分离、表征和总 DNA 提取以鉴定真菌异养植物中的内生真菌
Summary
本文旨在为植物相关内生真菌的分离、分离株的长期保存、形态学表征和总 DNA 提取提供详细和充分的方案,以便随后进行分子鉴定和宏基因组分析。
Abstract
霉菌异养植物是菌根依赖性最极端的形式之一,完全丧失了自养能力。与任何其他重要资源一样重要,与这些植物密切相关的真菌对它们来说是必不可少的。因此,研究真菌异养物种的一些最相关的技术是能够研究相关真菌的技术,尤其是那些栖息在根部和地下器官中的真菌。在这种情况下,通常应用鉴定培养依赖性和培养依赖性内生真菌的技术。分离真菌内生菌为形态学鉴定它们、分析它们的多样性和维持接种物提供了一种用于兰花种子共生萌发的方法。然而,众所周知,有大量不可培养的真菌栖息在植物组织中。因此,不依赖文化的分子鉴定技术提供了更广泛的物种多样性和丰度。本文旨在为启动两个调查程序提供必要的方法支持:一个是依赖于文化的调查程序,另一个是独立的调查程序。关于培养依赖性方案,详细介绍了从收集地点到实验室设施收集和维护植物样品的过程,以及从真菌异养植物的地下和地上器官中分离丝状真菌,保留分离物的集合,通过载玻片培养方法对菌丝进行形态学表征,以及通过总DNA提取对真菌进行分子鉴定。包括不依赖培养的方法,详细程序包括收集植物样本进行宏基因组分析和使用商业试剂盒从叶绿素植物器官中提取总DNA。最后,还建议使用连续性方案(例如,聚合酶链反应 [PCR]、测序)进行分析,并在此处介绍技术。
Introduction
根据定义,内生真菌是那些栖息在植物器官和组织内部的不明显感染(即不会对其宿主造成伤害)的真菌1,2。这些真菌可以与寄主植物进行中性或有益的相互作用,可能赋予对病原体和不利环境条件的抵抗力,并可能有助于植物有益化合物(例如生长因子和其他植物激素)的合成1,3。菌根内生菌是与植物建立菌根关联的真菌,参与养分转移4.在兰科中,与菌根内生菌的相互作用是绝大多数物种种子萌发和该科所有植物幼苗建立的基础5.在这种情况下,真菌异养兰花代表了对其菌根伙伴的完全依赖性,因为它们依赖于这些真菌在其整个生命周期中的矿物质营养物质和碳化合物转移6.因此,在研究真菌异养生命策略时,分离和鉴定关联真菌是基础。此外,人们对真菌内生菌在真菌异养植物中的作用知之甚少,甚至对这些真菌的真正多样性知之甚少 7,8.
内生真菌的研究 可以通过不同的技术 进行,传统上被描述为不依赖培养或依赖,例如:(a)直接观察,(b)真菌分离和形态学和/或分子鉴定,以及(c)植物组织的总DNA提取和分子鉴定9。在直接观察(a)中,内生真菌可以通过光学或电子显微镜9在植物细胞和组织内部进行研究,因为Pena-Passos等人10详细介绍了不同的显微镜方案。通过分离方法(b),可以根据真菌内生菌的菌落、菌丝以及生殖或抗性结构形态来表征真菌内生菌。此外, 通过 分离技术,可以通过DNA提取、分子识别序列(条形码或指纹图谱)的扩增和测序11来对分离株进行分子鉴定。后一种技术 (c) 能够在植物组织内部(元条形码)中通过 DNA 提取对内生真菌进行分子鉴定,然后进行文库制备和测序12。
此外,真菌分离株可以使用来自自养或真菌异养兰花的种子进行共生发芽试验。这种应用的一个例子是 Sisti 等人进行的研究13,描述了 Pogoniopsis schenckii(一种真菌异养兰)的原生球茎发育的萌发和初始阶段,与其一些分离株(包括非菌根内生真菌)相关。Pena-Passos等人10在视频中详细介绍了应用的共生萌发方案。分离与不同植物器官相关的真菌可以对植物-真菌相互作用的性质进行不同的研究(例如,理解这种关联的生态或生理方面,以及调查从真菌到植物的营养转移)9。
第 1 节中介绍的方法基于地下器官样本的收集,因为这些器官在收集方面存在最大的困难,并且由于菌根内生菌定植于它们,因此它们引起了人们的极大兴趣。然而,包含的方案(步骤1.1和1.2)都可以应用于其他真菌异养植物器官(例如,根茎,花茎和果实)。步骤1.1中描述的收集方法被指定用于分离内生真菌(第2节)进行形态学表征(第4节和第5节)和/或总DNA提取以进行分离物鉴定(第6节)。另一方面,步骤1.2中描述的收集方法专门用于植物组织的总DNA提取,用于元条形码技术(第7节)。在第 3 节中,介绍了四种丝状真菌储存和保存方法,其中两种用于短期储存(3-6 个月),另外两种适合长期储存(>1 年)。形态学特征(第 4 节和第 5 节)可能与分子鉴定相关联,以加强它并提供有关真菌宏观和微观形态学的重要信息。 图1 总结了随后描述的集体方法。
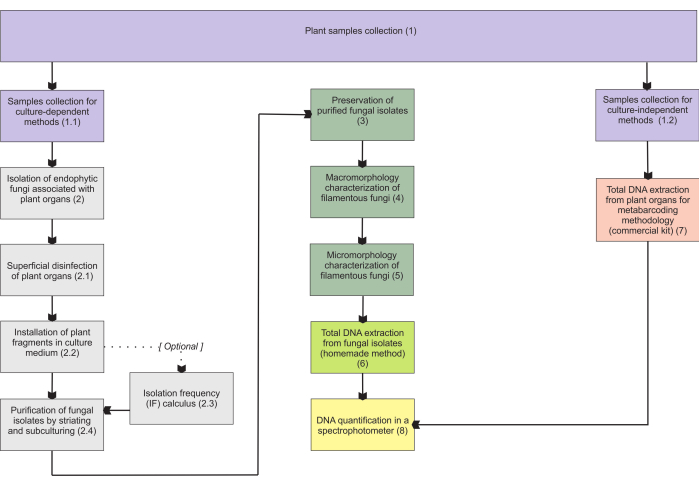
图1:所提出方法的示意图。 通过培养依赖性和独立性方法进行植物收集和真菌分离、保存和分子鉴定。 请点击这里查看此图的较大版本.
Protocol
Representative Results
Discussion
植物样品的表面灭虫是所提出的协议中最关键的阶段之一。PDA培养皿中不会受到上次洗涤的液滴污染,这是非常可取的。细菌经常在隔离皿中被观察到为污染物,通常比空气中的孢子真菌更多,因为内生细菌在植物组织中也很常见3,11。因此,在安装器官碎片时,在培养基中添加抗生素是必不可少的。当组合不同的抗生素类型时,可以获得更好的效果,?…
Divulgations
The authors have nothing to disclose.
Acknowledgements
我们感谢 FAPESP (2015/26479-6) 和 CNPq (447453/2014-9) 的资助。JLSM 感谢 CNPq 提供的生产力资助 (303664/2020-7)。MPP 感谢 Capes(硕士学位奖学金,流程 88887.600591/2021-00)和 CNPq。
Materials
| Adhesive tape | (from any company, for adhesive tape mount in micromorphological analyses) | ||
| Ampicillin | Sigma-Aldrich | A5354 | (for installation of plant fragments; other antibiotics may be used – check step 2.2.1) |
| Autoclave | (from any company, for materials sterilization in many steps) | ||
| Bacteriological agar | Sigma-Aldrich | A1296 | (for many steps) |
| C1, C2, C3, C4, C5, and C6 solutions | Qiagen | 12888-50 | (purchased with DNeasy PowerSoil kit) |
| Centrifuge | Merck/Eppendorf | 5810 G | (for total DNA extraction from fungal isolates) |
| Centrifuge tubes | Merck | CLS430828 | (for samples collection) |
| Chloroform | Sigma-Aldrich | C2432 | (for total DNA extraction from fungal isolates) |
| Congo red | Supelco | 75768 | (for hyphae staining) |
| Cryotubes | Merck | BR114831 | (for many steps) |
| Ethanol | Supelco | 100983 | It will be necessary to carry out the appropriate dilutions (for many steps) |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | 3609 | (for total DNA extraction from fungal isolates) |
| Filter paper | Merck | WHA10010155 | (for many steps) |
| Glass test tubes | Merck | CLS7082516 | (for cryopreservation in unhulled rice grains) |
| Glass wool | Supelco | 20411 | (for cryopreservation in unhulled rice grains) |
| Glucose | Sigma-Aldrich | G8270 | Or dextrose (for cryopreservation in vermiculite) |
| Glycerol | Sigma-Aldrich | G5516 | Or glycerin (for cryopreservation in vermiculite, for preparing LPCB) |
| Isopropanol | Sigma-Aldrich | 563935 | (for total DNA extraction from fungal isolates) |
| Lactic acid | Sigma-Aldrich | 252476 | (for preparing LPCB – hyphae staining) |
| Lactophenol blue solution (LPCB) | Sigma-Aldrich | 61335 | (for hyphae staining) |
| Laminar flow hood | (class I, from any company, for many steps) | ||
| Light microscope | (from any company, for hyphae observation) | ||
| MB Spin Columns | Qiagen | 12888-50 | (purchased with DNeasy PowerSoil kit) |
| Methyl blue (cotton blue) | Sigma-Aldrich | M5528 | (for preparing LPCB – hyphae staining) |
| Microcentrifuge tube (1.5 mL) | Merck | HS4323 | (for total DNA extraction from fungal isolates) |
| Microcentrifuge tube (2 mL) | Merck | BR780546 | (for many steps) |
| Mineral oil | (for preservation of fungal isolates) | ||
| Paper bags | Average size 150 mm x 200 mm (for samples collection) | ||
| Petri dish (Glass, 120 mm x 20 mm) | Merck/Pyrex | SLW1480/10D | (autoclavable, for fungi slide culture, prefer higher ones) |
| Petri dish (Glass, 50 mm x 17 mm) | Merck/Aldrich | Z740618 | (for purification of fungal isolates); alternatively: polystyrene petri dishes (sterile, γ-irradiated, non-autoclavable) |
| Petri dish (Glass, 80 mm x 15 mm) | Merck/Brand | BR455732 | (for installation of plant fragments); alternatively: polystyrene petri dishes (sterile, γ-irradiated, non-autoclavable) |
| Phenol | Sigma-Aldrich | P1037 | (for total DNA extraction from fungal isolates, for preparing LPCB) |
| Porcelain mortar | Sigma-Aldrich | Z247464 | (for total DNA extraction from fungal isolates) |
| Porcelain pestle | Sigma-Aldrich | Z247502 | (for total DNA extraction from fungal isolates) |
| Potato dextrose agar (PDA) | Millipore | P2182 | (for many steps) |
| PowerBead tubes | Qiagen | 12888-50 | (purchased with DNeasy PowerSoil kit) |
| Rapid mounting medium (Entellan) | Sigma-Aldrich | 1.0796 | (for fungi slide culture) |
| Silica gel | Supelco | 717185 | (for cryopreservation in unhulled rice grains) |
| Sodium chloride (NaCl) | Sigma-Aldrich | S9888 | (for total DNA extraction from fungal isolates) |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | L3771 | Lauryl sulfate sodium salt (for total DNA extraction from fungal isolates) |
| Sodium hypochlorite (w/ 2% active chlorine) | (commercial product, for superficial desinfestation) | ||
| Soil DNA extraction kit (DNeasy PowerSoil kit) | Qiagen | 12888-50 | (for total DNA extraction from plant organs) |
| Spectrophotometer – Nanodrop 2000/2000c | ThermoFisher Scientific | ND2000CLAPTOP | (for total DNA extraction from plant organs) |
| Stereomicroscope | (=dissecting microscope, from any company, for macromorphological analyses) | ||
| Tetracycline | Sigma-Aldrich | T7660 | (for installation of plant fragments) |
| Thermoblock | Merck/Eppendorf | EP5362000035 | (or from other companies) |
| Tissue homogenizer and cell lyzer | SPEX SamplePrep | 2010 Geno/Grinder – Automated Tissue Homogenizer and Cell Lyzer (for total DNA extraction from plant organs) | |
| Toluidine blue O | Sigma-Aldrich/Harleco | 364-M | (for hyphae staining) |
| Trehalose | Sigma-Aldrich | T9531 | (for cryopreservation in vermiculite) |
| Tris Base Solution (Tris) | Sigma-Aldrich | T1699 | (for total DNA extraction from fungal isolates) |
| Unhulled rice grains | (for cryopreservation) | ||
| U-shaped glass rod | (or an adaptation – check step 5.4.1, for fungi slide culture) | ||
| Vermiculite | Fine granulometry (for cryopreservation in vermiculite) | ||
| Vortexer | Sigma-Aldrich/BenchMixer | BMSBV1000 | (for total DNA extraction from fungal isolates) |
| Yeast extract | Sigma-Aldrich | Y1625 | (for cryopreservation in vermiculite) |
References
- de Azevedo, J. L. Endophytic microorganisms. Ecologia Microbiana. , 117-137 (1998).
- Stone, J. K., Bacon, C. W., White, J. F. An overview of endophytic microbes: endophytism defined. Microbial Endophytes. , 17-44 (2000).
- Schulz, B., Boyle, C. What are Endophytes. Microbial Root Endophytes. , 1-13 (2006).
- Smith, S. E., Read, D. J. . Mycorrhizal Symbiosis. , (2008).
- Rasmussen, H. N., Dixon, K. W., Jersáková, J., Těšitelová, T. Germination and seedling establishment in orchids: a complex of requirements. Annals of Botany. 116 (3), 391-402 (2015).
- Rasmussen, H. N., Rasmussen, F. N. Orchid mycorrhiza: implications of a mycophagous life style. Oikos. 118 (3), 334-345 (2009).
- Ma, X., Kang, J., Nontachaiyapoom, S., Wen, T., Hyde, K. D. Non-mycorrhizal endophytic fungi from orchids. Current Science. 109 (1), 72-87 (2015).
- Favre-Godal, Q., Gourguillon, L., Lordel-Madeleine, S., Gindro, K., Choisy, P. Orchids and their mycorrhizal fungi: an insufficiently explored relationship. Mycorrhiza. 30 (1), 5-22 (2020).
- Sun, X., Guo, L. -. D. Endophytic fungal diversity: review of traditional and molecular techniques. Mycology. 3 (1), 65-76 (2012).
- Pena-Passos, M., Sisti, L. S., Mayer, J. L. S. Microscopy techniques for interpreting fungal colonization in mycoheterotrophic plants tissues and symbiotic germination of seeds. Journal of Visualized Experiments. (183), e63777 (2022).
- Araújo, W. L., et al. . Endophytic microorganisms: Theoretical and Practical Aspects of Isolation and Characterization. 1st ed. 1, 257 (2014).
- de Souza, R. S. C., et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Scientific Reports. 6, 28774 (2016).
- Sisti, L. S., et al. The role of non-mycorrhizal fungi in germination of the mycoheterotrophic orchid Pogoniopsis schenckii Cogn. Frontiers in Plant Science. 10, 1589 (2019).
- Araújo, W. L., et al. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Canadian Journal of Microbiology. 47 (3), 229-236 (2001).
- Castellani, A. Further researches on the long viability and growth of many pathogenic fungi and some bacteria in sterile distilled water. Mycopathologia. 20 (1-2), 1-6 (1963).
- Currah, R. S., Zelmer, C. D., Hambleton, S., Richardson, K. A. Fungi from orchid mycorrhizas. Orchid Biology: Reviews and Perspectives, VII. , 117-170 (1997).
- Freitas, E. F. S., et al. Diversity of mycorrhizal Tulasnella associated with epiphytic and rupicolous orchids from the Brazilian Atlantic Forest, including four new species. Scientific Reports. 10 (1), 7069 (2020).
- Sato, M., Inaba, S., Noguchi, M., Nakagiri, A. Vermiculite as a culture substrate greatly improves the viability of frozen cultures of ectomycorrhizal basidiomycetes. Fungal Biology. 124 (8), 742-751 (2020).
- Pereira, O. L., Kasuya, M. C. M., Borges, A. C., Araújo, E. F. D. Morphological and molecular characterization of mycorrhizal fungi isolated from neotropical orchids in Brazil. Canadian Journal of Botany. 83 (1), 54-65 (2005).
- Riddell, R. W. Permanent stained mycological preparations obtained by slide culture. Mycologia. 42 (2), 265-270 (1950).
- Walsh, T. J., Hayden, R. T., Larone, D. H. . Larone’s Medically Important Fungi: A Guide to Identification. , (2018).
- Microscopy: Chemical Reagents. British Mycological Society Available from: https://www.britmycolsoc.org.uk/field_mycology/microscopy/reagents (2022)
- Senanayake, I. C., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 11 (1), 2678-2754 (2020).
- Slifkin, M., Cumbie, R. Congo red as a fluorochrome for the rapid detection of fungi. Journal of Clinical Microbiology. 26 (5), 827-830 (1988).
- Raeder, U., Broda, P. Rapid preparation of DNA from filamentous fungi. Letters in Applied Microbiology. 1 (1), 17-20 (1985).
- Martins, M. K., et al. Molecular characterization of endophytic microorganisms. Endophytic microorganisms: theoretical and practical aspects of isolation and characterization. 1st edition. , 189-211 (2014).
- Rayner, R. W. A Mycological Colour Chart. Commonwealth Mycological Institute. , (1970).
- Kornerup, A., Wanscher, J. H. . Methuen Handbook of Colour. Methuen handbook of colour. , (1967).
- Ridgway, R. . Color Standards and Color Nomenclature. , (1912).
- McGinnis, M. R. . Laboratory Handbook of Medical Mycology. , (2012).
- Webster, J., Weber, R. . Introduction to Fungi. , (2007).
- Sridharan, G., Shankar, A. A. Toluidine blue: A review of its chemistry and clinical utility. Journal of Oral and Maxillofacial Pathology. 16 (2), 251-255 (2012).
- Smith, D., Onions, A. H. S. A comparison of some preservation techniques for fungi. Transactions of the British Mycological Society. 81 (3), 535-540 (1983).
- Ryan, M. J., Smith, D., Jeffries, P. A decision-based key to determine the most appropriate protocol for the preservation of fungi. World Journal of Microbiology and Biotechnology. 16 (2), 183-186 (2000).
- Lalaymia, I., Cranenbrouck, S., Declerck, S. Maintenance and preservation of ectomycorrhizal and arbuscular mycorrhizal fungi. Mycorrhiza. 24 (5), 323-337 (2014).
- Zettler, L. W., Corey, L. L. Orchid mycorrhizal fungi: isolation and identification techniques. Orchid Propagation: From Laboratories to Greenhouses-Methods and Protocols. , 27-59 (2018).
- Yu, S., Wang, Y., Li, X., Yu, F., Li, W. The factors affecting the reproducibility of micro-volume DNA mass quantification in Nanodrop 2000 spectrophotometer. Optik. 145, 555-560 (2017).
- Martos, F., et al. Independent recruitment of saprotrophic fungi as mycorrhizal partners by tropical achlorophyllous orchids. New Phytologist. 184 (3), 668-681 (2009).
- Schoch, C. L., et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences. 109 (16), 6241-6246 (2012).
- White, T. J., Bruns, T., Lee, S., Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 18 (1), 315-322 (1990).
- Sanger, F., Nicklen, S., Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences. 74 (12), 5463-5467 (1977).
- Ranjard, L., et al. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Applied and Environmental Microbiology. 67 (10), 4479-4487 (2001).
- Metzker, M. L. Sequencing technologies-the next generation. Nature Reviews Genetics. 11 (1), 31-46 (2010).

