Construction of Adenoviral Vectors using DNA Assembly Technology
Summary
In this study, an infectious clone of human adenovirus type 7 (HAdV-7) was constructed, and an E3-deleted HAdV-7 vector system was established by modifying the infectious clone. This strategy used here can be generalized to make gene transfer vectors from other wild-type adenoviruses.
Abstract
Adenoviral vectors have been used as a gene transfer tool in gene therapy for more than three decades. Here, we introduce a protocol to construct an adenoviral vector by manipulating the genomic DNA of wild-type HAdV-7 by using a DNA assembly method. First, an infectious clone of HAdV-7, pKan-Ad7, was generated by fusing the viral genomic DNA with a PCR product from plasmid backbone, comprising of the kanamycin-resistant gene and the origin of replication (Kan-Ori), through DNA assembly. This was done by designing a pair of PCR primers, that contained ~25 nucleotides of the terminal sequence of HAdV-7 inverted terminal repeat (ITR) at the 5′ end, a non-cutter restriction enzyme site for HAdV-7 genome in the middle, and a template-specific sequence for PCR priming at the 3′ end. Second, an intermediate plasmid-based strategy was employed to replace the E3 region with transgene-expressing elements in the infectious clone to generate an adenoviral vector. Briefly, pKan-Ad7 was digested with dual-cutter restriction enzyme Hpa I, and the fragment containing the E3 region was ligated to another PCR product of plasmid backbone by Gibson assembly to construct an intermediate plasmid pKan-Ad7HpaI. For convenience, restriction-assembly was used to designate the plasmid cloning method of combined restriction digestion and assembly. Using restriction-assembly, the E3 genes in pKan-Ad7HpaI was replaced with a green fluorescent protein (GFP) expression cassette, and the modified E3 region was released from the intermediate plasmid and restored to the infectious clone to generate an adenoviral plasmid pKAd7-E3GFP. Finally, pKAd7-E3GFP was linearized by Pme I digestion and used to transfect HEK293 packaging cells to rescue recombinant HAdV-7 virus. To conclude, a DNA assembly-based strategy was introduced here for constructing adenoviral vectors in general laboratories of molecular biology without the need of specialized materials and instruments.
Introduction
Over the past three decades, recombinant adenoviral vectors have been widely used in vaccine development and gene therapy1,2,3,4 as well as in basic research due to their outstanding biological properties, such as high gene transduction efficiency, non-integration to the host genome, the manipulative viral genome, and the ease of large-scale production.
Currently, the most commonly used adenoviral vectors are constructed based on human adenovirus 5 (HAdV-5)5,6. Although HAdV-5 vector-mediated transduction provides encouraging results, preclinical and clinical applications have revealed several disadvantages, (e.g., high pre-existing anti-vector immunity within the human population and low transduction efficiency in cells lacking the coxsackievirus and adenovirus receptor (CAR)). To circumvent these problems, there has been a great interest to construct vectors based on other human or mammalian adenovirus types3,7,8.
Until now, the most popular method to construct an adenoviral vector is homologous recombination in bacteria5. Such bacterial strains must express recombinases, which can affect the stability or amplification of the plasmids they bear. Some strains are even commercially unavailable. Recently, methods based on other principles, including bacterial artificial chromosomes, direct cloning, or direct DNA assembly, have been employed to generate infectious clones of adenovirus or recombinant adenoviral vectors9,10,11,12. However, these methods are somewhat unfriendly to researchers with little experience in this field.
In 2018, the process of constructing an adenovirus infectious clone is simplified in the laboratory by directly ligating the virus genome with a PCR product carrying plasmid backbone through Gibson assembly13. After that, the methods of restriction digestion and Gibson assembly are combined together to load transgenes to existing adenoviral plasmids14,15,16,17,18. For the sake of convenience, restriction-assembly is used hereafter to refer to the method of combined restriction enzyme digestion and Gibson assembly. Strategies were further developed to construct adenoviral vectors from infectious clones by using restriction-assembly19. The essence of restriction-assembly is to include fragments excised from plasmids as much as possible in a DNA assembly reaction, while short PCR products serve as linkers or patches for plasmid modification. At the same time, the number of fragments included is kept as low as possible. Such efforts reflect the payoff; the possibility of unwanted mutations caused by PCR or DNA assembly can be minimized, and the success rate can be improved. Conclusively, a pipeline from a wild-type adenovirus to an adenoviral vector has been set up in the laboratory13,14,15,16,17,18,19.
Here, we attempt to introduce these methods by providing examples of constructing an HAdV-7 infectious clone and an E3-deleted replication-competent HAdV-7 vector.
Protocol
NOTE: Adenoviruses are classified as Biosafety Level 2 (BSL-2); all the steps to use the viruses were carried out in a biosafety level 2 laboratory. The wild-type HAdV-7 was isolated in 2017 from the nasopharyngeal aspirate specimen of a 10-month-old infant who was hospitalized with acute respiratory tract infection in Beijing Children's Hospital20. The virus stocks were stored at -80 °C.
1. Extraction of HAdV-7 genomic DNA
- Seed 2.0 x 106 of the HEK293 cells in one T-25 flask containing 5 mL of Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS) and culture at 37 °C in a humidified atmosphere supplemented with 5% CO2. Cells should be 80%-90% confluent and ready for infection in 18-24 h.
- Inoculate the cells with wild-type HAdV-7 for 2 h. Remove the virus-containing medium by aspiration and add 5 mL of fresh DMEM containing 2% FBS to the cells.
- Extract the viral genomic DNA with the modified Hirt's method21
- In 2 or 3 days post-infection, when cytopathic effect (CPE) is observed on 50%-90% of cells, discard the culture supernatant and rinse cells once with phosphate buffered saline (PBS).
NOTE: Cytopathic effect (CPE) is defined as a phenomenon where the virus-infected cells round up, swell, and loosely attach or detach from the culture flask into grape-like clusters22. - Lyse cells in the flask in 1.0 mL of lysis buffer containing 25 mM Tris-HCl, 0.5 mM EDTA, 50 µg/mL protease K, and 0.8% SDS (pH7.6) for 5 min on ice. Transfer the lysate to a 1.5 mL microcentrifuge tube and incubate the tube in a water bath at 50 °C for 30 min.
- Add 120 µL of precipitation buffer to the tube. Place the tube on ice for another 30 min after gently mixing.
NOTE: The precipitation buffer is composed of 3 M CsCl, 1 M potassium acetate, and 0.67 M acetic acid. It will help precipitate the cellular debris and cellular chromosome DNA after centrifugation while keeping the virus genome in the supernatant. - Centrifuge the tube for 25 min at 15,000 x g at 4 °C, and then transfer the supernatant (about 1.1 mL) to a 15 mL polypropylene tube.
- Add 2.2 mL of ice-cold absolute ethanol to the tube. Mix gently and let the tube stand on ice for 15 min. Centrifuge for 5 min at 2000 x g at room temperature.
- Aspirate the supernatant carefully and wash the precipitate (virus genomic DNA) once with 800 µL of 70% ethanol. Centrifuge the tube for 6 min at 15,000 x g at room temperature. Discard the supernatant and air-dry the DNA pellet at the bottom. Do not overdry the DNA pellet since it may cause difficulty in redissolving.
- Dissolve the pellet in 200 µL of TE buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA), add 1 µL of RNase A (10 mg/mL), and incubate the tube at 37 °C for 15 min. Purify the dissolved DNA by using a commercial genomic DNA clean and concentrator kit (see Table of Materials).
NOTE: Quantitate the viral genomic DNA with the commonly used method in the laboratory. Generally, 1-5 µg of genomic DNA can be obtained in a T-25 flask of HEK293 cells infected with adenovirus.
- In 2 or 3 days post-infection, when cytopathic effect (CPE) is observed on 50%-90% of cells, discard the culture supernatant and rinse cells once with phosphate buffered saline (PBS).
2. Construction of infectious clone of HAdV-7 by DNA assembly
- Design PCR primers to amplify the plasmid backbone (Figure 1). Add the overlap sequences that are needed for DNA assembly to the 5' ends of PCR primers. In addition, include non-cutter restriction sites (e.g., Pme I site) on both ends of HAdV-7 genome to help release the viral genome from the infectious clone.
NOTE: The plasmid backbone containing antibiotic resistant gene and replication origin is amplified by PCR and fused to HAdV-7 genome. Adenoviruses can be rescued from packaging cells with higher efficiency when linearized virus genome is used instead of a circular adenoviral plasmid. After considering above mentioned factors, primers are designed as shown in Figure 1. - Perform PCR to amplify a fragment (Kan-Ori) containing kanamycin resistance gene (Kan) and pBR322 origin (Ori) using pShuttle-CMV as the template with primers of Ad7KanF1 and Ad7KanR1 (Table S1). Set the PCR reaction as follows: one cycle at 98 °C for 30 s, then five cycles of denaturing at 98 °C for 8 s, annealing at 68 °C for 30 s, extension at 72 °C for 90 s, and finally 25 cycles of denaturing at 98 °C for 8 s, annealing and extension at 72 °C for 2 min.
- Run the PCR product in a 1% agarose gel by electrophoresis, and purify the fragment using a commercial DNA gel recovery kit.
- Add the recovered Kan-Ori and viral genomic DNA of HAdV-7 to the commercial DNA Assembly Master Mix. Set up the assembly reaction in a total volume of 20 µL and a molar ratio of vector to viral genomic DNA of 1:2. Incubate at 50 °C for 1 h.
NOTE: Set the reaction in a total volume of 20 µL with 1-3 µL of vector, 7-9 µL of viral genomic DNA, and 10 µL of DNA Assembly Master Mix. - Transform the chemically competent E. coli strain TOP10 cells with 10 µL of the assembly product using heat shock. Spread the transformation mix onto kanamycin-containing (50 µg/mL) Luria-Bertani (LB-Kan) plates. Incubate plates at 37 °C for at least 12 h.
- Inoculate at least three clones each in 5 mL of LB-Kan medium at 37 °C for 12 h with gentle shaking (200-250 rpm). Extract plasmid DNA using a commercial Plasmid Miniprep Kit.
- Digest the plasmid using restriction endonuclease, and then check the size of DNA fragments with agarose gel electrophoresis (Figure 2A). Moreover, confirm the fusion sites by sequencing. The correct assembly plasmid is named pKan-Ad7.
- Grow a 100 mL culture of bacteria from the correct clone. Extract a maxi-plasmid preparation using a commercial Plasmid Maxprep Kit.
3. In silico construction and modification of an intermediate plasmid
NOTE: Generally, an intermediate plasmid should be constructed, and how to construct an ideal intermediate plasmid is the key step. DNA analysis software with a restriction enzyme module, such as pDRAW32 (a free software, available at www.acaclone.com), is needed for the design of an intermediate plasmid.
- Import the DNA sequence of pKan-Ad7 to pDRAW32 to draw a plasmid map. Annotate the regions of interest on the map. For pKan-Ad7, E1, E3 regions, and plasmid backbone (Kan-Ori) were annotated.
- Operate the pDRAW32 program. Click Settings > Enzyme Selection firstly, and then click Min/Max Cuts. Input 1 in the box Min and input 2 in the box Max, and tick the box before in the entire sequence. Lastly, click OK to show the unique and dual cutter restriction enzyme sites on the map.
NOTE: Generally, only the restriction enzymes that recognize a sequence of 6 bp or longer are selected. As shown in Figure 3A, 17 enzymes match the enzyme selection criteria. - Consider the first restriction site that flanks the E3 region since the E3 is planned to be modified. Hpa I, Sal I, Avr II-Mlu I, and Spe I satisfied the requirement.
NOTE: For this experiment, Hpa I is selected because digestion of pKan-Ad7 with it would generate the shortest fragment carrying the E3 region, and a smaller intermediate plasmid would make future modification easier. The selection of Sal I sites also has advantages-such an intermediate plasmid is suitable for the modification of both E1 and E3 regions (Figure S1). - Create a final sequence file displaying both DNA strands of the intermediate plasmid pKan-Ad7HpaI. This virtual sequence can be used as a template to design overlapping primers.
- Copy the sequence between the two Hpa I restriction sites including the E3 region. For subsequent assembly, it is necessary to copy each ~20 bp sequence (Tm equal to or greater than 48°C) outside the Hpa I restriction site (Figure 3C, overlap-Final 1 and overlap-Final2).
- Add the recognition sequence of Pme I (Figure 3C, gtttaaac) at both ends of the above sequence.
- Add the sequence of backbone plasmid containing antibiotic-resistant gene and replication origin (Figure 3C, Kan-Ori) outside of the Pme I restriction site.
- Import the final sequence to pDRAW32 to draw a plasmid map. Annotate the E3 region on the map.
- Operate the pDRAW32 program. Click Settings > Enzyme Selection firstly, and then click Min/Max Cuts. Input 1 in both the box Min and Max, and tick the box before in entire sequence. Click OK lastly to show the unique restriction enzyme sites on the map. It can be seen that a lot of unique cutters are available for E3 modification.
NOTE: For this experiment, BstZ17 I and Mlu I are selected for the replacement of partial E3. A dual cutter can also be useful. For example, dual cutter Ssp I together with unique cutter BssH II can be selected for the replacement of the whole E3 region by using overlap extension PCR.
- Design primers using the virtual sequence as a template for the construction of intermediate plasmid (Figure 3C).
NOTE: To shorten the length of each primer, four primers were designed. The PCR product, which is amplified with primers of Ad7HpaIF1 and Ad7HpaIR1, would be used as the template to obtain the final PCR fragment with primers of Ad7HpaIF2 and Ad7HpaIR2 (Figure 3C).
4. Rescue of infectious recombinant adenovirus in HEK293 cells
NOTE: The three adenoviruses generated in this article are all rescued according to the following protocol.
- Digest 10 µg of recombinant adenoviral plasmid with 2 µL of Pme I for 5 h at 37 °C in a total volume of 100 µL to linearize plasmid. Check 1 µL of the digested DNA on a 0.8% agarose gel. Digestion should yield a ~34 kb fragment of the adenoviral genome and a ~2.5 kb plasmid backbone of Kan-Ori.
- Purify the remnant linearized adenoviral plasmid using the genomic DNA Clean and Concentrator kit.
- Culture HEK293 cells in DMEM plus 10% FBS. Seed HEK293 cells (2 x 106 cells) in a T-25 flask 1 day before transfection so that they reach 70%-80% confluency the next day. Use 5 µg of Pme I digested adenoviral plasmid to transfect HEK293 cells by using commercially available transfection reagents.
NOTE: The medium of the transfected cells should be replaced every 3 days. - Check the cells every day, collect the cells, and culture medium in a 15 mL polypropylene tube when the comet-foci form (Figure 2B, indicating virus replication).
- Lyse the cells via three freeze-thaw cycles. After centrifugation for 12 min at 1200 x g at room temperature, collect the supernatant and store it at -80 °C for subsequent virus propagation.
NOTE: When the recombinant adenovirus expresses GFP or other fluorescent proteins, it is easier to identify whether the virus is rescued successfully, as the foci formed by fluorescent protein-positive cells could be observed under a fluorescence microscope intuitively.
5. Construction of E3-deleted HAdV-7 genome plasmid
NOTE: The genome of HAdV-7 is 35,239 bp in length, and the E3 region is located between 27,354 bp and 31,057 bp of the genome. To construct an intermediate plasmid, find an endonuclease with two cutting sites on the infectious clone plasmid, or two endonucleases with a single cutting site on the plasmid. There are Hpa I, Sal I, or Avr II and Mlu I available. The sequence between BstZ17 I (28,312 bp) and Mlu I (30,757 bp) is to be deleted and replaced with the CMV-GFP-PA cassette or CMV-mCherry-PA cassette.
- Digest 600 ng of pKan-Ad7 with Hpa I. Purify the large fragment pKAd7-HpaI-FL (~30 kb) and small fragment pKAd7-HpaI-FS (~7 kb) respectively, using a DNA gel recovery kit after gel electrophoresis. Store the pKAd7-HpaI-FL at -20 °C.
- Amplify the fragment containing Kan and Ori using pShuttle-CMV as the template with primers of Ad7HpaIF1 and Ad7HpaIR1 (Table S1) by PCR. Recover and dilute the PCR product (2541 bp) to be used as a template for the second round of PCR reaction to amplify Kan-Ori2 with primers of Ad7HpaIF2 and Ad7HpaIR2.
- Purify the PCR product (2584 bp) after resolving in a 1% agarose gel by electrophoresis. Set the PCR reaction as follows: one cycle at 98 °C for 30 s, then 30 cycles of denaturing at 98 °C for 8 s, annealing and extension at 72 °C for 2 min.
NOTE: The central part of Kan-Ori2 is the fragment containing Kan and Ori, followed by the sequences of a unique restriction endonuclease which does not exist in the small fragment pKAd7-HpaI-FS (e.g., Pme I), and the overlapping region with the large fragment pKAd7-HpaI-FL; the outermost sequences is the overlapping region with small fragment pKAd7-HpaI-FS.
- Purify the PCR product (2584 bp) after resolving in a 1% agarose gel by electrophoresis. Set the PCR reaction as follows: one cycle at 98 °C for 30 s, then 30 cycles of denaturing at 98 °C for 8 s, annealing and extension at 72 °C for 2 min.
- Add the PCR product Kan-Ori2 and the recovered pKAd7-HpaI-FS in step 5.1 to DNA Assembly Master Mix. Incubate at 50 °C for 1 h. Follow steps 2.4- 2.6. The correct plasmid is named pKan-Ad7HpaI (~10 kb).
- Amplify the CMV-GFP-PA fragment (~1.7 kb) containing CMV promoter, GFP CDS, and SV40 polyA signal (PA) using pShuttle-GFP23 as the template with primers of Ad7E3GFPF1 and Ad7E3GFPR1 (Table S1). Purify the PCR product (1650 bp) after resolving in a 1% agarose gel by electrophoresis.
NOTE: Overlapping regions that can be assembled with BstZ17 I and Mlu I digested pKan-Ad7HpaI should be included in the primers, and a unique restriction site (e.g., Sbf I) which does not exist in the plasmid pKan-Ad7 is added into the primers for further restriction-assembly. - Digest 300 ng of pKan-Ad7HpaI with BstZ17 I and Mlu I. Purify the fragment of 7.3 kb (most of the E3 regions are deleted) after resolving in a 1% agarose gel by electrophoresis. Add the recovered 7.3 kb fragment and the recovered CMV-GFP-PA fragment (~1.6 kb) to DNA Assembly Master Mix. Follow steps 2.4-2.6. The correct plasmid is named pKAd7-E3GFP (~9 kb).
NOTE: All the sequences between BstZ17 I and Mlu I belong to the E3 region. - Digest 300 ng of pKAd7-HE3GFP with Pme I. Purify the large fragment (~6.4 kb) using a DNA gel recovery kit after gel electrophoresis. Add the purified fragment and pKAd7-HpaI-FL recovered in step 5.1 to DNA Assembly Master Mix. Incubate at 50 °C for 1 h. Follow steps 2.4- 2.6. The correct plasmid is named pKAd7-E3GFP.
- Transfect HEK293 cells with Pme I-linearized pKAd7-E3GFP as described in section 3. The rescued adenovirus is named Ad7-E3GFP.
- Amplify the CMV-mCherry-PA fragment (~1.6 kb) containing CMV promoter, mCherry CDS, and SV40 polyA signal using pKFAV4-CX19A as the template with the primers of Ad7E3CHEF1 and Ad7E3CHER1. Purify the PCR product after resolving in a 1% agarose gel by electrophoresis.
- Digest 600 ng of pKAd7-E3GFP with Sbf I, and purify the fragment of ~35 kb. Add the 35 kb fragment and the recovered CMV-mCherry-PA fragment to DNA Assembly Master Mix. Incubate at 50 °C for 1 h. Follow steps 2.4-2.6. The correct plasmid is named pKAd7-E3CHE.
- Transfect HEK293 cells with Pme I-linearized pKAd7-E3CHE as described in section 3. The rescued adenovirus is named Ad7-E3CHE.
6. Amplification and purification of the recombinant adenovirus in HEK293 cells
- Inoculate an 80%-90% confluent monolayer of HEK293 cells grown in eight 150 mm diameter TC-treated culture dishes with adenovirus. Incubate at 37 °C for 2 h under mild agitation. Replace the supernatant with 25 mL of DMEM containing 2% FBS.
- Incubate the cells at 37 °C for 2-3 days.
- Discard most of the supernatant of the eight 150 mm diameter dishes when a CPE affects the majority of cells. Harvest infected cells using cell lifters with a final elution volume of ~6.0 mL into 15 mL polypropylene tubes.
- Disrupt the cells by three freeze-thaw cycles to release the virus. Clear the lysate by centrifugation for 12 min at 1200 x g at room temperature. Collect the virus-containing supernatant.
- Add MgCl2 (0.1 mM) and benzonase nuclease (50 U/mL) to the supernatant. Vortex gently for 40 min at 37 °C. Add NaCl (600 mM) to the supernatant and vortex gently for another 40 min at 37 °C. Centrifuge for 12 min at 1200 x g at room temperature. Collect the virus-containing supernatant for purification.
- Prepare cesium chloride (CsCl) with different concentrations of 1.35 g/mL, 1.30 g/mL, and 1.25 g/mL with 10 mM Tris-HCl.
- Add 0.8 mL of 1.35 g/mL CsCl solution into the bottom of a 5.1 mL Ultra-Clear tube using a pipette, and then slowly add 0.8 mL of 1.30 g/mL CsCl solution and 0.8 mL of 1.25 g/mL CsCl solution on top of the first solution sequentially.
- Lastly, carefully overlay ~2.5 mL of the supernatant collected in step 6.3 on the top of 1.25 g/mL CsCl solution, and fill the tube to 2-3 mm from the top using 10 mM Tris-HCl.
- Centrifuge for 2 h at 200,000 x g and 8 °C in a swinging-bucket rotor with slow acceleration and deceleration to separate the intact viral particles from the defective viral particles.
NOTE: After centrifugation, several white bands can be clearly seen in the CsCl solution. - Carefully remove the bands of the empty particles and the layer of cell debris using the pipette, and collect the lowest band containing mature virus using a 2 mL syringe.
- Transfer the collected virus suspension to a 20 K MWCO dialysis cassette, and dialyze in dialysis buffer containing 10 mM Tris-HCl, 150 mM NaCl, 1 mM MgCl2, and 2% glycerol (pH 8.0) overnight at 4 °C.
- Exchange the dialysis buffer containing 10 mM Tris-Cl, 150 mM NaCl, 1 mM MgCl2, and 5% glycerol (pH 8.0) the next day and dialyze 5 h at 4 °C.
- Collect the dialyzed virus particles using a 2 mL syringe. Prepare multiple aliquots of desired small volumes (20-200 µL) and store the purified virus at -80 °C.
NOTE: Particle titer of the purified virus can be determined by measuring the content of genomic DNA, and infectivity titer can be determined on HEK293 cells by counting GFP-positive cells with the limiting dilution assay.
7. Identification of recombinant adenovirus genome by restriction enzyme digestion
- Infect an 80%-90% confluent monolayer of HEK293 cells grown in one T-25 flask with recombinant adenovirus. Incubate for 2 h at 37 °C. Replace the culture medium with fresh DMEM containing 2% FBS.
- Remove the culture supernatant and extract the viral genomic DNA with the modified Hirt's method described in step 1.3 when CPE occurrs within 2-3 days.
- Digest the genomic DNA of the recombinant adenovirus with indicated enzymes, and analyze on agarose gel electrophoresis. The recombinant adenoviral plasmid served as a control.
Representative Results
The strategy for the construction of an infectious clone of HAdV-7 is shown in Figure 1 and Figure 2. Two infectious clone plasmids were randomly selected and identified by BstZ17 I, BamH I, and EcoR V, respectively. The results showed that the fragments were consistent with the expected size (Figure 2A), indicating that the plasmids were constructed correctly. Comet-foci could be seen in the cells 12 days after the Pme I-linearized plasmid was transfected into HEK293 cells (Figure 2B), indicating that the virus was rescued successfully.
In this study, the E3-deleted adenoviral vector was generated to overexpress the GFP (Figure 4) or mCherry as examples. During the modification of the E3 region, an intermediate plasmid pKan-Ad7HpaI (Figure 3) was constructed first. Plasmids from three colonies were analyzed with EcoR V and Hind III digestion, the electrophoresis results showed that all the plasmids were correct (Figure 5A). Moreover, the sequencing chromatogram showed that there is no mutation at the assembly sites (Figure 5C, partial results were shown). After the E3 region between BstZ17 I and Mlu I was deleted and replaced with the CMV-GFP-PA cassette, three of the modified plasmid pKAd7-HE3GFPs were identified by restriction enzyme digestion and sequencing (data not shown). The results showed that the digested fragments were exactly as expected, indicating that the plasmids were constructed correctly (Figure 5B).
After modification, the resulting plasmid pKAd7-E3GFP contains the CMV-GFP-PA cassette flanked by two Sbf I sites (Figure 4), which could be used to replace the CMV-GFP-PA cassette with other genes expediently. GFP expression could be seen under a fluorescence microscope at 48 h after HEK 293 cells were transfected with Pme I-linearized pKAd7-E3GFP, and fluorescent foci and CPE could be observed at 13 days post-transfection (Figure 6). The cells and medium were harvested and used to inoculate HEK293 cells after three freeze-thaw cycles; GFP expression and CPE could be observed at 48 h post-infection (Figure 6). Following the purification protocol described, concentrated viral particles appeared as several opalescent virion bands within the CsCl gradient (Figure 7A). The lowest band below 1.35 g/cm3 CsCl was collected and used for negative staining with phosphotungstic acid (PTA), and the intact icosahedral adenovirus could be observed under an electron microscope (Figure 7B). The top (lighter) band contained empty, noninfectious particles and had to be avoided. In this protocol, eight T-150 cm2 tissue culture dishes were used, for which 1.5 mL of Ad7-E3GFP stock with the infectious titer of 2.3 x 1011 IU/mL and viral particle titer of 6.0 x 1012 vp/mL were produced. To identify the genomic integrity of recombinant Ad7-E3GFP, the genomic DNA was digested with indicated restriction endonuclease and separated by electrophoresis; all the fragments were consistent with expectations (Figure 7C).
Using this protocol, the CMV-GFP-PA in pKAd7-E3GFP could be replaced with the gene of interest within 2 to 3 days. Herein, the CMV-GFP-PA was substituted for CMV-mCherry-PA by using restriction-assembly, and the new recombinant adenoviral plasmid pKAd7-E3CHE was used to transfect HEK293 cells after being linearized by Pme I. The mCherry expression could be detected in 24 h post-transfection, and the fluorescent foci were formed at 15 days after transfection (Figure 8A). Expression of mCherry and CPE could be observed in HEK293 cells at 48 h after inoculation with the rescued Ad7-E3CHE. The results of electrophoresis after restriction endonuclease digestion showed that the genome of Ad7-E3CHE was completely consistent with expectations (Figure 8B).
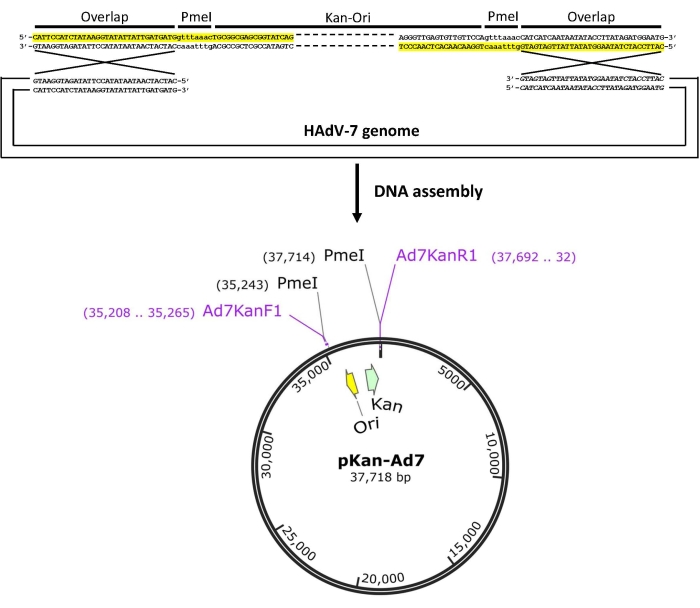
Figure 1: Schematic diagram of the construction of infectious clones of human adenovirus 7 (HAdV-7) by using DNA assembly. The primers (marked in yellow) contain not only the sequences paired with the template, but also the overlapping regions (overlap) that can be assembled with the HAdV-7 genome. Moreover, the sequence of a restriction endonuclease site (e.g., Pme I) that is absent in both of the genome of HAdV-7 and the plasmid backbone should also be included in the primers to facilitate the release of the genome. The amplified plasmid backbone Kan-Ori was assembled with viral genomic DNA of HAdV-7 in vitro to form the infectious clone plasmid pKan-Ad7. Please click here to view a larger version of this figure.
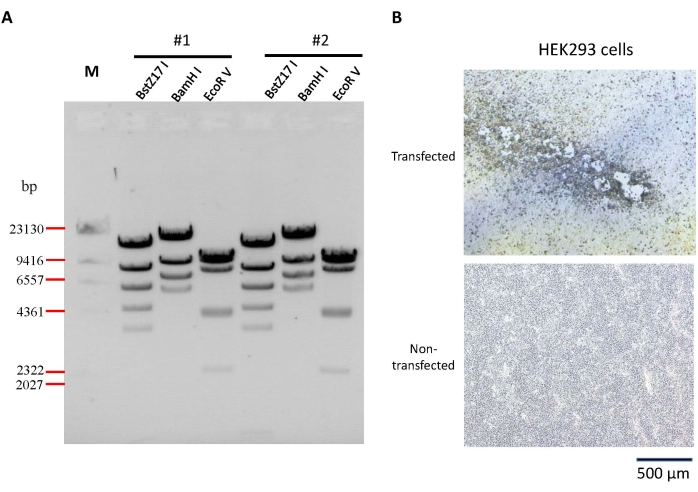
Figure 2: Identification of infectious clone of HAdV-7. (A) Two of the infectious clone plasmid pKan-Ad7s were digested with BstZ17 I, BamH I, and EcoR V, respectively. The predicted molecular weights of digested fragments were 716, 947, 1164, 3460, 4349, 5828, 8116, and 13,138 bp for BstZ17 I; 5713, 6945, 9058, and 16,002 bp for BamH I; 2338, 4048, 4162, 7792, 9306, and 10,072 bp for EcoR V. (B) Rescue of HAdV-7. Pme I-linearized pKan-Ad7 was used to transfect HEK293 cells. CPE occurred at day 12 post-transfection, indicating the rescue of HAdV-7 successfully. Please click here to view a larger version of this figure.
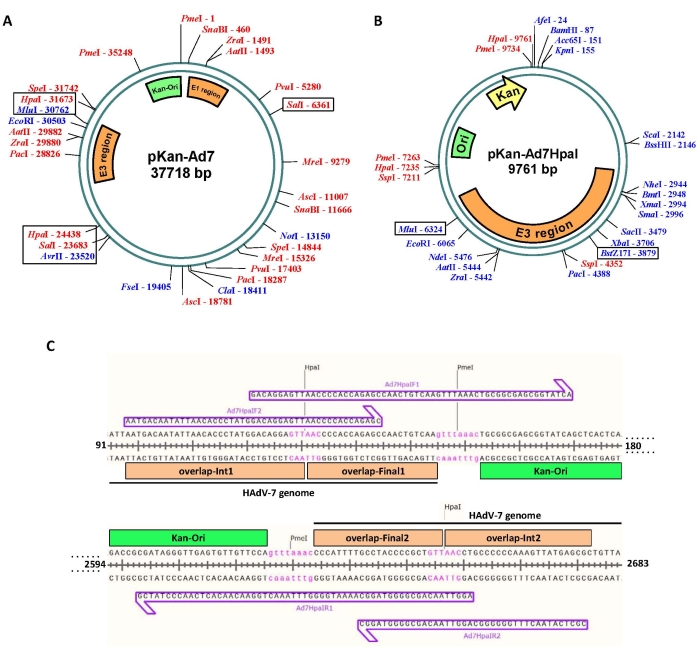
Figure 3: In silico construction of the intermediate plasmid. (A) Plasmid map of infectious clone pKan-Ad7. Shown are the unique and dual cutter restriction enzyme sites. Hpa I, Sal I, and Avr II-Mlu I can be selected for constructing an intermediate plasmid containing the E3 region. To construct a smallest intermediate plasmid, Hpa I was finally chosen to conduct the experiments. (B) Map of the intermediate plasmid pKan-Ad7HpaI. Shown are the restriction sites of all the unique cutters and some dual ones. E3 region between the BstZ17 I and Mlu I sites were finally replaced with a CMV promoter-controlled GFP expression cassette. (C) Primer design for amplifying plasmid backbone. The PCR product of plasmid backbone would be fused to the short fragment from Hpa I-digested pKan-Ad7 to generate the intermediate plasmid pKan-Ad7HpaI by Gibson assembly. Shown are the designed primers. Overlaps for Gibson assembly and Pme I sites would be integrated to the product through two rounds of PCR by using primers of Ad7HpaIF1/R1 and Ad7HpaIF2/R2 in sequence. Please click here to view a larger version of this figure.
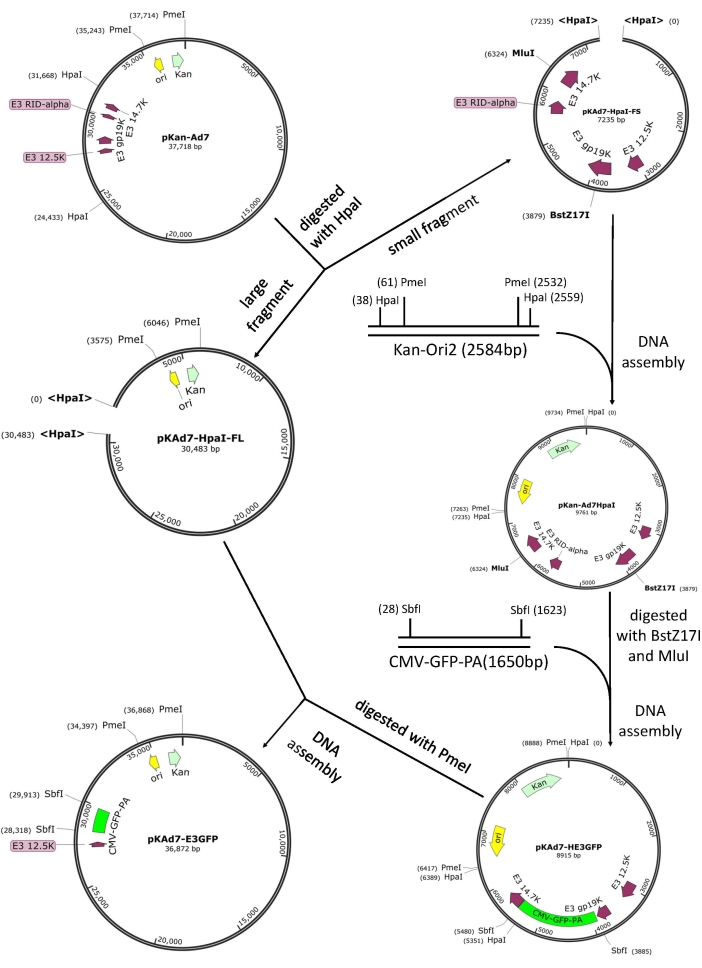
Figure 4: Schematic diagram of construction of recombinant adenoviral plasmid pKAd7-E3GFP by using restriction-assembly. Please click here to view a larger version of this figure.
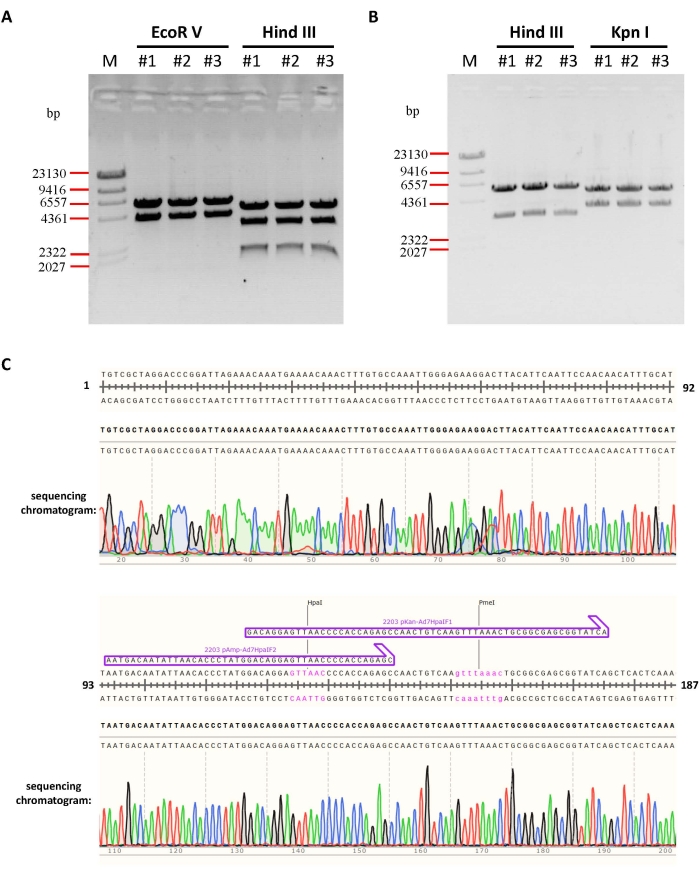
Figure 5: Identification of intermediate plasmid by digestion and sequencing. (A) Three of the intermediate plasmid pKan-Ad7HpaI were extracted and identified by indicated restriction endonuclease digestion. The predicted molecular weights of digested fragments were 4048 and 5713 bp for EcoR V, and 1710, 3206, and 4845 bp for Hind III. (B) Three of the plasmid pKAd7-HE3GFP were extracted and identified by restriction endonuclease digestion. The predicted molecular weights of digested fragments were 3206 and 5709 bp for Hind III, and 3725 and 5190 bp for Kpn I. (C) The fusion sites of plasmid pKan-Ad7HpaI were confirmed by sequencing. Due to limited space, only the sequencing results of one fusion site of the plasmid were shown here. Please click here to view a larger version of this figure.
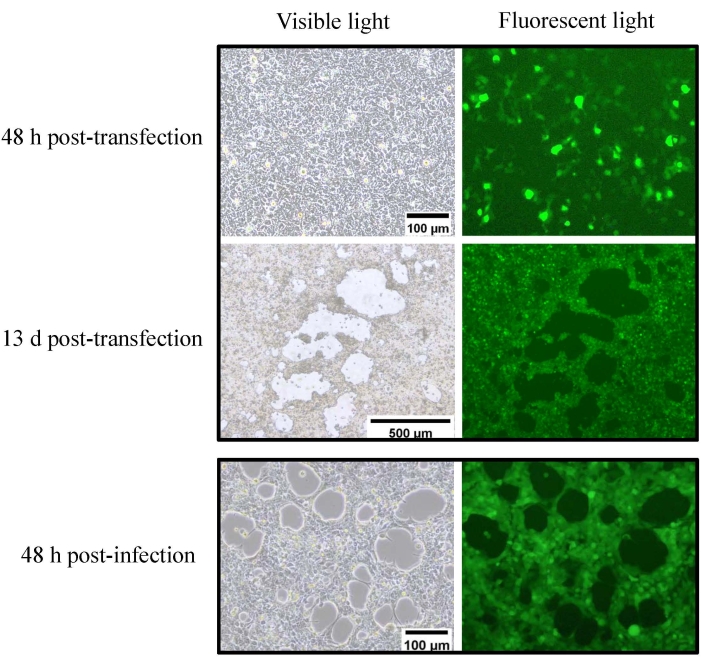
Figure 6: Rescue and amplification of recombinant adenovirus Ad7-E3GFP. HEK293 cells at 90% confluence (T-25 flask) were transfected with 5 µg of Pme I-linearized pKAd7-E3GFP using commercial transfection reagents. GFP expression and CPE could be observed under fluorescence microscope at 48 h and 13 days post-transfection, respectively. The cells and medium were collected and used to inoculate HEK293 cells; CPE and GFP expression could be seen at 48 h post-infection. Please click here to view a larger version of this figure.
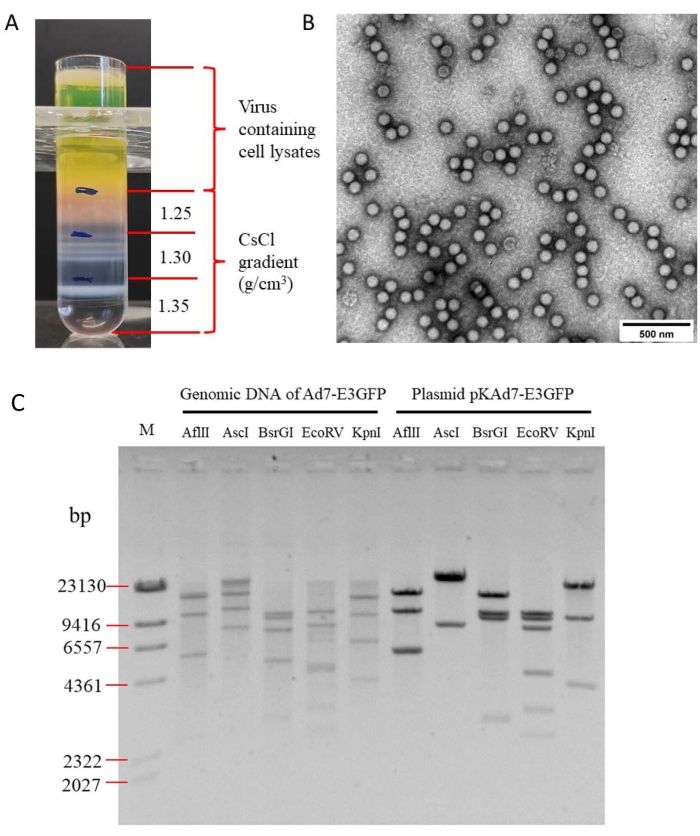
Figure 7: Purification and identification of recombinant adenovirus Ad7-E3GFP. (A) Purification of Ad7-E3GFP. Virus-containing cell lysates were loaded on CsCl gradient, and virus bands could be seen after ultracentrifugation. (B) Observation of the purified Ad7-E3GFP under EM. (C) Identification of Ad7-E3GFP genomic DNA. The genomic DNA was digested with indicated restriction endonuclease after extraction from the infected cells. Plasmid pKAd7-E3GFP was served as a control. The predicted molecular weights of digested fragments of Ad7-E3GFP genomic DNA were 530, 1191, 2368, 5310, 10,065, and 14,937 bp for Afl II; 7774, 11,006, and 15,621 bp for Asc I; 531, 2623, 4789, 7413, 9163, and 9882 bp for BsrG I; 342, 2338, 2432, 2860, 4162, 4403, 7792, and 10,072 bp for EcoR V; 1005, 3725, 6084, 9570, and 14,017 bp for Kpn I. The predicted molecular weights of digested fragments of the pKAd7-E3GFP plasmid were 1191, 5310, 5369, 10,065, and 14,937 bp for Afl II; 7774 and 29,098 bp for Asc I; 531, 2623, 9163, 9882, and 14,673 bp for BsrG I; 342, 2338, 2860, 4162, 7792, 9306, and 10,072 bp for EcoR V; 1005, 3725, 9570, and 22,572 bp for Kpn I. M: lambda/HindIII DNA marker. Please click here to view a larger version of this figure.
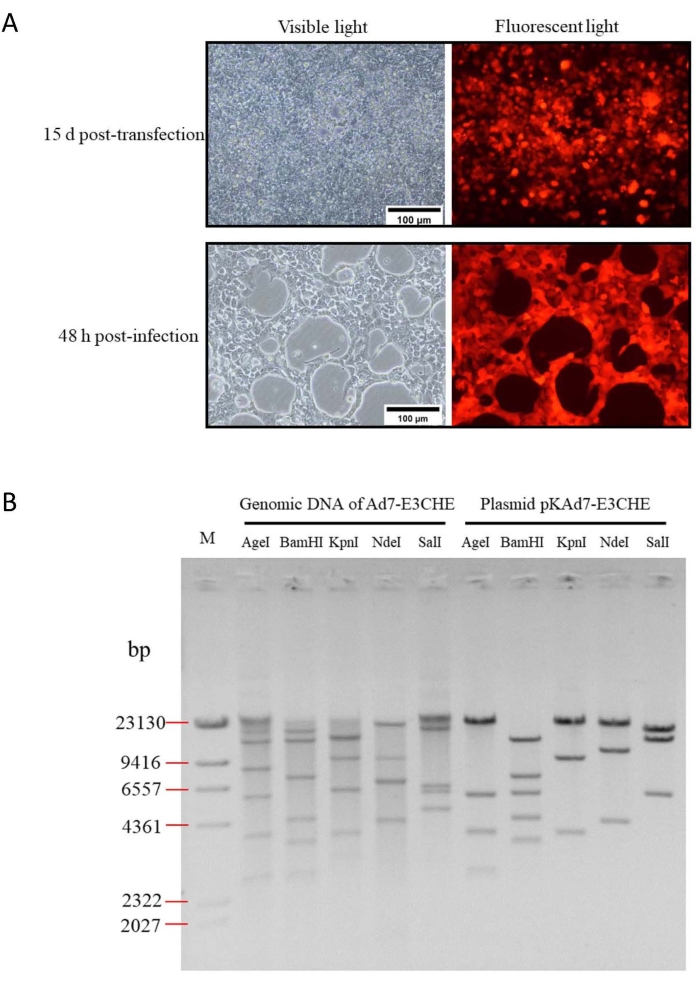
Figure 8: Generation, amplification, and identification of recombinant adenovirus Ad7-E3CHE. (A) Rescue and amplification of recombinant adenovirus Ad7-E3CHE. HEK293 cells at 90% confluence (T-25 flask) were transfected with 5 of µg Pme I-linearized pKAd7-E3CHE using transfection reagent. Fluorescent foci and CPE could be observed under fluorescence microscope at 15 days post-transfection. The cells and medium were collected and used to inoculate HEK293 cells. CPE and mCherry expression could be seen at 48 h post-infection. (B) Identification of Ad7-E3CHE genomic DNA. The genomic DNA was digested with indicated restriction endonuclease after extraction from the infected cells. Plasmid pKAd7-E3 CHE served as a control. The predicted molecular weights of digested fragments of Ad7-E3CHE genomic DNA were 379, 653, 2619, 3766, 5721, 8335, and 12,939 bp for Age I; 716, 750, 947, 1164, 2607, 3460, 4349, 7281, and 13,138 bp for BamH I; 1005, 3725, 6095, 9570, and 14,017 bp for Kpn I; 1635, 4256, 6853, and 21,668 bp for Nde I; 4810, 5920, 6360, and 17,322 bp for Sal I. The predicted molecular weights of digested fragments of pKAd7-E3CHE plasmid were 379, 653, 2619, 3766, 5721, and 23,745 bp for Age I; 716, 947, 1164, 3460, 4349, 5828, 7281, and 13,138 bp for BamH I; 1005, 3725, 9570, and 22,583 bp for Kpn I; 4256, 10,959, and 21,668 bp for Nde I; 5920, 13,641, and 17,322 bp for Sal I. M: lambda/HindIII DNA marker. Please click here to view a larger version of this figure.
Table S1: Summary information of primers. Please click here to download this Table.
| Product plasmid | Size (bp) | DNA assembly reaction | Plasmid extraction | Restriction analysis | Sanger sequencing | |
| large fragment (bp) | small fragment (bp) | (colonies picked/with expected size) | (digested/correct) | (sequenced/correct) | ||
| pKan-Ad7 | 37718 | 35239 | 2543 | 6/5 | 2/2 | 2/2 |
| pKan-Ad7HpaI | 9761 | 7235 | 2584 | 6/6 | 3/3 | 3/3 |
| pKAd7-HE3GFP | 8915 | 7316 | 1650 | 6/6 | 3/3 | 3/3 |
| pKAd7-E3GFP | 36872 | 30483 | 6444 | 3/2 | 2/2 | 2/2 |
| pKAd7-E3CHE | 36883 | 35277 | 1655 | 3/3 | 2/2 | 2/2 |
Table 1: Summary data of plasmid construction. The restriction-assembly procedure of plasmid construction included fragment preparation, DNA assembly reaction, bacterial transformation, plasmid extraction, restriction analysis, and sequencing confirmation. Generally, dozens or hundreds of colonies could be produced on a LB plate after transforming E. coli TOP10 competent cells with DNA assembly product. The colonies were randomly picked up and propagated, and plasmid DNA was extracted and further digested with endonuclease and the bands pattern of digested plasmid was photographed and analyzed after electrophoresis. Finally, the whole plasmid or PCR products covering the fusion sites or PCR-amplified regions in the plasmid were subjected into Sanger sequencing.
Figure S1: Virtual intermediate plasmid for modification of both E1 and E3 regions. The long fragment from Sal I-digested pKan-Ad7 can be used to construct the intermediate plasmid pKan-Ad7SalI. Shown are the restriction sites of all unique cutters and some dual ones. E1 region, which is between the SnaB I and MauB I sites, can be precisely modified by overlap extension PCR. Similarly, Spe I together with BsrG I can help modify the E3 region. Please click here to download this File.
Discussion
Different adenoviruses have various tissue tropisms, and the prevalence of host pre-existing immunity against different adenoviruses can fluctuate intensively in human beings24, which attracts the interest in constructing novel adenoviral vectors for gene therapy or vaccine development. However, the establishment of a new adenoviral vector system remains cumbersome for generic laboratories of molecular biology.
Here, we introduced a protocol for generating vectors from wild-type adenoviruses by using restriction digestion and Gibson assembly techniques13,14,15,16,17,18,19. In general, this process can be divided into three steps. Firstly, the PCR product of the plasmid backbone is mixed with the adenovirus genomic DNA, and Gibson assembly is employed to generate an infectious clone. Designing PCR primers is the key factor: the 3' ends of the primers are specific sequences for amplifying the plasmid backbone which contains replication origin and antibiotic resistant gene, and the 5' ends are the reverse-complement sequences homologous to the terminus of adenovirus ITR. In addition, a non-cutter restriction site of the viral genome is added in the middle, which is used to release the viral genome from the infectious DNA clone. Secondly, an intermediate plasmid-based strategy is carried out to construct an adenoviral vector system. A fragment containing the region to be modified is excised from the infectious clone and used to construct an intermediate plasmid. Overlap extension PCR is the major approach to take for site-directed modification such as deletion of unessential genes and insertion of the exogenous gene expression cassette. The modified virus fragment is further excised from the intermediate plasmid by restriction digestion and restored to the corresponding region in the infectious DNA clone to generate an adenoviral vector plasmid. For this step, sequence analysis is of great importance to determine the region to be deleted and the ideal insertion site for the gene of interest. Generally, a dual cutter is selected to cut the infectious DNA clone to generate one small and one large fragment, while the target region for further modification is located at the small fragment, with which the intermediate plasmid is constructed. The adenoviral plasmid is linearized and used to transfect packaging cells for the rescue of recombinant viruses. Finally, the transgene loading strategy has already been considered in advance and integrated into the vector system. The transgene expression cassette in the adenoviral plasmid can be easily removed and replaced with other transgenes by restriction-assembly thanks to the preserved dual cutter sites flanking this region.
An infectious clone provides a solid starting point for the construction of a new adenoviral vector system. Generally, the method of homologous recombination in bacteria is used to construct infectious clones of adenoviruses5. PCR is performed to amplify the sequences at both ends of the adenoviral genome as homologous arms of 500-2000 bp in length, and the PCR products are ligated with a plasmid backbone to form a small plasmid. The small plasmid is linearized, mixed with adenovirus genomic DNA, and used to transform E. coli BJ5183 strain by electroporation. The recombination product, an infectious clone plasmid, needs to be identified and transferred into a recombinase-negative E. coli strain for plasmid amplification. The whole procedure contains at least three rounds of bacterial transformation and takes approximately 1 week, assuming that no unexpected events occur. Gibson assembly substantially simplifies the construction of adenovirus infectious clones. Compared with homologous recombination, it has the following advantages13: Firstly, DNA assembly is much simpler and more efficient as only one round of bacterial transformation is needed. Secondly, DNA assembly preserves the adenovirus genome in an infectious clone with higher fidelity because recombinase-positive bacteria, in which plasmids can be unstable, are excluded from the whole procedure. Finally, less sequence information is needed for DNA assembly since overlaps as short as 15-30 bp are enough for efficient assembly, which means that the genome can be cloned from an unknown adenovirus even if there is only sequence information at the end of the ITR.
A limitation of this approach is that finding the proper endonuclease is often complicated and cumbersome. This process runs through the whole protocol, including the construction of infectious clone plasmid, intermediate plasmid, and modification of region of interest (ROI). Sometimes, a suitable enzyme to modify the ROI cannot be found in the intermediate plasmid. In that case, it is necessary to construct another intermediate plasmid that also depends on the DNA sequence analysis software to make the plasmid smaller and facilitate the search for suitable restriction endonucleases. Occasionally, DNA assembly reaction may yield a few clones without the insert, as some restriction enzymes do not cut the plasmid to completion. In that case, select two different restriction endonucleases rather than a unique enzyme to digest the vector, which would avoid the self-anneal of overhangs. Another limitation of DNA assembly is that sequencing verification is required after each reaction, as there is the potential for mismatches in this method. However, incorrect assembly reactions do not occur very often. Dozens or hundreds of colonies can be grown in each LB plate after transforming E. coli TOP10 competent cells with DNA assembly product, and plasmids with the wrong assembly are rarely found (Table 1).
In summary, we showed here how to generate an E3-deleted adenoviral vector from an isolated wild-type HAdV-7, and the restriction assembly we introduced is a highly efficient, easy-to-use, and multipurpose method for adenoviral vector construction and modification.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research was funded by Beijing Natural Science foundation (7204258), National Natural Science Foundation of China (82161138001, 82072266), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-026), and the research and application on molecular tracing of essential respiratory pathogens in Beijing, by the Capital Health Development and Research of Special (2021-1G-3012).
Materials
| 1.5 mL polypropylene microcentrifuge Tube | Axygen | MCT-150-C | Storage of virus |
| 15 mL polypropylene centrifuge tubes | Corning | 430790 | Storage of virus |
| 150 mm TC-treated culture dishes | Corning | 430599 | Growth of HEK29E cells |
| 20 K MWCO dialysis cassette | ThermoFisher Scientific | 66005 | Dialysis of virus |
| Acetic acid | Amresco | 714 | Extraction of DNA |
| Afl II | NEB | R0520 | Digestion |
| Agarose | Takara | 5260 | Electrophoresis |
| Age I | NEB | R0552 | Digestion |
| Asc I | NEB | R0558 | Digestion |
| BamH I | NEB | R0136 | Digestion |
| Benzonase Nuclease | Sigma | E8263-25KU | Purification of virus |
| BsrG I | NEB | R0575 | Digestion |
| Cell lifter | Corning | 3008 | Scrape off the cells |
| CsCl | Sigma | C3032 | Purification of virus |
| DNA gel recovery kit | Zymo | D4045 | Recovery of DNA |
| Dulbecco’s modified Eagle’s medium (DMEM) | Cytiva | SH30022.01 | HEK293 cells medium |
| E.coli TOP10 competent cells | TIANGEN BIOTECH (BEIJING) CO.,LTD. | CB-104 | Transformation of assembly product |
| EcoR V | NEB | R3195 | Digestion |
| EDTA | Thermo Fisher Scientific | R3104 | Extraction of DNA |
| Fetal bovine serum (FBS) | Cytiva | SV30208.02 | HEK293 cells culture |
| Genomic DNA Clean and concentrator kit | Zymo | D4065 | Purification of DNA |
| Glycerol | Shanghai Macklin Biochemical Co., Ltd | G810575 | Dialysis of virus |
| HEK293 cells | ATCC | CRL-1573 | Amplification of virus |
| High-Fidelity DNA Polymerase | NEB | M0491 | PCR |
| Hind III | NEB | R3104 | Digestion |
| Kanamycin sulfate | Amresco | 408 | Selection of plasmid |
| Kpn I | NEB | R3142 | Digestion |
| Lambda/HindIII DNA marker | Takara | 3403 | Electrophoresis |
| LB broth | BD | 240230 | LB plate for bacteria |
| LB medium | Solarbio Life Science | L1010 | Medium for bacteria |
| MgCl2 | Sigma | 63068 | Dialysis of virus |
| Microcentrifuge | Thermo Fisher Scientific | Sorvall Legend Micro 21R | Extraction of DNA |
| NaCl | Sigma | S5886 | Dialysis of virus |
| Nde I | NEB | R0111 | Digestion |
| NEBuilder HiFi DNA Assembly Master Mix | NEB | E2621 | DNA assembly |
| Nhe I | NEB | R0131 | Digestion |
| Phosphate Buffered Saline | Cytiva | SH30256.01 | Washing of cells |
| Pipette | Thermo Fisher Scientific | Matrix | Aspirate the medium |
| Plasmid Maxprep Kit | Vigorous Biotechnology Beijing Co., Ltd. | N001 | Extraction of DNA |
| Plasmid Miniprep Kit | TIANGEN BIOTECH (BEIJING) CO.,LTD. | DP103 | Extraction of DNA |
| Pme I | NEB | R0560 | Digestion |
| Potassium acetate | Amresco | 698 | Extraction of DNA |
| Protease K | Thermo Fisher Scientific | AM2542 | Extraction of DNA |
| pShuttle-CMV | Stratagene | 240007 | PCR template |
| RNase | Beyotime | D7089 | Extraction of DNA |
| Sal I | NEB | R0138 | Digestion |
| Sbf I | NEB | R3642 | Digestion |
| SDS | Amresco | 227 | Extraction of DNA |
| Swinging-bucket rotor | HITACHI | S52ST | Purification of virus |
| T-25 cell flask | Corning | 430639 | Growth of HEK29E cells |
| T-75 cell flask | Corning | 430641 | Growth of HEK29E cells |
| Transfection reagent | Polyplus-transfection | 114-15 | Transfection |
| Transmission electron microscope | FEI | TECNAI 12 | Obsevation of virus |
| Tris-HCl | Amresco | 234 | Dialysis of virus |
| Ultracentrifuge | HITACHI | Himac CS120GXII | Purification of virus |
References
- Lasaro, M. O., Ertl, H. C. New insights on adenovirus as vaccine vectors. Molecular Therapy. 17 (8), 1333-1339 (2009).
- Fuchs, J. D., et al. Safety and immunogenicity of a recombinant adenovirus serotype 35-Vectored HIV-1 vaccine in adenovirus serotype 5 seronegative and seropositive individuals. Journal of AIDS & Clinical Research. 6 (5), (2015).
- Milligan, I. D., et al. Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia ankara-vectored ebola vaccines: a randomized clinical trial. JAMA. 315 (15), 1610-1623 (2016).
- Zhu, F. C., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 395 (10240), 1845-1854 (2020).
- He, T. C., et al. A simplified system for generating recombinant adenoviruses. Proceedings of the National Academy of Sciences of the United States of America. 95 (5), 2509-2514 (1998).
- Danthinne, X., Imperiale, M. J. Production of first generation adenovirus vectors: a review. Gene Therapy. 7 (20), 1707-1714 (2000).
- Guo, X., et al. Systemic and mucosal immunity in mice elicited by a single immunization with human adenovirus type 5 or 41 vector-based vaccines carrying the spike protein of Middle East respiratory syndrome coronavirus. Immunology. 145 (4), 476-484 (2015).
- Folegatti, P. M., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 396 (10249), 467-478 (2020).
- Jang, Y., Bunz, F. AdenoBuilder: A platform for the modular assembly of recombinant adenoviruses. STAR Protocols. 3 (1), 101123 (2022).
- Zhou, X., Sena-Esteves, M., Gao, G. Construction of recombinant adenovirus genomes by direct cloning. Cold Spring Harbor Protocols. 2019 (5), (2019).
- Ruzsics, Z., Lemnitzer, F., Thirion, C. Engineering adenovirus genome by bacterial artificial chromosome (BAC) technology. Methods in Molecular Biology. 1089, 143-158 (2014).
- Zhou, D., et al. An efficient method of directly cloning chimpanzee adenovirus as a vaccine vector. Nature Protocols. 5 (11), 1775-1785 (2010).
- Zou, X. H., et al. DNA assembly technique simplifies the construction of infectious clone of fowl adenovirus. Journal of Virological Methods. 257, 85-92 (2018).
- Guo, X., et al. Site-directed modification of adenoviral vector with combined DNA assembly and restriction-ligation cloning. Journal of Biotechnology. 307, 193-201 (2020).
- Liu, H., et al. Single plasmid-based, upgradable, and backward-compatible adenoviral vector systems. Human Gene Therapy. 30 (6), 777-791 (2019).
- Yan, B., et al. User-friendly reverse genetics system for modification of the right end of fowl adenovirus 4 genome. Viruses. 12 (3), 301 (2020).
- Zhang, W., et al. Fiber modifications enable fowl adenovirus 4 vectors to transduce human cells. Journal of Gene Medicine. 23 (10), 3368 (2021).
- Zou, X., et al. Fiber1, but not fiber2, is the essential fiber gene for fowl adenovirus 4 (FAdV-4). Journal of General Virology. 102 (3), 001559 (2021).
- Guo, X., et al. Restriction-assembly: a solution to construct novel adenovirus vector. Viruses. 14 (3), 546 (2022).
- Duan, Y., et al. Genetic analysis of human adenovirus type 7 strains circulating in different parts of China. Virologica Sinica. 36 (3), 382-392 (2021).
- Arad, U. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques. 24 (5), 760-762 (1998).
- Knipe, D. M. H. . P. M. Fields’ Virology. 2, 1733 (2013).
- Liu, H. Y., Han, B. J., Zhong, Y. X., Lu, Z. Z. A three-plasmid system for construction of armed oncolytic adenovirus. Journal of Virological Methods. 162 (1-2), 8-13 (2009).
- Mennechet, F. J. D., et al. A review of 65 years of human adenovirus seroprevalence. Expert Review of Vaccines. 18 (6), 597-613 (2019).

.
