Bringing the Clinic Home: An At-Home Multi-Modal Data Collection Ecosystem to Support Adaptive Deep Brain Stimulation
Summary
The protocol shows a prototype of the at-home multi-modal data collection platform that supports research optimizing adaptive deep brain stimulation (aDBS) for people with neurological movement disorders. We also present key findings from deploying the platform for over a year to the home of an individual with Parkinson’s disease.
Abstract
Adaptive deep brain stimulation (aDBS) shows promise for improving treatment for neurological disorders such as Parkinson’s disease (PD). aDBS uses symptom-related biomarkers to adjust stimulation parameters in real-time to target symptoms more precisely. To enable these dynamic adjustments, parameters for an aDBS algorithm must be determined for each individual patient. This requires time-consuming manual tuning by clinical researchers, making it difficult to find an optimal configuration for a single patient or to scale to many patients. Furthermore, the long-term effectiveness of aDBS algorithms configured in-clinic while the patient is at home remains an open question. To implement this therapy at large scale, a methodology to automatically configure aDBS algorithm parameters while remotely monitoring therapy outcomes is needed. In this paper, we share a design for an at-home data collection platform to help the field address both issues. The platform is composed of an integrated hardware and software ecosystem that is open-source and allows for at-home collection of neural, inertial, and multi-camera video data. To ensure privacy for patient-identifiable data, the platform encrypts and transfers data through a virtual private network. The methods include time-aligning data streams and extracting pose estimates from video recordings. To demonstrate the use of this system, we deployed this platform to the home of an individual with PD and collected data during self-guided clinical tasks and periods of free behavior over the course of 1.5 years. Data were recorded at sub-therapeutic, therapeutic, and supra-therapeutic stimulation amplitudes to evaluate motor symptom severity under different therapeutic conditions. These time-aligned data show the platform is capable of synchronized at-home multi-modal data collection for therapeutic evaluation. This system architecture may be used to support automated aDBS research, to collect new datasets and to study the long-term effects of DBS therapy outside the clinic for those suffering from neurological disorders.
Introduction
Deep brain stimulation (DBS) treats neurological disorders such as Parkinson's disease (PD) by delivering electrical current directly to specific regions in the brain. There are an estimated 8.5 million cases of PD worldwide, and DBS has proved to be a critical therapy when medication is insufficient for managing symptoms1,2. However, DBS effectiveness can be constrained by side-effects that sometimes occur from stimulation that is conventionally delivered at fixed amplitude, frequency, and pulse width3. This open-loop implementation is not responsive to fluctuations in symptom state, resulting in stimulation settings that are not appropriately matched to the changing needs of the patient. DBS is further hindered by the time-consuming process of tuning stimulation parameters, which is currently performed manually by clinicians for each individual patient.
Adaptive DBS (aDBS) is a closed-loop approach shown to be an effective next iteration of DBS by adjusting stimulation parameters in real time whenever symptom-related biomarkers are detected3,4,5. Studies have shown beta oscillations (10-30 Hz) in the subthalamic nucleus (STN) occur consistently during bradykinesia, a slowing of movement that is characteristic of PD6,7. Similarly, high-gamma oscillations (50-120 Hz) in the cortex are known to occur during periods of dyskinesia, an excessive and involuntary movement also commonly seen in PD8. Recent work has successfully administered aDBS outside the clinic for prolonged periods5, however the long-term effectiveness of aDBS algorithms that were configured in-clinic while a patient is home has not been established.
Remote systems are needed to capture the time-varying effectiveness of these dynamic algorithms in suppressing symptoms encountered during daily living. While the dynamic stimulation approach of aDBS potentially enables a more precise treatment with reduced side-effects3,9, aDBS still suffers from a high burden on clinicians to manually identify stimulation parameters for each patient. In addition to the already large set of parameters to program during conventional DBS, aDBS algorithms introduce many new parameters which must also be carefully adjusted. This combination of stimulation and algorithm parameters yields a vast parameter space with an unmanageable number of possible combinations, prohibiting aDBS from scaling to many patients10. Even in research settings, the additional time required to configure and assess aDBS systems make it difficult to adequately optimize algorithms solely in the clinic, and remote updating of parameters is needed. To make aDBS a treatment that can scale, stimulation and algorithm parameter tuning must be automated. In addition, outcomes from therapy must be analyzed across repeated trials to establish aDBS as a viable long-term treatment outside the clinic. There is a need for a platform that can collect data for remote evaluation of therapy effectiveness, and to remotely deploy updates to aDBS algorithm parameters.
The goal of this protocol is to provide a reusable design for a multi-modal at-home data collection platform to improve aDBS effectiveness outside the clinic, and to enable this treatment to scale to a greater number of individuals. To our knowledge, it is the first data collection platform design that remotely evaluates therapeutic outcomes using in-home video cameras, wearable sensors, chronic neural signal recording, and patient-driven feedback to evaluate aDBS systems during controlled tasks and naturalistic behavior.
The platform is an ecosystem of hardware and software components built upon previously developed systems5. It is maintainable entirely through remote access after an initial installation of minimal hardware to allow multi-modal data collection from a person in the comfort of their home. A key component is the implantable neurostimulation system (INS)11 which senses neural activity and delivers stimulation to the STN, and records acceleration from chest implants. For the implant used in the initial deployment, neural activity is recorded from bilateral leads implanted in the STN and from electrocorticography electrodes implanted over the motor cortex. A video recording system helps clinicians monitor symptom severity and therapy effectiveness, which includes a graphical user interface (GUI) to allow easy cancellation of ongoing recordings to protect patient privacy. Videos are processed to extract kinematic trajectories of position in two dimensional (2D) or three dimensional (3D), and smart watches are worn on both wrists to capture angular velocity and acceleration information. Importantly, all data is encrypted before being transferred to long-term cloud storage, and the computer with patient-identifiable videos can only be accessed through a virtual private network (VPN). The system includes two approaches for post-hoc time-aligning of all data streams, and data is used to remotely monitor the patient's quality of movement, and to identify symptom-related biomarkers for refining aDBS algorithms. The video portion of this work shows the data collection process and animations of kinematic trajectories extracted from collected videos.
A number of design considerations guided the development of the protocol:
Ensuring data security and patient privacy: Collecting identifiable patient data requires utmost care in transmission and storage in order to be health insurance portability and accountability act (HIPAA)12, 13 compliant and to respect the patient's privacy in their own home. In this project, this was achieved by setting up a custom VPN to ensure privacy of all sensitive traffic between system computers.
Stimulation parameter safety boundaries: It is critical to ensure that the patient remains safe while trying out aDBS algorithms that may have unintended effects. The patient's INS must be configured by a clinician to have safe boundaries for stimulation parameters that do not allow for unsafe effects from over-stimulation or under-stimulation. With the INS system11 used in this study, this feature is enabled by a clinician programmer.
Ensuring the patient veto: Even within safe parameter limits, the daily variability of symptoms and stimulation responses may result in unpleasant situations for the patient where they dislike an algorithm under test and wish to return to normal clinical open-loop DBS. The selected INS system includes a patient telemetry module (PTM) that allows the patient to manually change their stimulation group and stimulation amplitude in mA. There is also an INS-connected research application that is used for remote configuration of the INS prior to data collection14, which also enables the patient to abort aDBS trials and control their therapy.
Capturing complex and natural behavior: Video data was incorporated in the platform to enable clinicians to remotely monitor therapy effectiveness, and to extract kinematic trajectories from pose estimates for use in research analyses15. While wearable sensors are less intrusive, it is difficult to capture the full dynamic range of motion of an entire body using wearable systems alone. Videos enable the simultaneous recording of the patient's full range of motion and their symptoms over time.
System usability for patients: Collecting at-home multi-modal data requires multiple devices to be installed and utilized in a patient's home, which could become burdensome for patients to navigate. To make the system easy to use while ensuring patient control, only the devices that are implanted or physically attached to the patient (in this case it included the INS system and smart watches) must be manually turned ON prior to initiating a recording. For devices that are separate from the patient (in this case it includes data recorded from video cameras), recordings start and end automatically without requiring any patient interaction. Care was taken during GUI design to minimize the number of buttons and to avoid deep menu trees so that interactions were simple. After all devices are installed, a research coordinator showed the patient how to interact with all devices through patient-facing GUIs that are a part of each device, such as how to terminate recordings on any device and how to enter their medication history and symptom reports.
Data collection transparency: Clearly indicating when cameras are turned ON is imperative so that people know when they are being recorded and can suspend recording if they need a moment of privacy. To achieve this, a camera-system application is used to control video recordings with a patient-facing GUI. The GUI automatically opens when the application is started and lists the time and date of the next scheduled recording. When a recording is ongoing, a message states when the recording is scheduled to end. In the center of the GUI, a large image of a red light is displayed. The image shows the light being brightly lit whenever a recording is ongoing, and changes to a non-lit image when recordings are OFF.
The protocol details methods for designing, building, and deploying an at-home data collection platform, for quality-checking the data collected for completeness and robustness, and for post-processing data for use in future research.
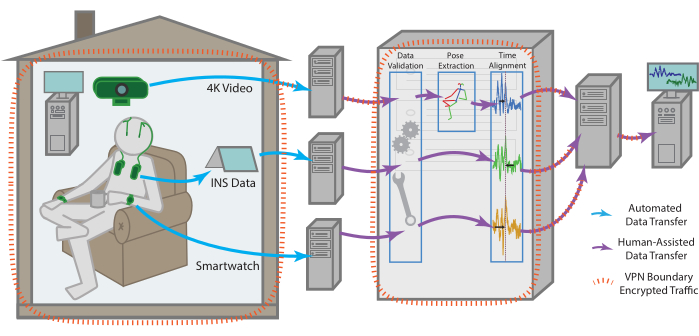
Figure 1: Data flow. Data for each modality is collected independently from the patient's residence before being processed and aggregated into a single remote storage endpoint. The data for each modality is sent automatically to a remote storage endpoint. With the help of one of the team members, it can then be retrieved, checked for validity, time aligned across modalities, as well as subjected to more modality-specific pre-processing. The compiled dataset is uploaded then to a remote storage endpoint that can be securely accessed by all team members for continued analysis. All machines with data access, especially for sensitive data such as raw video, are enclosed within a VPN that ensures all data is transferred securely and stored data is always encrypted. Please click here to view a larger version of this figure.
Protocol
Patients are enrolled through a larger IRB and IDE approved study into the aDBS at the University of California, San Francisco, protocol # G1800975. The patient enrolled in this study additionally provided informed consent specifically for this study.
1. At-home system components
- Central server and VPN
- Acquire a personal computer (PC) running a Linux-based operating system (OS) dedicated to serving a VPN. House the machine in a secure room. Disk encrypt the machine to ensure data security.
- Configure the VPN server to be publicly accessible on at least one port.
NOTE: In this case, this was achieved by collaborating with the IT department to give the server an externally facing static-IP address and a custom URL by the university's DNS hosting options. - For server installation complete the following steps once on the PC selected for serving the VPN.
- Firewall configuration: Run the following commands in the PC terminal to install and configure uncomplicated firewall:
sudo apt install ufw
sudo ufw allow ssh
sudo ufw allow <port-number>/udp
sudo ufw enable - Server VPN installation: Install the open-source WireGuard VPN protocol16 on the PC and navigate to the installation directory. In the PC terminal, run umask 007 to update directory access rules.
- Key generation: In the PC terminal, run
wg genkey | tee privatekey | wg pubkey > publickey
This generates a public/private key pair for the VPN server. This public key will be shared to any client PC that connects to the VPN. - VPN configuration: In the PC terminal, run touch <interface_name>.conf to create a configuration file, where the file name should match the name of the interface. Paste the following server rules into this file:
[Interface]
PrivateKey = <contents-of-server-privatekey>
Address = ##.#.#.#/##
PostUp = iptables -A FORWARD -i interface_name -j ACCEPT; iptables -t nat -A POSTROUTING -o network_interface_name -j MASQUERADE
PostDown = iptables -D FORWARD -i interface_name -j ACCEPT; iptables -t nat -D POSTROUTING -o network_interface_name -j MASQUERADE
ListenPort = #####
[Peer]
PublicKey = <contents-of-client-publickey>
AllowedIPs = ##.#.#.#/## - Activating the VPN: Start the VPN by entering wg-quick up <interface_name> in the terminal. To enable the VPN protocol to automatically start whenever the PC reboots, run the following in the terminal:
systemctl enable wg-quick@ <interface_name>
- Firewall configuration: Run the following commands in the PC terminal to install and configure uncomplicated firewall:
- For client installation complete the following steps for each new machine that needs access to the VPN.
- Client VPN installation: Install the VPN protocol according to the OS-specific instructions on the WireGuard16 download page.
- Adding a client to the VPN: Take the public key from the configuration file generated during installation. Paste this key into the peer section of the server's configuration file.
- Activating the VPN: Start the VPN per the OS-specific instructions on the WireGuard16 download page.
- Cloud storage
- Select a cloud storage site to enable all recorded data streams to be stored long-term in one place. Here, an Amazon web service-based cloud storage site that was compatible with the selected data transfer protocol was used.
- Implantable neuromodulation system
- Following IRB and IDE guidelines, select an implantable neuromodulation system (INS)11 that allows patients to manually change their stimulation settings.
- Acquire a tablet PC and install the open-source UCSF DBS application to allow for INS recordings, reporting medications and symptoms or any other patient comments14. Configure INS data that is streamed to the tablet to be uploaded to a temporary HIPPA-compliant cloud storage endpoint, for temporary storage prior to data de-identification and offloading to long-term cloud storage.
- Video collection system
- Acquire a PC capable of collecting and storing the desired amount of video files prior to transferring them to cloud storage. Ensure that the PC motherboard includes a trusted platform module (TPM) chip.
NOTE: In this case, a PC with a 500 GB SSD, a 2 TB HDD and a 6 GB GPU was selected. A 2 TB disk ensures that videos can be buffered after a lengthy recording session or in the case of losing internet connection for a couple of days, while the single PC keeps hardware minimally intrusive in the home. - Install the desired OS and follow prompts to enable automatic disk encryption to ensure patient privacy and to avoid data leakage. In this case a Linux-based OS with an Ubuntu distribution was chosen for its ease of use and reliability.
- Separately encrypt any hard disks after the OS is installed. Be sure to enable automatic disk re-mounting upon system reboots.
- Configure the PC's on-board TPM chip to maintain access to the disk-encrypted PC after a system reboot17.
NOTE: If using a Linux OS, be sure to select a motherboard with a TPM2 chip installed to enable this step. If a Windows OS is used, automatic disk encryption and unlocking can be handled by the Bitlocker program. - Configure the PC as a VPN client by following the installation steps in 1.1.4. Enable the VPN protocol to automatically start whenever the PC is rebooted as in section 1.1.3.5 to ensure that researcher computers can always remotely access the PC (recommended).
- Create a GitHub machine user account to easily automate updates to software installed on the PC. This account serves as a webhook to automate pulling from the remote git endpoint and helps identify any updates pushed from the remote machine.
- Select software to schedule and control video recordings and install this on the PC. To maximize patient privacy and comfort, the selected software should include a graphical user interface (GUI) to clearly indicate when recordings are ongoing, and to enable easy termination of recordings at any point in time.
NOTE: If desired, the authors' custom video recording application with a patient-facing GUI can be installed by downloading the application and following instructions on GitHub (https://github.com/Weill-Neurohub-OPTiMaL/VideoRecordingApp). - Select a monitor to indicate when videos are being recorded and to enable people to easily terminate recordings. Select a monitor with touchscreen capability so that recordings can be terminated without needing to operate a keyboard or mouse.
- Install a remote desktop application on the PC. This enables running an application with a GUI such that the GUI remains visible on both the patient side and the remote researcher side.
NOTE: The open-source NoMachine remote desktop application worked best for a Linux OS. - Select USB-compatible webcams with sufficiently high-resolution for calculating poses in the given space.
NOTE: In this case 4k-compatible webcams were chosen, which offer multiple resolution and framerate combinations including 4k resolution at 30 fps or HD resolution at 60 fps. - Select robust hardware for mounting webcams in the patient's home. Use gooseneck mounts with clips to secure them to the furniture to prevent the cameras from shaking.
- Select a data transfer protocol with encryption capability and install this on the PC. Create a configuration to access the cloud storage site, then create an encryption configuration to wrap the first configuration prior to data transfer.
NOTE: In this case an open-source data transfer and file syncing protocol with encryption capability was installed18. The data transfer protocol documentation explains how to configure data transfer to cloud storage. The protocol was first installed on the VPN server and an encryption configuration was created that transfers data to the offsite cloud storage site.
- Acquire a PC capable of collecting and storing the desired amount of video files prior to transferring them to cloud storage. Ensure that the PC motherboard includes a trusted platform module (TPM) chip.
- Wearable-sensor data components
- Select smart watches to be worn on each wrist of the patient to track signals including movement, accelerometry and heart rate.
NOTE: The Apple watch series 3 was selected with a built-in movement disorder symptom monitor that generates PD symptom scores such as dyskinesia and tremor scores. - Select and install software on each smart watch that can start and end recordings and can transfer data to cloud storage. Select an application which uploads all data streams to its associated online portal for researchers and clinicians to analyze19.
- Select smart watches to be worn on each wrist of the patient to track signals including movement, accelerometry and heart rate.

Figure 2: Video recording components. The hardware components to support video data collection are minimal, including a single tower PC, USB-connected webcams, and a small monitor to display the patient-facing GUI. The monitor is touchscreen-enabled to allow easy termination of any ongoing or scheduled recordings by pressing the buttons visible on the GUI. The center of the GUI shows an image of a recording light that turns to a bright red color when video cameras are actively recording. Please click here to view a larger version of this figure.
2. In-home configuration
- Hardware installation
- Determine an appropriate space for mounting webcams that minimizes disruptions to the home. Determine the space through discussions with the patient; here the home office area was chosen as the optimal site for balancing recording volume against privacy.
- Mount webcams in the identified area on the selected mounting hardware. Clipping gooseneck mounts to nearby heavy furniture prevents cameras from shaking whenever someone steps nearby.
- Place the PC sufficiently close to the mounted webcams such that their USB cables can connect to the PC.
- Place the tablet PC, INS components, smart watches, and smart phones near a power outlet such that all devices can stay plugged in and are ready to use at any time.
- Confirm that the VPN is ON by running route -n in the PC terminal. If not, follow instructions to activate the VPN in section 1.1.3.5.
- Start the video recording application
- Video recording schedule: Prior to collecting any data, discuss an appropriate recording schedule with the patient. Configure this schedule on the video recording software.
NOTE: If using the authors' custom video recording application, instructions for setting a schedule can be found on GitHub (https://github.com/Weill-Neurohub-OPTiMaL/VideoRecordingApp#installation-guide). - Update recording software: Ensure that the latest version of the selected video recording software has been uploaded to the PC using the GitHub machine user account installed in 1.4.6.
- Start video recordings: Log into the PC through the installed remote desktop software and start the video recording software.
NOTE: If using the authors' custom video recording application, instructions for starting the application can be found on GitHub (https://github.com/Weill-Neurohub-OPTiMaL/VideoRecordingApp#installation-guide).
- Video recording schedule: Prior to collecting any data, discuss an appropriate recording schedule with the patient. Configure this schedule on the video recording software.
- Video camera calibration
- Disable autofocus: For computing intrinsic parameters such as lens and perspective distortion, follow the instructions based on the selected OS and webcams to turn off the autofocus.
NOTE: On Linux, webcams are accessed via the video for Linux API, which by default turns on autofocus every time the computer connected to the cameras is restarted. Configuring a script to automatically disable this is necessary to preserve the focus acquired during camera calibration for processing 3D pose. - Intrinsic calibration: Acquire a 6 x 8 checkerboard pattern with 100 mm squares to support 3D calibration of pose estimation software20. Record a video from each individual webcam while a researcher angles the checkerboard in-frame of all cameras. Ensure that the checkerboard has an even number of rows and an uneven number of columns (or vice versa). This will remove ambiguity regarding rotation.
- Extrinsic calibration: Record a video from all three webcams simultaneously. Be sure that videos are recorded at the same resolution as any videos to be processed for 3D pose estimates. To ensure exact time synchronization across all videos, flash an IR LED light at the beginning and end of the recording. Use video editing software to manually sync the videos by marking frames at the onset of the LED and trimming the videos to an equal length.
- Calibration matrices: Pass the videos recorded in the previous two steps through OpenPose21 to generate intrinsic and extrinsic calibration matrices.
NOTE: OpenPose uses the OpenCV library for camera calibration, and further instructions can be found through the documentation on the OpenPose GitHub20, 22.
- Disable autofocus: For computing intrinsic parameters such as lens and perspective distortion, follow the instructions based on the selected OS and webcams to turn off the autofocus.
3. Data collection
- Patient instructions to start recording
- Check device battery and power: The INS device is always ON to provide constant stimulation for the subject. To start recording of neural data, ask the patient to turn on the tablet PC and ensure that the clinician telemetry modules (CTMs) for both left and right INS devices are ON and fully charged.
- CTM placement: Place the CTMs on both sides of the chest. For maximum connectivity and to reduce packet loss, position the CTMs close to the chest implants during recordings. Additional locations to place CTMs are chest pockets of a jacket or using a specialized scarf.
- Activate tablet connection: Once the tablet has booted up, ask the patient to open the DBS application and select Connect, which prompts a Bluetooth connection to the CTMs and subsequently the INS devices14.
- Camera activation: Ask the patient to confirm that video cameras are connected to the PC through their USB cables, and that the cameras have turned ON.
NOTE: If using the authors' custom video recording application, ongoing recordings are clearly indicated on the patient-facing GUI by a large image of a red light that is brightly lit. This changes to a non-lit red light when recordings are OFF. The selected webcams also have a small white indicator light. - Smart watch activation: Ask the patient to turn ON smart watches and smart phones by holding down the Power button. Next, ask them to open the smart watch application to initiate data recording and PD symptom tracking.
- Gesture-based data-alignment and recording scenarios
- Write out any desired tasks for the patient to perform during data recordings prior to starting a data collection.
- As multi-device clock-based synchronization for aligning time stamps can be unreliable, ask the patient to perform a gesture that can be used to synchronize the time stamps from recorded data at the onset of every new recording, even when planning to record during periods of free behavior.
NOTE: The authors designed a simple gesture where the patient tapped both implanted INS devices while keeping their hands within view of the cameras. This tapping creates distinctive patterns in the inertial recordings from the smart watches and the INS accelerometer and is easy to observe in videos.
- Patient instructions to end recording
- Switch the stimulation group back to the patient's preferred clinically assigned group.
- In the patient-facing GUI of the DBS application, enter a symptom report.
- Close the DBS application, which will disconnect the CTMs and conclude INS streaming.
- Close the smart watch recording application and return the CTMs, smartphones and smart watch devices back to their charging ports.
- Data offloading
- Transfer raw videos to cloud storage through the data transfer protocol using an encrypted configuration. Create a cron job on the video recording PC to automatically transfer recorded videos to cloud storage through the data transfer protocol18.
NOTE: Depending on the resolution of videos and the number of hours recorded each day, internet speed must be sufficiently high to enable all videos to be transferred to cloud storage within 24 hours. If data transfer is too slow, disk space could run out, causing additional video recordings scheduled for the following day to fail. - Save INS data to the HIPAA-secure cloud endpoint configured in step 1.3.2. Download INS data from the HIPAA-secure cloud endpoint and deidentify the data. Save the deidentified data to external cloud storage.
NOTE: The open source OpenMind preprocessing code23 was utilized to deidentify data and convert it from json files to a table format. The patient's tablet was configured with a HIPAA-secure cloud endpoint for temporary storage of the raw INS data; however conceivably the same cloud storage site used for long-term storage could also be used for this step provided it is HIPAA compliant, and data are encrypted prior to offloading. - If desired, save a copy of the smart watch data on an external cloud storage so all data streams are accessible in one location.
- Transfer raw videos to cloud storage through the data transfer protocol using an encrypted configuration. Create a cron job on the video recording PC to automatically transfer recorded videos to cloud storage through the data transfer protocol18.
4. System characterization
- Raw data visualization: In the desired coding environment, visualize all raw data streams to ensure data was recorded and transferred appropriately without loss or corruption.
NOTE: The application that was selected to manage smart watch recordings has a browser app that is helpful for visualizing smart watch data24. - Video frame and timestamp lags: Inspect any lags between timestamps generated from different webcams. Analyze lags by recording videos with a programmable LED light placed in-frame of all webcams.
NOTE: Analysis revealed that a video segmenting function25 imported by the custom video recording app was the source of increasing timestamp lags. Recording videos without the segmenting function resulted in between-webcam frame and timestamp lags that did not increase over time (See Supplementary File 1 and Supplementary Figure 1).
5. Post-hoc data pre-processing and alignment
- Pose data
- Install software to calculate joint position estimates from recorded videos.
NOTE: The OpenPose library was selected since it includes hand and face tracking in both 2D and 3D. - The OpenPose library does not automatically handle cases where multiple people are in-frame, so use a post-processing script to ensure that each person's pose estimates are continuous from one frame to the next. OpenPose provides code to easily generate animations, either in 2D or 3D, for visual checks on pose estimation quality.
- Install software to calculate joint position estimates from recorded videos.
- Gesture-based time alignment
- For each INS device (left and right), follow the steps described below using the authors' data-alignment GUI (https://github.com/Weill-Neurohub-OPTiMaL/ManualTimeAlignerGUI).
- Read in data: Access the saved INS and smart watch accelerometry data from cloud storage for the desired data session.
NOTE: An additional time series can be added if desired. Figure 3 shows the pose position of the right middle fingertip in green. - Visualize data streams in the GUI: Use the manual time align GUI to overlay the INS accelerometry, smart watch accelerometry, and pose data.
- Zooming in on alignment artifacts: Zoom in to the time axis and move the viewing window to the chest tapping section of the recording. Shift the aligning time series so that the peak from the chest taps on both the INS and smart watch time series signals overlap as closely as possible.
NOTE: The GUI is designed to facilitate manual alignment of arbitrary time series to a common true time. Figure 3 shows the true time series in blue, while the aligning time series are shown in orange and green. Key guides for GUI alignment are stated on the GitHub ReadMe (https://github.com/Weill-Neurohub-OPTiMaL/ManualTimeAlignerGUI#time-alignment). - Alignment confirmation: Move the GUI window to each of the chest tapping tasks in the recording and confirm the alignment remains consistent throughout the time series. Press the Switch Aligning button and repeat alignments on remaining data streams.
- Warning flags: To indicate whether data was missing, shifted, or other general warnings regarding data quality, set warning flags in the GUI using the D, S and F keys respectively.
- Read in data: Access the saved INS and smart watch accelerometry data from cloud storage for the desired data session.
- For each INS device (left and right), follow the steps described below using the authors' data-alignment GUI (https://github.com/Weill-Neurohub-OPTiMaL/ManualTimeAlignerGUI).
- Zero-normalized cross correlation (ZNCC) time alignment
- Identify the signal most likely to be closest to true time. Usually this is either the one with the highest sample frequency or the fastest internet time refresh.
- Resample the two signals to have the same temporal sampling frequency, and individually z-score both signals. This ensures that the resulting ZNCC scores will be normalized to be between -1 and 1, giving an estimate of the level of similarity between the two signals, useful for catching errors.
- Calculate the cross correlation of the second signal and the first signal at every time lag.
- If phase information of the two signals is not important, take the absolute value of the measured cross correlation curve.
NOTE: If the behavior is significantly a-periodic then the phase information is not necessary, as in this case. - Analyze the ZNCC curve. If there is a single clear peak, with a peak ZNCC score above 0.3 then the time of this peak corresponds to the time lag between the two signals. If there are multiple peaks, no clear peak, or the ZNCC score is low across all time lags, then the two signals need to be manually aligned.
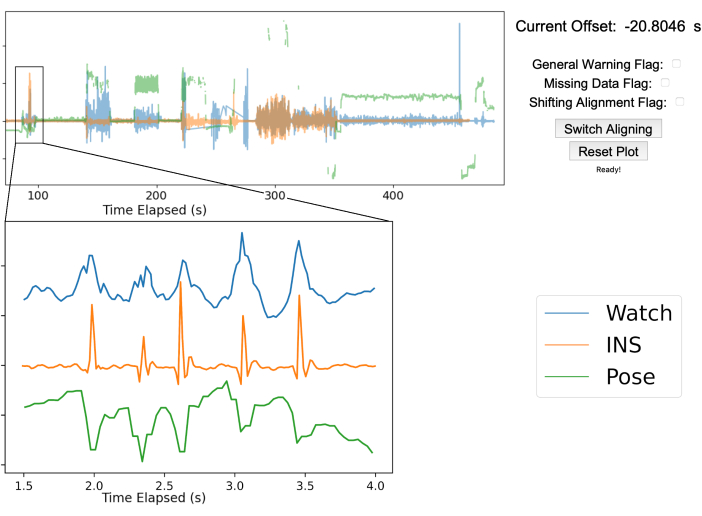
Figure 3: Gesture-based data alignment. The top half of the figure showcases the manual alignment GUI after aligning the three streams of data. The blue line is the smartwatch accelerometry data, the orange line is the accelerometry data from the INS, and the green line is the 2D pose position of the right middle fingertip from a single webcam. The top right shows the offset between the true time from the smart watch and INS as well as various warning flags to mark any issues that arise. In this example, the INS was 20.8 s ahead of the smartwatch. The bottom left graph is zoomed in to show the five chest taps performed by the patient for data alignment. The five peaks are sufficiently clear in each data stream to ensure proper alignment. Please click here to view a larger version of this figure.
Representative Results
Prototype platform design and deployment
We designed a prototype platform and deployed it to the home of a single patient (Figure 1). After the first installation of hardware in the home, the platform can be maintained, and data collected entirely through remote access. The INS devices, smart watches, and cameras have patient-facing applications allowing patients to start and stop recordings. The video collection hardware enables automatic video recordings after an approved schedule has been configured. Patients can easily cancel an ongoing recording by simply pressing a button on the video recording application GUI (Figure 2). All collected data was encrypted and transferred to a cloud storage site for researchers to process and analyze.
Data collection
For the first deployments and data collection cycles, we asked the patient to conduct self-guided clinical tasks. The tasks were taken from the unified Parkinson's disease rating scale (UPDRS)26, namely resting tremor, thumb-to-index finger tapping, hand opening and closing, wrist pronation-supination, sit-to-stand movement and walking, and a typing task. All tasks were repeated three times for each recording day. For each repetition, a different stimulation amplitude was set to expose potential stimulation-related symptoms of PD. Figure 4 shows a schematized example of what a week of data collected with the system might look like.
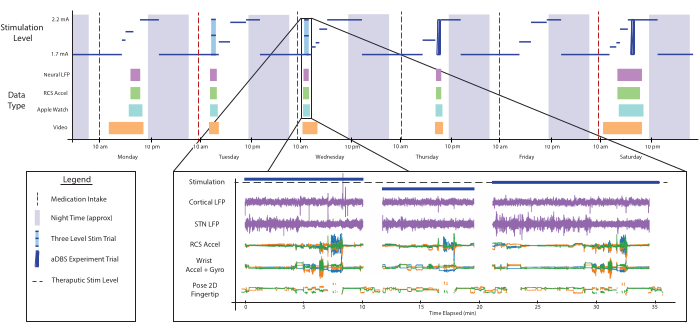
Figure 4: Data availability. A schematized demonstration of what a week of data collected with the system might look like. The top plot shows the stimulation level (blue) over the course of several day/night cycles. Stimulation changes for this patient are dependent on their sleep schedule and the times of medication intake (vertical red lines). At arbitrary times throughout the day, the data collection system can be enabled remotely to collect data for multiple modalities, shown as colored boxes. One example of all the parallel, time-aligned data-streams, just down-selected to the left side of the body, is shown in the bottom plot. During this recording, the patient was asked to perform a series of clinical assessments during low, therapeutic, and high amplitude stimulation conditions. All data shown here corresponds to real data collected but has been compressed across separate experiments for ease of visualization and to show variety. Abbreviations: LFP=local field potential, STN=Subthalamic nucleus, Accel=accelerometer, Gyro=gyroscope, 2D=two-dimensional. Please click here to view a larger version of this figure.
Manual alignment
The manual alignment GUI provides an easy-to-use platform for aligning multiple streams of data. As shown in Figure 3, chest taps provide a clearly identifiable artifact in all data modalities (INS, smart watches, videos) that can be used in manual alignment. The GUI was a useful means of aligning the data, but this could be exchanged for any other alignment tool that researchers would like to use. In some instances, the data streams have a slight drift. A potential future solution to this problem would be to divide the session data into different trials, each with its own chest tap sequence. Each trial can then be individually aligned to minimize the impact of drift.
Zero-normalized cross correlation (ZNCC) time alignment
The method for ZNCC works well in some cases but it has a few critical vulnerabilities. For example, for some movements, the two accelerometer signals can be phase shifted with respect to one another. If a phase aligned and phase shifted movement are both included in the analyzed epochs, then the ZNCC can have either multiple or even no clear peak. The normalization of ZNCC allows these alignments to be automatically identified and discarded as needed. This method works best if both signals are relatively noise free and windowed to an epoch with large, synchronized effects in both traces. The best results were achieved when the patient was asked to perform a series of strong taps with both hands against their chest. In practice however, manual verification of automated alignment was necessary for enough cases that the advantage of using the automated method was negligible.
Data quality
Data loss during automated transfer was negligible since the data transfer protocol process backs up raw copies to ensure that any losses are recoverable. Data loss from connectivity issues occurred regularly, since Bluetooth and radio frequency sometimes have unexpected connection dropouts and are range limited. Short gaps of up to 2 seconds occurred approximately a few times per hour, and longer gaps of up to 2 minutes occurred approximately once every couple of hours. Beyond data loss, significant stimulation artifacts were observed in neural data, the severity of which depended on the recording and the stimulation groups chosen. The largest artifacts occur near the stimulation frequency, well outside ranges of interest. No artifacts were observed in data from smart watches. Videos were recorded at a constant frame rate; however duplicate frames were identified in videos. This yielded an actual frame rate to be a few frames less than the theoretical frame rate as stated by the webcam specifications. More noticeable than the duplicate frames however were freezing periods that were identified in videos at varying intervals depending on the recording day. Freeze periods of approximately 10 frames or less were regularly observed; however longer sections of approximately 2 to 30 seconds long were also observed at irregular periods.
Longitudinal data collection
Table 1 shows the data that the platform prototype has periodically collected over the course of 1.5 years. In that time, hundreds of hours of data were collected, with a total of 293 hours of INS data across both sides of the body, 224 hours of smart watch data for both watches, and 2,037 hours of video data across three webcams. This demonstrates that the platform supports at-home data collection over extended periods of time while offering a rare opportunity to observe longitudinal changes in neural data and corresponding stimulation requirements.
| Data Type | Total Duration (hh:mm:ss) | Total Days | Storage Size |
| Neural | 293:17:33 | 90 | 28.94 GB |
| Watch | 224:06:05 | 89 | 35.67 GB |
| Video | 2037:06:11 | 228 | 146,073.77 GB |
Table 1: Longitudinal overview of collected data. The deployed platform collected data during several experiments over the course of 1.5 years. Approximately 90 days were recorded with neural, video, and smart watch data streams being collected.
2D and 3D pose estimates
Several pose estimation software packages are now available. Pose estimation was tested using OpenPose, an open-source software package21. This was successfully installed following the documentation provided by the organization's GitHub, as well as many other unofficial tutorials found on the web. The processing time for OpenPose varies significantly based on how the OpenPose library and its extensive dependencies are installed, the size of the GPU used, and whether the optional hands and face key points are processed. 2D pose was relatively easy to implement, however 3D pose was notably more difficult and preliminary 3D results yielded inconsistent quality equal to that of 2D pose. The low-quality 3D pose estimation may have been impacted negatively by suboptimal camera calibration, periods where camera autofocus was erroneously turned on, or inherent in the OpenPose software itself. However, synchronized high-quality videos from multiple angles may provide rich inputs for a variety of available pose estimation software packages. It is recommended that a test setup be completed outside of the patient's home, with manual benchmarking of different available pose estimation software packages.
Supplementary Figure 1: Video frame lag analysis. Lags in timestamps generated from the video recording app were detected during system characterization. To investigate the cause of the lags, the frame number and timestamp generated from each camera was determined by recording a red LED light that blinked at random intervals, then the variations in timestamp lags across cameras was computed. (Top) LED intensities (in RGB units) measured on each of the three cameras, demonstrating the time offsets observed between the three cameras (denoted with red arrows). (Bottom) Three plots show the between-camera timestamp lags in number of frames for a series of LED blinks over the entire recording. Each recording was broken up into multiple segments and the frame lag was approximately constant over time. Please click here to download this File.
Supplementary File 1: Video frame and timestamp analysis method. Please click here to download this File.
Discussion
We share the design for an at-home prototype of a multi-modal data collection platform to support future research in neuromodulation research. The design is open-source and modular, such that any piece of hardware can be replaced, and any software component can be updated or changed without the overall platform collapsing. While the methods for collecting and deidentifying neural data are specific to the selected INS, the remaining methods and overall approach to behavioral data collection are agnostic to which implantable device is used. We deployed the platform to the home of an individual with PD and collected data during both experimental and naturalistic periods. During deployments, data collections and post-hoc data processing, several aspects were discovered that were particularly crucial to enabling successful research iterations.
A valuable member of our team was the research coordinator who traveled to the patient's home to install hardware, set up the VPN, perform camera calibration for 3D pose, and walk the patient through how to use each device's patient-facing GUI. Importantly, the research coordinator additionally served as the main point of contact between the patient and the research team. The patient preferred to use their email chat function to quickly send messages back and forth. Having a consistent and accessible point of contact was particularly helpful in two ways:
To establish a familiar communication channel for the patient to request changes to scheduled recordings and to communicate any difficulties in system use. This helped the research coordinator to identify convenient times for the patient to conduct recording experiments. The main difficulty in system use reported was the need to keep track of battery life for several devices.
To allow system troubleshooting to be minimally disruptive to the patient. Most troubleshooting stemmed from network connectivity problems which occurred on average once every couple of weeks. While rebooting devices typically resolved these issues, the watches frequently required multiple restarts, which the patient reported was burdensome.
It is essential to ensure robust remote access to the hardware placed in the patient's home. To accomplish this, having a stable internet connection is crucial. It is also necessary to configure a disk-encrypted machine to automatically unlock whenever a machine reboots. Unsurprisingly, an ethernet cable consistently yielded the fastest and most reliable network connections. Less expected was the need to configure a TPM chip, necessary due to choosing Linux as the OS. If a Windows OS is used, their Bitlocker program will take care of this automatically. Finally, configuring the deployed PC to automatically enable the VPN and re-mount the hard disk drive upon system reboots ensured continued remote access without needing to repeatedly re-visit the patient's home. Incorporating a VPN and a data encryption protocol in the platform design was pivotal for data security and integrity. The VPN allows a network of computers to be connected without needing custom port forwarding to be configured on a patient's private router. The open-source data encryption protocol Rclone program provided with an off-the-shelf data encryption and an easily automatable means of transferring data from patient devices to cloud storage18. The data encryption protocol makes back-up copies of raw data during its data transfer steps to ensure that losses are recoverable. These steps ensured that the patient's private data was kept secure and uncorrupted.
To be able to conduct meaningful data analysis, it is essential that the data collected from multiple devices be time aligned. The clocks on each device are likely not perfectly aligned to a common internet time, even if manufacturers suggest that they are. Additionally, some devices can experience drifting at unpredictable times, changing their offsets relative to the other devices. This creates difficulty in working towards fully automated, real-time adaptive algorithms, and future research will need to carefully consider solutions to this problem. Methods of automatic alignment were explored using normalized cross-correlation. This works reasonably well in many cases; however, time drifts must be minimal, and the data should contain clearly identifiable signals. Because both large drift and periods where data had too much noise or packet loss were encountered, this fully automated method cannot be fully relied upon. To minimize the burden of manually aligning data, we created a simple GUI to allow researchers to visually check data streams with relative ease and rapidity.
The inclusion of video data to the system enables clinicians to measure symptom severity through remote observation, and researchers can obtain event labels. In addition, pose estimates can be calculated from videos as a continuous metric of movement quality such as measuring the speed and smoothness of finger movements over time. However, collecting high resolution videos from multiple cameras requires extensive storage space. For example, collecting 8 hours of 4k videos in the MJPEG format from three cameras takes approximately 0.5 TB of storage space. Recording and storing large quantities of data quickly becomes expensive, creating an economic bottleneck for deploying this system to many patients. In order to make such platforms scale to many patients, future system designers need to reduce the amount of data required for long-term storage. Future systems should consider including real-time pose processing so that videos can be promptly deleted after the pose is processed. Real-time pose could also provide feedback about fine motor skills in closed-loop algorithms, which is outside of the scope of this work. If preserving some video data is needed for clinician review or event labeling, these can be downsampled to a lower resolution before they are saved on the cloud storage.
Finally, to efficiently address the design flaws and implementation errors that invariably arise when building an integrated system, acquiring a replica of the hardware to be deployed for use as a test-rig is extremely valuable. This was true for testing the hardware and software that was selected for collecting videos and processing pose data. The entire process of acquiring videos and pose estimates in both 2D and 3D space was significantly more challenging than anticipated. A test rig allows for troubleshooting and stress-testing a number of important steps prior to deployment, including:
Properly calibrating cameras within the layout constraints of a given room.
Identifying the appropriate video resolution and framerate to support high quality pose estimation. For small rooms or office-like environments, HD video recording is likely sufficient, as the size of individuals on the recorded video is large enough so the pose can be easily computed while requiring significantly less storage space than 4k video.
Discovering bugs in recorded videos, such as freezing frames or time lags between sequentially written video files.
Exposing unexpected software defaults such as re-setting the camera autofocus upon machine reboots, which occludes the benefit of camera calibration.
Trial and error to find compatible versions of the software libraries that must be pre-installed to enable OpenPose to run on a medium-sized GPU.
A particular limitation of this work is deploying the platform in a single pilot study to the home of one individual, preventing us from discovering any cross-participant generalizations from being discovered. However, throughout the design and development process, the system was designed to be scalable and support multiple deployments to support remote studies, and the purpose of this pilot study was to establish the technological feasibility of a sophisticated at-home monitoring platform. Modifying this pilot design based upon some of the discussed crucial findings and deploying the platform to more homes will allow further refinement of the design to support future research in at-home aDBS. In addition, collecting data during additional time points when an individual is not performing pre-determined experiments will offer insights to improve analyses and overall therapy effectiveness. aDBS may provide a preferable method for treating neurological diseases including PD compared to conventional DBS that can have unacceptable side-effects. Bringing this important therapy to many individuals requires automating parameter tuning and analyzing therapy effectiveness outside the clinic across time. The platform provides a novel approach to collect in-home video camera, smart watch, neural recording, and patient-report data during experimental and natural activities from the comfort of the patient's own home. The system will further contribute to creating novel multi-modal datasets to support future discoveries in the treatment of neurological diseases15.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program (DGE-2140004), the Weill Neurohub, and the National Institute of Health (UH3NS100544). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, the Weill Neurohub, or the National Institute of Health. We thank Tianjiao Zhang for his expert consultations on platform design and the incorporation of video data. We especially thank the patient for their participation in this study and for the feedback and advice on network security and platform design.
Materials
| Analysis RCS Data Processing | OpenMind | https://github.com/openmind-consortium/Analysis-rcs-data, open-source | |
| Apple Watches | Apple, Inc | Use 2 watches for each patient, one on each wrist | |
| BRIO ULTRA HD PRO BUSINESS WEBCAM | Logitech | 960-001105 | Used 3 in our platform design |
| DaVinci Resolve video editing software | DaVinci Resolve | used to support camera calibration | |
| Dell XPS PC | Dell | 2T hard disk drive, 500GB SSD | |
| Dropbox | Dropbox | ||
| ffmpeg | N/A | open-source, install to run the Video Recording App | |
| Gooseneck mounts for webcams | N/A | ||
| GPU | Nvidia | A minimum of 8GB GPU memory is recommended to run OpenPose, 12GB is ideal | |
| Java 11 | Oracle | Install to run the Video Recording App | |
| Microsoft Surface tablet | Microsoft | ||
| NoMachine | NoMachine | Ideal when using a Linux OS, open-source | |
| OpenPose | N/A | open-source | |
| Rclone file transfer program | Rclone | Encrypts data and copies or moves data to offsite storage, open-source | |
| StrivePD app | RuneLabs | We installed the app on the Apple Watches to start recordings and upload data to an online portal. | |
| Summit RC+S neuromodulation system | Medtronic | For investigational use only | |
| touchscreen-compatible monitor | N/A | ||
| Video for Linux 2 API | The Linux Kernel | Install if using a Linux OS for video recording | |
| Wasabi | Wasabi | Longterm cloud data storage | |
| WireGuard VPN Protocol | WireGuard | open-source |
References
- World Health Organization. . Parkinson disease. , (2022).
- Herrington, T. M., Cheng, J. J., Eskandar, E. N. Mechanisms of deep brain stimulation. Journal of Neurophysiology. 115 (1), 19-38 (2016).
- Little, S., et al. Adaptive deep brain stimulation for Parkinson’s disease demonstrates reduced speech side effects compared to conventional stimulation in the acute setting. Journal of Neurology, Neurosurgery & Psychiatry. 87 (12), 1388-1389 (2016).
- Mohammed, A., Bayford, R., Demosthenous, A. Toward adaptive deep brain stimulation in Parkinson’s disease: a review. Neurodegenerative Disease Management. 8 (2), 115-136 (2018).
- Gilron, R., et al. Long-term wireless streaming of neural recordings for circuit discovery and adaptive stimulation in individuals with Parkinson’s disease. Nature Biotechnology. 39 (9), 1078-1085 (2021).
- Ray, N. J., et al. Local field potential beta activity in the subthalamic nucleus of patients with Parkinson’s disease is associated with improvements in bradykinesia after dopamine and deep brain stimulation. Experimental Neurology. 213 (1), 108-113 (2008).
- Neumann, W. J., et al. Subthalamic synchronized oscillatory activity correlates with motor impairment in patients with Parkinson’s disease. Movement Disorders: Official Journal of the Movement Disorder Society. 31 (11), 1748-1751 (2016).
- Swann, N. C., et al. Adaptive deep brain stimulation for Parkinson’s disease using motor cortex sensing. J. Neural Eng. 11, (2018).
- Little, S., et al. Adaptive deep brain stimulation in advanced Parkinson disease. Annals of Neurology. 74 (3), 449-457 (2013).
- Connolly, M. J., et al. Multi-objective data-driven optimization for improving deep brain stimulation in Parkinson’s disease. Journal of Neural Engineering. 18 (4), 046046 (2021).
- Stanslaski, S., et al. Creating neural "co-processors" to explore treatments for neurological disorders. 2018 IEEE International Solid – State Circuits Conference – (ISSCC). , 460-462 (2018).
- . Rights (OCR), O. for C. The HIPAA Privacy Rule. HHS.gov. , (2008).
- . Rights (OCR), O. for C. The Security Rule. HHS.gov. , (2009).
- Perrone, R., Gilron, R. . SCBS Patient Facing App (Summit Continuous Bilateral Streaming). , (2021).
- Strandquist, G., et al. . In-Home Video and IMU Kinematics of Self Guided Tasks Correlate with Clinical Bradykinesia Scores. , (2023).
- Donenfeld, J. A. WireGuard: Next Generation Kernel Network Tunnel. Proceedings 2017 Network and Distributed System Security Symposium. , (2017).
- etzion. . Ubuntu 20.04 and TPM2 encrypted system disk – Running Systems. , (2021).
- Craig-Wood, N. . Rclone. , (2023).
- Chen, W., et al. The Role of Large-Scale Data Infrastructure in Developing Next-Generation Deep Brain Stimulation Therapies. Frontiers in Human Neuroscience. 15, 717401 (2021).
- Hidalgo, G. i. n. &. #. 2. 3. 3. ;. s., et al. . CMU-Perceptual-Computing-Lab/openpose. , (2023).
- Cao, Z., Hidalgo, G., Simon, T., Wei, S. E., Sheikh, Y. OpenPose: Realtime Multi-Person 2D Pose Estimation Using Part Affinity Fields. IEEE Transactions on Pattern Analysis and Machine Intelligence. 43 (1), 172-186 (2021).
- Cao, Z. . openpose/calibration_module.md at master · CMU-Perceptual-Computing-Lab/openpose. , (2023).
- Sellers, K. K., et al. Analysis-rcs-data: Open-Source Toolbox for the Ingestion, Time-Alignment, and Visualization of Sense and Stimulation Data From the Medtronic Summit RC+S System. Frontiers in Human Neuroscience. 15, 398 (2021).
- Ranti, C. . Rune Labs Stream API. , (2023).
- . ffmpeg development community. FFmpeg Formats Documentation. , (2023).
- Goetz, C. G., et al. Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): Scale presentation and clinimetric testing results: MDS-UPDRS: Clinimetric Assessment. Movement Disorders. 23 (15), 2129-2170 (2008).

