protein ubiquitylation is a dynamic and intricate modification that creates an assortment of different signals resulting in several cellular outcomes in eukaryotic organisms. Ubiquitylation is a reversible process consisting in the attachment of a ubiquitin protein containing 76 amino acids to the substrate, occurring in an enzymatic cascade composed by three distinct reactions1. The first step is characterized by ubiquitin activation, which depends on an ATP hydrolysis to form a high-energy thioester-linked ubiquitin between the ubiquitin C-terminus and the cysteine residue present in the active site of the E1 enzyme. Subsequently, the ubiquitin is transferred to the E2 enzyme forming a thioester-liked complex with the ubiquitin. Afterward, the ubiquitin is covalently attached to the substrate by the E2, or more often, by the E3 enzyme, which recognizes and interacts with the substrate2,3. Occasionally, E4 enzymes (Ubiquitin-chain elongation factors) are necessary to promote multiubiquitin chain assembly3.
Ubiquitin has seven lysine residues (K6, K11, K27, K29, K33, K48, and K63), allowing the formation of polyubiquitin chains that generate distinct linkages to produce different tridimensional structures that are going to be recognized by several effector proteins4,5. Hence, the kind of polyubiquitin chain introduced in the substrate is essential to decide its cell fate6,7,8. Moreover, the substrate could also be ubiquitinated through its N-terminal residues called N-degrons. Specific E3 ubiquitin-ligases are responsible for N-degron recognition, allowing the polyubiquitylation of nearby lysine residue9.
Nowadays, there are more than 40 different SCF-specific substrates characterized. Among those, key regulators of several biological pathways, including cell differentiation and development as well as cell survival and death, can be found10,11,12,13. Thus, the identification of specific substrates of each E3 ubiquitin-ligase is essential to design a comprehensive map of various biological events. Even though the identification of true substrates is biochemically challenging, the use of biochemistry-based methods is very suitable to evaluate chain specificity and the distinction between mono- and polyubiquitylation14. This study describes a complete protocol for ubiquitylation assay using the mammalian cell line HEK293T overexpressing the substrate UXT-V2 (Ubiquitously expressed prefoldin-like chaperone isoform 2) with the E3 ubiquitin-ligase complex SCF(Fbxo7). UXT-V2 is an essential co-factor for NF-κB signaling, and once this protein is knocked down in cells, it inhibits TNF-α-induced NF-κB activation11. Thus, to detect polyubiquitylated UXT-V2, the proteasome inhibitor MG132 is used since it has the ability to block the proteolytic activity of the 26S subunit of the proteasome complex15. Furthermore, the cell extract is submitted to a small-scale IP to purify the substrate, utilizing a specific antibody immobilized to agarose resin for subsequent detection by WB using selected antibodies. This protocol is very useful to validate substrate ubiquitylation in the cellular environment, and it can also be adapted for different types of mammalian cells and other E3 ubiquitin-ligase complexes. However, it is necessary to validate the substrate tested through an in vitro ubiquitylation assay as well, since both protocols complement each other regarding the identification of true substrates.
Protocol
NOTE: An overview of ubiquitylation assay protocol in mammalian cells is represented in Figure 1.
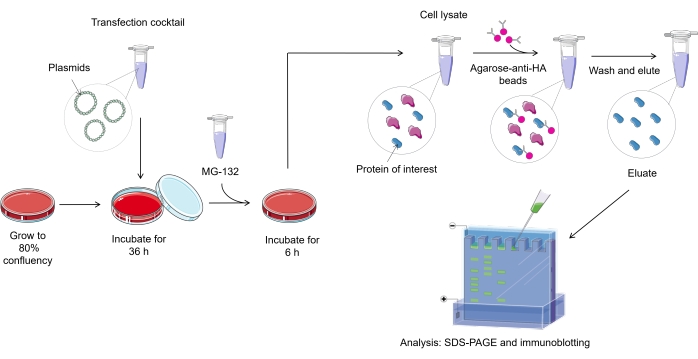
Figure 1. Overview of the ubiquitylation assay procedure. Please click here to view a larger version of this figure.
1. Cell culture
- Grow HEK293T cell line in a 100 mm TC-treated culture dish to 80%-90% confluence in growth media (Dulbecco's Modified Eagle's Medium (DMEM) high glucose supplemented with 10% fetal bovine serum (FBS) and penicillin (100 units), streptomycin (100 µg) and L-glutamine (0.292 mg/mL)). Incubate the culture at 37 °C in a humidified cell culture incubator at 5% CO2.
- To passage the cells, aspirate the media from the culture dish using a serological pipette. Wash the cells once with 1 mL of sterilized 1x phosphate-buffered saline (PBS 1x).
- Detach the cells by adding 1 mL of trypsin/EDTA (ethylenediaminetetraacetic acid) solution. Incubate the dish at 37 °C for 5 min. Resuspend the cells using a serological pipette in 2 mL of growth media.
- Transfer the cell suspension to a fresh and clean 15 mL tube and centrifuge at 500 x g for 5 min at room temperature (RT). Remove the supernatant by pouring it out carefully. Gently resuspend the cell pellet in 3 mL of growth media by pipetting up and down to obtain a homogenous cell suspension.
- Transfer 1 mL of the cell suspension to a 100 mm TC-treated culture dish containing 9 mL of growth media.
NOTE: If the cells are in good conditions, each culture dish of HEK293T, wherein the confluence is 80%-90%, can generate three culture dishes with 80% confluence 2 days after the passage.
2. Cell transfection
NOTE: It is not recommended to transfect the cell culture if the confluence reached is less than 80%.
- Before transfection, certify if the cells are free from contamination and at an adequate confluence for transient transfections.
- For each transfection sample, prepare the DNA-Polyethylenimine (PEI 1 µg/µL at pH 7.2) complexes as follows:
- Dilute 3 µg of each plasmid in 100 µL of opti-MEM I reduced serum medium without supplementation and mix it gently by pipetting the solution up and down.
NOTE: Here, the cells were transfected with 4 µg of each plasmid: Empty vector (pcDNA3) or FLAG-Fbxo7 constructs and UXT-V2-HA, with or without 6xHis-myc-ubiquitin. The total DNA content was 12 µg. - Thaw the PEI at RT and add it into the solution following a proportion of 3 µL of PEI per 1 µg of DNA. Homogenize the solution by pipetting up and down. Then incubate it for 15 min at RT to allow the formation of DNA-PEI complexes.
NOTE: The optimal proportion of PEI volume per DNA quantity differs according to the cell line selected.
- Dilute 3 µg of each plasmid in 100 µL of opti-MEM I reduced serum medium without supplementation and mix it gently by pipetting the solution up and down.
- Add the total volume of DNA-PEI complexes to each dish containing the cell culture and mix it gently by rocking the plate back and forth. Incubate the cells at 37 °C in a humidified cell culture incubator at 5% CO2.
- Replace the growth medium after 5 h, since prolonged PEI exposure can be toxic to HEK293T cells. Incubate the cells at 37 °C in a humidified cell culture incubator at 5% CO2 for 36 h.
3. Cell lysis and immunoprecipitation
- After the incubation period and 6 h prior to cell lysis, treat the transfected cells with 10 µM of the proteasome inhibitor MG-132. Once again, incubate the cells at 37 °C in a humidified cell culture incubator at 5% CO2.
- Aspirate the media from each culture dish using a serological pipette and wash it once with 1 mL of 1x PBS. Detach the cells by adding 1 mL of trypsin and incubating the dish at 37 °C for 5 min. Resuspend the cells in 1 mL of growth media.
- Transfer the cell suspension into a fresh and clean 15 mL tube and centrifuge it at 500 x g for 5 min at RT.
- Remove the supernatant by pouring it out carefully. Gently resuspend the cell pellet in 200 µL of ice-cold NP-40 lysis buffer (50 mM Tris-HCl pH 7.2, 225 mM KCl, and 1% NP-40), supplemented with the protease and phosphatase inhibitors cocktail (10 mM NaF and 1 mM Na3VO4), and transfer the solution into a clean 1.5 mL microtube.
- Incubate the cell lysate for 30 min on ice. After incubation, centrifugate the cell lysates at 16,900 x g for 20 min at 4 °C.
- Meanwhile, equilibrate the agarose-anti-HA beads with ice-cold NP-40 lysis buffer. Use 15 µL of agarose-anti-HA beads for each sample. Wash the beads with 200 µL of NP-40 lysis buffer by pulsing it in a microcentrifuge tube at 3,000 x g for 1 min at 4 °C. With a pipette, aspirate and discard the supernatant very carefully; repeat this process three times. Afterward, keep the beads equilibrated on ice until use.
- After centrifuging the cell lysates, recover the supernatant. Quantify the protein content in the total lysate using the Bradford method16.
- Ensure that each sample subjected to immunoprecipitation presents an equal amount of protein. Incubate the necessary volume of cell lysate with the equilibrated agarose-anti-HA beads for 4 h, gently rotating in a rotating incubator at 4 °C, which allows the UXT-V2-HA to bind to the agarose-anti-HA beads.
- Collect the agarose-anti-HA beads by pulsing them in a microcentrifuge tube at 3,000 x g for 1 min at 4 °C. Carefully aspirate and discard the supernatant. Wash the beads three times with ice-cold NP-40 cell lysis buffer and twice with ice-cold FLAG/HA buffer (10 mM Hepes pH 7.9, 15 mM MgCl2, 225 mM KCl, and 0.1% NP-40).
- After the final wash, remove all the supernatant carefully using a pipette and elute the polyubiquitylated protein with HA peptide (300 µg/mL) diluted on FLAG/HA buffer. Incubate the agarose-anti-HA beads with HA peptide for 1 h at 4 °C in a rocking shaker platform.
- Spin down the beads at 3,000 x g for 2 min at 4 °C and carefully pipette the supernatant containing the polyubiquitinated proteins. If necessary, store the eluate in a fresh and clean microtube at -20 °C.
- Resolve the eluates and the cell lysates in 10% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis)17 and immunoblotting.
- In this study, a wet transfer WB was performed. For this kind of transfer, place the gel in a transfer sandwich composed of filter paper-gel-membrane-filter paper, cushion it with pads, and press it together by a support grid. Place this system vertically in a tank filled with transfer buffer and between stainless steel/platinum wire electrodes. The transfer occurs for 90 min at 150 V in wet transfer buffer (glycine 192 mM, tris-base 25 mM, 0.025% SDS, 20% Methanol).
NOTE: Since the cell extracts were quantified (step 3.7), run an SDS-PAGE with an equal quantity of protein for each sample. Also, to resolve the eluate, run equal volumes of each sample. - Probe the immunoblot membrane using selected antibodies 11. Ensure that a smear signal is detected in the eluate from the polyubiquitylated substrate pulled in the IP process by using anti-myc antibody in the samples containing the wild-type E3 ligase, the substrate, and myc-ub. In the cell lysate (input), ensure that the signal from the chosen substrate, the Fbxo7 protein, ubiquitylated proteins, and a housekeeping protein (e.g., GAPDH and β-actin) is detected to guarantee the same quantity of proteins in each lane.
NOTE: The dilution for each antibody used was prepared according to the manufacturer's instructions.
Representative Results
UXT (ubiquitously expressed transcript) is a prefoldin-like protein that forms ubiquitously expressed protein-folding complexes in mouse and human tissues such as heart, brain, skeletal muscle, placenta, pancreas, kidney, and liver18. Two splicing isoforms of UXT, which are named UXT-V1 and UXT-V2, have been described performing distinct functions and subcellular locations. UXT-V1 is predominantly localized in the cytoplasm and inside the mitochondria, and it is implicated in TNF-α-induced apoptosis and antiviral signalosome formation19,20. Most research has focused on UXT-V2 that is mainly localized in the nucleus and acts as a co-factor for multiple transcriptional factors involved in cell proliferation regulation, differentiation, and in inflammatory pathways. UXT-V2 is involved in the NF-κB signaling pathway working as an essential coactivator in the NF-κB enhanceosome21. It was demonstrated that UXT-V2 is a canonical substrate of the E3 ubiquitin-ligase SCF(Fbxo7), mediating its polyubiquitylation and degradation by the proteasome, and consequently inhibiting the NF-κB signaling pathway11.
The specific polyubiquitylation of UXT-V2-HA mediated by Fbxo7 in cells was observed after probing the eluate from anti-HA IP with an anti-myc antibody, which detects the myc-ub conjugated to the substrate (Figure 2). The specificity of anti-myc for polyubiquitylated substrate was visualized in lanes 1 and 2 (Figure 2), wherein smear of polyubiquitylated proteins was not detected when cells were co-transfected with Fbxo7 and myc-ub in the absence of UXT-V2-HA plasmid (Figure 2, lane 1). Additionally, no smear corresponding to protein polyubiquitination was visualized in the combination of Fbxo7 and UXT-V2-HA without the myc-ub plasmid (Figure 2, lane 2). To prove that UXT-V2 polyubiquitylation was mediated by Fbxo7, an empty vector pcDNA3, wild type Fbxo7 or Fbxo7-ΔF-box mutant, which is unable to assemble active SCF complex due to the absence of the F-box domain that is responsible for interaction with SKP1, was transfected in combination with UXT-V2-HA and myc-ub. A strong smear signal of polyubiquitylated UXT-V2 was observed only in the presence of the wild-type Fbxo7 (Figure 2, lane 4), suggesting that UXT-V2 was polyubiquitylated by SCF(Fbxo7) complex in cells compared to the controls (Figure 2, lanes 3 and 5). The presence of a faint smear signal of polyubiquitylated UXT-V2 in the presence of these negative controls might indicate the action of endogenous Fbxo7 or other E3 ubiquitin-ligase activity.
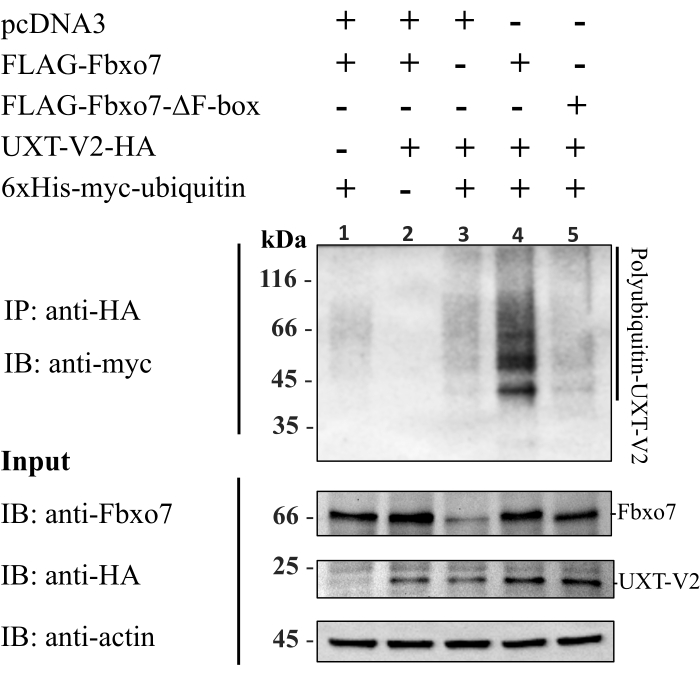
Figure 2. Ubiquitylation assay in cells: Transient transfection of HEK293T cells with the indicated plasmids. After cell lysis, the cell extracts (inputs) were immunoprecipitated with agarose-anti-HA beads. The eluted and input samples were resolved by SDS-PAGE, and the western blot was probed with specific antibodies. Please click here to view a larger version of this figure.
Discussion
Ubiquitylation is an essential post-translational modification that regulates the levels of several proteins and plays a crucial role in many signaling pathways and biological processes, ensuring a healthy intracellular environment. The ubiquitin-proteasome system (UPS) is one of the main focuses of recent pharmaceutical research, providing the possibility of stabilizing tumor suppressors or inducing the degradation of oncogenic products22. For instance, the aberrant proliferation of plasma cell neoplasms responsible for monoclonal immunoglobulin secretion in multiple myeloma (MM) promotes a pathophysiological pathway due to misfolded and/or unfolded protein accumulation in the endoplasmic reticulum (ER)23. Once this ER stress happens, it activates several processes, including the activation of the ubiquitin-proteasome system. The use of proteasome inhibitors (bortezomib, carfilzomib, and ixazomib) to induce protein accumulation and consequent cell death for multiple myeloma treatment showed promising results as an anti-tumor drug by reducing disease progression in high-risk patients24,25. Moreover, other efforts focused on protein stability are aimed, such as inhibitors for the E3 ubiquitin-ligase MDM2 that showed great ability in avoiding the ubiquitylation of p53, hence preventing its degradation through the proteasome26. Thus, a new era of pharmaceutical/biomedical research centered on understanding and controlling the UPS as a method to combat the disease has arisen.
This study describes a method to analyze the ubiquitylation of a specific target protein by a given E3 ubiquitin-ligase in a cellular environment. In this assay, a significant number of cells are transiently co-transfected with selected plasmids encoding for a tagged protein substrate, an E3 ligase, and a tagged ubiquitin. Before lysis, the cells must be treated with a proteasome inhibitor to allow the accumulation of polyubiquitylated proteins that would undergo degradation in the proteasome. In order to obtain a less complex sample, the target protein was immunoprecipitated, and its polyubiquitylation was analyzed by WB using specific antibodies based on the ubiquitin tag.
The amount of protein in each sample and agarose beads must be normalized to allow the data comparison among those samples. A high-intensity smear signal of UXT-V2 polyubiquitylation in the presence of the wild-type E3 ubiquitin-ligase was detected. Even though the signal obtained from the target protein, the E3 ligase, and the ubiquitin could be from an endogenous origin, the use of recombinant proteins has the advantage of higher yield and signal detection in WB analysis. Furthermore, using the wild-type E3 ligase and its mutant allows the specific analysis of their activity in the substrate, which is an essential control for this experiment. It was observed that the Fbxo7 mutant (Fbxo7-ΔF-box) could not promote the polyubiquitylation of UXT-V2, suggesting the smear signal detected in the presence of the wild-type Fbxo7 was specific. In addition, it is important to observe that in the absence of the tagged substrate or myc-ub, the polyubiquitylated smear was not detected, indicating the specificity of agarose-anti-HA used. Using an anti-ubiquitin antibody to probe the eluate from substrate IP might detect a polyubiquitylated smear signal even in the negative controls, since unspecific proteins or indirect protein partners of the target can also be eluted together with the substrate. Once these unspecific proteins might be polyubiquitylated, the high sensitivity of anti-ubiquitin can detect them.
There are also some variations for this protocol regarding plasmid transfection, cell lysis buffer, immunoprecipitation antibody, and WB probing. The IP presented here was developed with anti-HA antibody to precipitate the substrate, and the WB membrane was probed with anti-myc antibody to visualize the ubiquitin signal. The transfection of ubiquitin-HA and IP with anti-HA and WB probing the membrane with an anti-substrate or anti-substrate tag is also an alternative for this methodology. However, the immunoprecipitation of ubiquitin might bring a huge number of unspecific polyubiquitylated proteins in addition to ubiquitin-binding proteins. In this case, it is recommended to use RIPA buffer (radioimmunoprecipitation assay buffer) during cell lysis to minimize these undesired proteins27. However, it is essential to evaluate whether the IP protocol is compatible with RIPA buffer. Furthermore, substrate polyubiquitylation might impair the binding of the antibody to this specific target by blocking the conformational epitope. Thus, the application of antibodies with anti- tag is more reliable.
While this approach is very useful for soluble protein purification, it presents some limitations. First, the action of unspecific endogenous E3 ubiquitin-ligases could be observed. To overcome this issue and to confirm the specificity of the target ubiquitylation in cells, it is essential to also perform an in vitro ubiquitylation assay using purified E3 ligase complex and selected substrate in addition to E1 and E2 enzymes, ubiquitin, ubiquitin buffer, and ATP, which are commercially available. Another limitation of this method occurs when the overexpression of the target protein causes a cytotoxic effect that culminates in reduced cellular viability. In this situation, using the endogenous protein target for immunoprecipitation is recommended. Additionally, as mentioned above, this protocol is suitable for soluble protein extraction; therefore, it cannot extract membrane-bound proteins, for instance. Whether the substrate tested is a membrane protein, the RIPA buffer could replace the NP-40 lysis buffer described here if the IP protocol is not compromised by it. In this protocol, the focus of protein clearance is the ubiquitin-proteasome system, which explains that only the proteasome inhibitor was used. However, numerous soluble proteins undergo degradation through autophagy in the lysosome. Consequently, using a lysosome inhibitor, such as bafilomycin A1, could also be necessary to block late-phase autophagy due to vacuolar H+-ATPase28.
Since ubiquitylation is a reversible process, the ubiquitin can be removed from the substrate by the catalytic activity of deubiquitinases (DUBs); this is one of the major obstacles when characterizing E3 ubiquitin-ligase substrates29. The action of DUBs increases the substrate discrimination in this kind of experiment once it decreases polyubiquitylation dwell times on some targets30. Therefore, it would be beneficial to treat the cells with deubiquitinase inhibitors to avoid this problem. Positive or negative crosstalk could also influence the fate of a specific substrate. It is already established that post-translational modifications (PTMs) can crosstalk in various scenarios, and phosphorylation, for example, can regulate ubiquitylation by modulation of E3 ubiquitin-ligase activity, promoting substrate recognition by E3 ligases or through regulating substrate and E3 ligase interaction by affecting its cellular localization31. This could be an issue when choosing the cell line in which the experiment will be executed. If the signals that are prerequisites for ubiquitylation are absent in the desirable cell line, it is required to select an alternative cell line to conduct the analysis or induce the signal in the cell line selected. A common problem that may occur in ubiquitylation assays performed in cells is massive cell death after MG132 treatment, which is mostly caused by the generation of reactive oxygen species (ROS) that induce apoptotic cell death. After several tests to standardize the protocol, it was found an ideal MG132 treatment for HEK293T cells to not exceed 6 h before lysis at 10 µM of concentration. This protocol is also suitable for other mammalian cell types to identify substrate-E3 ubiquitin-ligase pairs; although, it is crucial to standardize the volume of PEI used in the transfection and the MG132 treatment period for the chosen cell line.
Divulgations
The authors have nothing to disclose.
Acknowledgements
F.R.T is supported by FAPESP grant number 2020/15771-6 and CNPq Universal 405836/2018-0. P.M.S.P and V.S are supported by CAPES. C.R.S.T.B.C was supported by FAPESP scholarship number 2019/23466-1. We thank Sandra R. C. Maruyama (FAPESP 2016/20258-0) for the material support.
Materials
| 1.5 mL microtube | Axygen | PMI110-06A | |
| 100 mm TC-treated culture dish | Corning | 430167 | |
| 15 mL tube | Corning | 430766 | |
| 96-well plate | Cralplast | 655111 | |
| Agarose-anti-HA beads | Sigma-Aldrich | E6779 | |
| Anti Mouse antibody | Seracare | 5220-0341 | Goat anti-Mouse IgG |
| Anti Rabbit antibody | Seracare | 5220-0337 | Goat anti-Rabbit IgG |
| Anti-Actin antibody | Sigma-Aldrich | A3853 | Dilution used: 1:2000 |
| Anti-Fbxo7 antibody | Sigma-Aldrich | SAB1407251 | Dilution used: 1:1000 |
| Anti-HA antibody | Sigma-Aldrich | H3663 | Dilution used: 1:1000 |
| Anti-Myc antibody | Cell Signalling | 2272 | Dilution used: 1:1000 |
| Bradford reagent | Sigma-Aldrich | B6916-500ML | |
| BSA | Sigma-Aldrich | A9647-100G | Bovine Serum Albumin |
| Cell incubator | Nuaire | NU-4850 | |
| Centrifuge | Eppendorf | 5804R | 500 x g for 5 min |
| ChemiDoc | BioRad | ||
| Digital pH meter | Kasvi | K39-2014B | |
| Dulbecco’s Modified Eagle’s Medium | Corning | 10-017-CRV | High glucose |
| Fetal bovine serum | Gibco | F4135 | Filtrate prior use |
| HA peptide | Sigma-Aldrich | I2149 | |
| HEK293T cells | ATCC | CRL-3216 | |
| Hepes | Gibco | 15630080 | |
| KCl | VWR Life Science | 0365-500G | |
| Kline rotator | Global Trade Technology | GT-2OIBD | |
| MG-132 | Boston Biochem | I-130 | |
| Microcentrifuge | Eppendorf | 5418R | |
| Na3VO4 (Ortovanadato) | |||
| NaF | |||
| Nitrocellulose blotting membrane | GE Healthcare | 10600016 | |
| NP40 (IGEPAL CA-630) | Sigma-Aldrich | I8896-100ML | |
| Optical microscope | OPTIKA microscopes | SN510768 | |
| Opti-MEM | Gibco | 31985-070 | |
| pcDNA3 | Invitrogen | V79020 | For mammalian expression |
| pcDNA3-2xFlag-Fbxo7 | Kindly donated by Dr. Marcelo Damário | Tag 2xFlag (N-terminal). Restriction enzymes: EcoRI and XhoI | |
| pcDNA3-2xFlag-Fbxo7-ΔF-box | Kindly donated by Dr. Marcelo Damário | Tag 2xFlag (N-terminal). Restriction enzymes: EcoRI and XhoI. Δ335-367 | |
| pcDNA3-UXTV2-HA | Kindly donated by Dr. Marcelo Damário | Tag HA (C-terminal). Restriction enzymes: EcoRI and XhoI | |
| pCMV-6xHis-Myc-Ubiquitin | Kindly donated by Dr. Marcelo Damário | Tag 6x-His-Myc (N-terminal). Restriction enzymes: EcoRI and KpnI | |
| Pen Strep Glutamine 100x | Gibco | 10378-016 | |
| Phosphate buffered saline 10x | AccuGENE | 51226 | To obtain a 1x PBS, dilute the 10x PBS into ultrapure water |
| Polyethylenimine (PEI) | Sigma-Aldrich | 9002-98-6 | |
| Ponceau S | VWR Life Science | 0860-50G | |
| Protease inhibitor cocktail SIGMAFAST | Sigma-Aldrich | S8820 | |
| Rocking Shaker | Kasvi | 19010005 | |
| SDS-PAGE system | BioRad | 165-8004 | |
| Solution Homogenizer | Phoenix Luferco | AP-22 | |
| Trizma base | Sigma-Aldrich | T6066-500G | |
| Trypsine (TrypLe Express) | Gibco | 12605-028 | |
| Western Blotting Luminol Reagent | Santa Cruz Biotechnology | SC-2048 |
References
- Popovic, D., Vucic, D., Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nature Medicine. 20 (11), 1242-1253 (2014).
- Callis, J. The ubiquitination machinery of the ubiquitin system. The Arabidopsis Book. 12, 0174 (2014).
- Koegl, M., et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 96 (5), 635-644 (1999).
- French, M. E., Koehler, C. F., Hunter, T. Emerging functions of branched ubiquitin chains. Cell Discovery. 7 (1), 6 (2021).
- Komander, D., et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Reports. 10 (5), 466-473 (2009).
- Clague, M. J., Urbé, S. Ubiquitin: Same molecule, different degradation pathways. Cell. 143 (5), 682-685 (2010).
- Davies, B. A., et al. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. Journal of Biological Chemistry. 278 (22), 19826-19833 (2003).
- Raasi, S., Wolf, D. H. Ubiquitin receptors and ERAD: A network of pathways to the proteasome. Seminars in Cell and Developmental Biology. 18 (6), 780-791 (2007).
- Pan, M., et al. Structural insights into Ubr1-mediated N-degron polyubiquitination. Nature. 600 (7888), 334-338 (2021).
- Raducu, M., et al. SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development. The EMBO Journal. 35 (13), 1400-1416 (2016).
- Spagnol, V., et al. The E3 ubiquitin ligase SCF(Fbxo7) mediates proteasomal degradation of UXT isoform 2 (UXT-V2) to inhibit the NF-κB signaling pathway. Biochimica et Biophysica Acta – General Subjects. 1865 (1), 129754 (2021).
- Teixeira, F. R., et al. Gsk3β and Tomm20 are substrates of the SCFFbxo7/PARK15 ubiquitin ligase associated with Parkinson’s disease. Biochemical Journal. 473 (20), 3563-3580 (2016).
- Tan, M. K. M., Lim, H. J., Bennett, E. J., Shi, Y., Harper, J. W. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Molecular Cell. 52 (1), 9-24 (2013).
- van Wijk, S. J., Fulda, S., Dikic, I., Heilemann, M. Visualizing ubiquitination in mammalian cells. EMBO Reports. 20 (2), 1-18 (2019).
- Kisselev, A. F., Goldberg, A. L. Proteasome inhibitors: From research tools to drug candidates. Chemistry and Biology. 8 (8), 739-758 (2001).
- Bradford, M. A. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 72 (1-2), 248-254 (1976).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 228, 726-734 (1970).
- Schröer, A., Schneider, S., Ropers, H. -. H., Nothwang, H. G. Cloning and characterization of UXT, a novel gene in human Xp11, which is widely and abundantly expressed in tumor tissue. Genomics. 56 (3), 340-343 (1999).
- Huang, Y., et al. UXT-V1 facilitates the formation of MAVS antiviral signalosome on mitochondria. The Journal of Immunology. 188 (1), 358-366 (2012).
- Huang, Y., et al. UXT-V1 protects cells against TNF-induced apoptosis through modulating complex II formation. Molecular Biology of the Cell. 22 (8), 1389-1397 (2011).
- Sun, S., et al. UXT is a novel and essential co-factor in the NF-κB transcriptional enhanceosome. The Journal of Cell Biology. 178 (2), 231-244 (2007).
- Huang, X., Dixit, V. M. Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Research. 26 (4), 484-498 (2016).
- Rajkumar, S. V. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. American Journal of Hematology. 95 (5), 548-567 (2020).
- Hideshima, T., et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Recherche en cancérologie. 61 (7), 3071-3076 (2001).
- Tietsche, V., et al. New proteasome inhibitors in the treatment of multiple myeloma. Hematology, Transfusion and Cell Therapy. 41 (1), 76-83 (2018).
- Vassilev, L. T., et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 303 (5659), 844-848 (2004).
- Kuiken, H. J., et al. Identification of F-box only protein 7 as a negative regulator of NF-kappaB signalling. Journal of Cellular and Molecular Medicine. 16 (9), 2140-2149 (2012).
- Yuan, N., et al. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica. 100 (3), 345-356 (2015).
- Iconomou, M., Saunders, D. N. Systematic approaches to identify E3 ligase Substrates. Biochemical Journal. 473 (22), 4083-4101 (2016).
- Zhang, Z. R., Bonifacino, J. S., Hegde, R. S. Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell. 154 (3), 609-622 (2013).
- Hunter, T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Molecular Cell. 28 (5), 730-738 (2007).

