Single-Molecule Surface-Enhanced Raman Scattering Measurements Enabled by Plasmonic DNA Origami Nanoantennas
Summary
This protocol demonstrates single-molecule surface-enhanced Raman scattering (SERS) measurements using a DNA origami nanoantenna (DONA) combined with colocalized atomic force microscopy (AFM) and Raman measurements.
Abstract
Surface-enhanced Raman scattering (SERS) has the capability to detect single molecules for which high field enhancement is required. Single-molecule (SM) SERS is able to provide molecule-specific spectroscopic information about individual molecules and therefore yields more detailed chemical information than other SM detection techniques. At the same time, there is the potential to unravel information from SM measurements that remain hidden in Raman measurements of bulk material. This protocol outlines the SM SERS measurements using a DNA origami nanoantenna (DONA) in combination with atomic force microscopy (AFM) and Raman spectroscopy. A DNA origami fork structure and two gold nanoparticles are combined to form the DONAs, with a 1.2-2.0 nm gap between them. This allows an up to 1011-fold SERS signal enhancement, enabling measurements of single molecules. The protocol further demonstrates the placement of a single analyte molecule in a SERS hot spot, the process of AFM imaging, and the subsequent overlaying of Raman imaging to measure an analyte in a single DONA.
Introduction
DNA origami is a nanotechnology technique that involves folding DNA strands into specific shapes and patterns. The ability to create structures with precise control at the nanoscale is one of the key advantages of DNA origami1. The ability to manipulate matter at such a small scale has the potential to revolutionize a wide range of fields, including medicine, electronics, and materials science2. For example, DNA origami structures have been used to deliver drugs directly to cancer cells3,4, create nanoscale sensors for detecting diseases5,6, and create intricate patterns on the surfaces of materials6,7. Furthermore, the ability to use DNA origami to create complex nanoscale structures has created new opportunities for studying fundamental biological processes at the nanoscale8.
Surface enhanced Raman scattering (SERS) is a robust analytical technique that detects and identifies molecules at extremely low concentrations9. It is based on the Raman effect, which is a change in the wavelength of scattered light10. SERS requires a plasmonic metal substrate to enhance the Raman signal of molecules adsorbed onto it. This enhancement can be up to 1011 times greater than the signal obtained from the same molecule measured by conventional Raman spectroscopy, making SERS a highly sensitive method for analyzing trace amounts of substances11.
The Raman signal enhancement of nanoparticles is mostly due to electromagnetic enhancement based on the excitation of localized surface plasmon resonance (LSPR)12. In this phenomenon, electrons within the metal nanoparticle oscillate collectively around the surface of the metal nanoparticle during light incidence. This results in the creation of a standing wave of electrons known as a surface plasmon, which can resonate with the incident light. The LSPR greatly enhances the electric field near the particle's surface, and the optical absorption of the particle is maximum at the plasmon resonant frequency. The surface plasmon's energy depends on the shape and size of the metal nanoparticle, as well as the properties of the surrounding medium13. A higher enhancement could be further achieved by plasmonic couplings, such as when two nanoparticles are close to each other at a distance of approximately 2.5 times the diameter length or less14. The proximity causes the LSPR of both nanoparticles to interact with each other, enhancing the electric field in the gap between the particles by several orders of magnitude, far exceeding the enhancement of a single nanoparticle15,16,17. The magnitude of the enhancement is inversely proportional to the distance between the nanoparticles; as the distance decreases, a critical point appears where the enhancement is sufficient to detect single molecules (SMs)18.
DNA origami represents a key technology that can efficiently arrange plasmonic nanoparticles to exploit and optimize the plasmonic coupling19. At the same time, molecules of interest can be placed precisely at the place where optical detection is most efficient. This has been demonstrated with DNA origami-based plasmonic nanoantennas for fluorescence detection20. In contrast to the detection of fluorescent labels, SERS provides the possibility to detect a direct chemical fingerprint of a molecule, making the single-molecule SERS very attractive for sensing as well as monitoring chemical reactions and mechanistic studies. DNA origami can also be used as a mask to fabricate plasmonic nanostructures with well-defined shapes21, although the possibility to position target molecules precisely into the hot spot is then lost.
Using colocalized atomic force microscopy (AFM) and Raman measurements, we can obtain the Raman spectra from a single DNA origami nanoantenna (DONA) and potentially a single molecule if placed at the position of highest signal enhancement. The DONA comprises a DNA origami fork and two precisely positioned nanoparticles, fully coated with DNA complementary to extended staple strands attached to the DNA origami. Upon DNA hybridization, the nanoparticles are bound to the DNA origami fork with a 1.2-2.0 nm gap between the nanoparticle surfaces22. Such assembly creates a hot spot in between the nanoparticles with a signal enhancement of up to 1011-fold, as calculated from finite difference time domain (FDTD) simulations22, thus permitting SM SERS measurements. However, the volume of the highest enhancement is small (in the 1-10 nm3 range), and consequently the target molecules need to be positioned precisely into this hot spot. The DNA origami fork allows the positioning of a single molecule between the two nanoparticles using a DNA fork bridge and suitable coupling chemistry. Nevertheless, observing such SMs in Raman spectra is very challenging22. Alternatively, the nanoparticles can be fully coated with the target molecule, such as a TAMRA dye, to allow for single DONA measurements with a higher SERS intensity21. In this case, the TAMRA is covalently attached to the DNA coating strand (Figure 1).
Protocol
1. DNA origami fork assembly
- Self-assemble the DNA origami structure in one pot. First, add 2.5 nM circular scaffold strand M13mp18 (7,249 nucleotides, see Table of Materials) and 100 nM of 201 short oligonucleotides (Table 1) in 1x TAE (tris[hydroxymethyl]aminomethane [tris], acetic acid, ethylenediamine acetic acid [EDTA]) supplemented with 15 mM MgCl2 buffer. Then, adjust the total volume to 100 µL using ultrapure water.
- Anneal the solution subsequently by a temperature gradient in a thermocycler, first by rapidly heating to 80 °C, then by cooling down from 80 °C to 20 °C at 1 °C/12 min, then from 20 °C to 16 °C at 1 °C/6 min, followed by rapidly cooling down from 16 °C to 8 °C.
- Use 100 kDa molecular weight cut-off (MWCO) Amicon filters (see Table of Materials) to purify the mixture from excess staples.
- Add 100 µL of the DNA origami solution to the Amicon filters, as well as 400 µL of ultrapure water, then centrifuge at 6,000 x g for 8 min at room temperature (RT).
- Discard the filtrate by removing the filter and flipping the tube to remove the wash-out in the sink, then add 400 µL of ultrapure water to the filter and centrifuge again. Repeat this step one more time.
- To collect the purified nanostructure solution, flip the filter upside down in a new tube and centrifuge (1,000 x g, 2 min, RT) following the manufacturer's instructions (see Table of Materials). This solution can be stored in a fridge at 8 °C for up to 2 weeks.
NOTE: Use 1x TAE buffer or 1x TAE supplemented with 15 mM MgCl2 buffer instead of ultrapure water for the Amicon filter purification step, since water decreases the stability and lowers the concentration of the DNA origami forks obtained. For a single dye molecule measurement, the DNA staple strand mixture is replaced with a modified strand mixture containing a TAMRA dye on the 5' end. This modified strand is in the middle of the nanofork bridge (Table 2).
2. Gold nanoparticle (AuNP) coating
NOTE: A modified version of the Liu et al.23 protocol was used to coat the AuNPs, and the coating process involved freezing the AuNP-DNA solution.
- Centrifuge (3,500 x g, 5 min, RT) 400 µL of 60 nm diameter AuNP solution (commercially obtained; see Table of Materials), and using a pipette, remove the supernatant. Then, resolubilize the pellet in 25 µL of ultrapure water. The final concentration of the AuNPs is ~0.3 nM.
- Add 1 µL of 100 mM tris-(2-carboxyethyl)phosphine (TCEP) solution to 4 µL of thiol-modified DNA (100 µM as supplied by the manufacturer; see Table of Materials) and incubate for 10 min at RT.
- Following incubation, add the 5 µL mixture to the concentrated AuNP solution (step 2.1), vortex for 5 s, and freeze for at least 2 h at -20 °C.
- After thawing at RT, centrifuge (3,500 x g, 5 min, RT) the mixture to remove the excessively added coating DNA. Using a pipette, remove the supernatant, and resolublize the pellet in 10 µL of water.
NOTE: There are two coating strands for each of the AuNPs on the nanofork (Table 2). The steps are the same except for the sequence of the DNA coating strand. Once the particles are coated, they are very stable; they can stay as they are for months in the fridge and can even be frozen. For fully coated dye AuNPs, the coating DNA strands have an internal TAMRA dye.
3. DONA assembly
- Add the coated AuNP solution to the nanofork solution, with a concentration molar ratio of 1.5:1.
- Add MgCl2 to a final concentration of 4 mM using a 50 mM MgCl2 stock solution. Adjust the final volume to 20 µL using ultrapure water.
- Hybridize the DONAs using a temperature gradient in a thermocycler. First, rapidly heat to 40 °C, then cool from 40 °C to 20 °C at 1 °C/10 min, and then rapidly cool from 20 °C to 8 °C.
4. Gel electrophoresis
NOTE: The unbound nanoparticles in the DONA solution are removed by agarose gel electrophoresis.
- Prepare 1% agarose gel. For this, dissolve 0.8 g of agarose in 80 mL of 1x TAE supplemented with 5 mM MgCl2.
- Add 2.25 µL of loading buffer (30% glycerol, 13 mM MgCl2; see Table of Materials) to 18 µL of DONA solution to reach a final concentration of 5 mM MgCl2 from 4 mM in step 3.2. The loading buffer also ensures that the sample stays inside the pocket of the gel.
- Run the gel for 60 min at 70 V in an ice water bath. The running buffer is 1x TAE supplemented with 5 mM MgCl2.
- Cut out the band of interest and place it on a paraffin plastic film-wrapped microscopy slide, and then squeeze out the solution using a second paraffin plastic film-wrapped microscopy slide. Using a pipette, collect the squeezed liquid into a 500 µL tube.
5. Colocalization of AFM and Raman measurements
- Plasma treat a silicon chip (see Table of Materials) for 10 min, then incubate 10 µL of the purified DONA solution plus 10 µL of 100 mM MgCl2 on the chip for 3 h. Then, wash the chip two times with a 1:1 mixture (by volume) of ethanol and water and blow-dry with compressed air. Then, tape the chip on a magnetic disc and insert it into the instrument for imaging.
NOTE: Before starting the measurements, the position of the Raman excitation laser is adjusted to be on top of the AFM probe tip. In the present work a HORIBA LabRAM HR Evolution Raman microscope coupled to a HORIBA Omegascope AFM is used. The instrument is controlled using the software LabSpec and AIST. - For the AFM measurement, use tapping mode (AC mode) with ACCESS-NC-A (resonance frequency: 300 kHz; spring constant: 45 N/m) tips24. The AC mode automatically controls all the parameters except for the scan rate, which is set to 1 Hz.
NOTE: Following AFM imaging, the AFM tip is withdrawn so that it is not in the path of the Raman laser. This is done using a macro function, "Probe away", programmed in the system. In this way, for any chosen point in the AFM image, the laser will be in the same position with no offset compared to AFM. - Choose the desired laser wavelength, power, and accumulation time for the Raman measurements, as all these parameters depend on the measured sample.
- Use the mentioned parameters for a fully coated TAMRA AuNP measurement: 633 nm laser, 100 µW power, and 1 s integration time.
- Use the mentioned parameters for a single TAMRA measurement: 633 nm laser, 400 µW power, and 4 s integration time.
- After adjusting the parameters, place the cursor on top of the desired DONA in the AFM image; the "move cursor" function in the software does this. Then, start the Raman measurement.
NOTE: There are different modes for acquiring the spectra: single point measurement-one-dimension time mapping-where a single point is measured over time25; and two-dimension XY area mapping, where a motorized stage is moved under the laser to acquire spectra from an array of points in an XY grid26.
Representative Results
Following the protocol, one should ensure that the DNA origami fork is correctly assembled; the preferred method for examining the structure of the forks is AFM imaging. Most forks are expected to be solid, with no broken arms. On the other hand, the bridge between the arms is difficult to image because of its small diameter and high flexibility; it also requires a very sharp AFM tip (Figure 2).
The solution color change at each step of the AuNP coating process indicates that everything works correctly. The color starts deep red with just the AuNPs, but as soon as the DNA is added, it changes to dark-purplish red. Freezing changes the color to purple and, after thawing, returns it to deep red (Figure 3A). Figure 3B depicts the absorbance spectra of bare AuNPs and DNA-coated AuNPs.
After that, in the DONA assembly steps, the color stays deep red throughout the whole process. During the agarose gel purification, a dimer band appears above the free AuNP band, the fastest-running band in the sample. This dimer band corresponds to the DONAs and is subsequently cut out and squeezed to extract the sample (Figure 4).
Finally, for colocalization measurements, AFM imaging of the sample is performed in search of DONAs (Figure 5). Following that, Raman spectra from single DONAs are collected and compared to ensure that the spectra obtained are from TAMRA molecules (Figure 6).
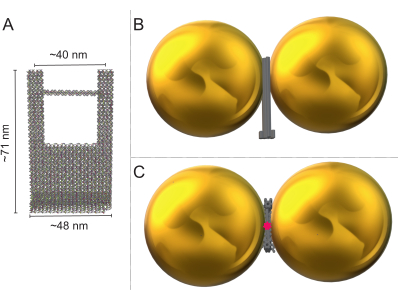
Figure 1: Schematic representation of the DNA origami fork and the fully assembled DONA. (A) Dimensions of the DNA origami fork with a bridge 90 nucleotides long. (B) Schematic side-view of an assembled DONA, with two AuNPs and the DNA origami fork in between. (C) Schematic top-view of the assembled DONA showing the placement position of the SM in the middle of the bridge. Please click here to view a larger version of this figure.
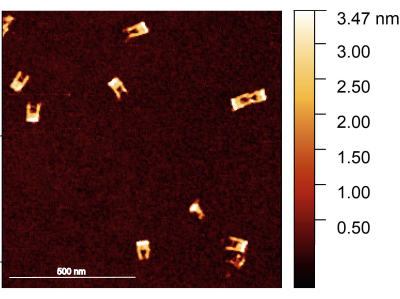
Figure 2: AFM image of the DNA origami forks after assembly. The forks are well formed, with the bridge visible in some of the forks. The forks have a height between 1.5-2 nm. Scale bar = 500 nm. Please click here to view a larger version of this figure.
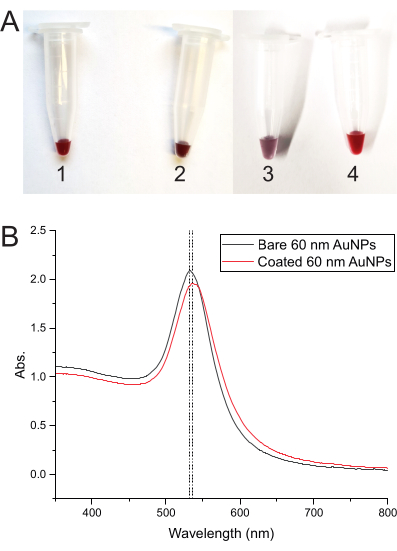
Figure 3: Color development and absorbance spectra of AuNPs. (A) Tubes showing the color of the AuNP solution in different steps. (1) Deep red color of the bare AuNP solution. (2) Dark-purplish red after the addition of the coating DNA to the AuNPs. (3) Purple color after freezing the coating DNA-AuNP mixture. (4) The color returns to deep red after thawing of the mixture. (B) Absorbance spectra of the 60 nm AuNPs, showing the shift in absorbance peak from bare AuNPs (tube 1) to DNA-coated AuNPs (tube 4). Please click here to view a larger version of this figure.
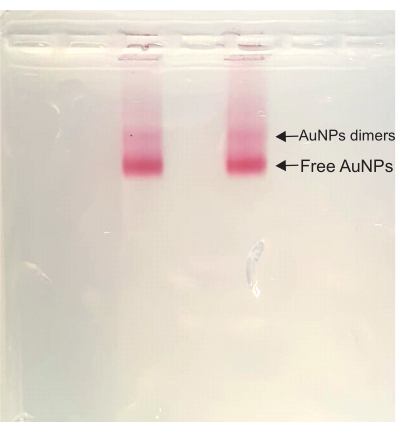
Figure 4: Agarose gel of the DONA solution. Both lanes have the same sample, and both the dimer band (DONA) and the free AuNP band are clearly visible. Please click here to view a larger version of this figure.
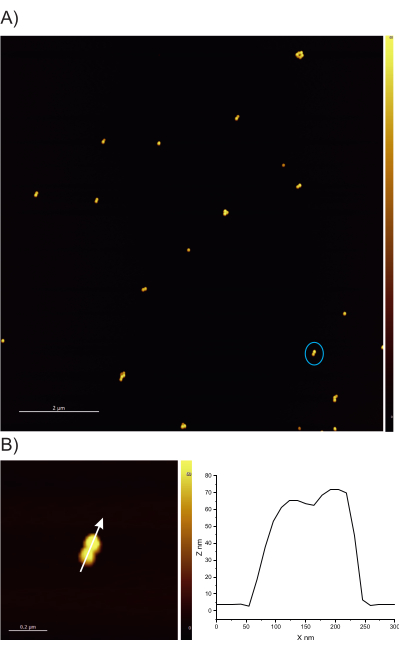
Figure 5: AFM image of DONAs. (A) The image shows multiple DONA structures; this AFM image is used for the colocalization measurements. (B) Zoomed-in image of the circled DONA and vertical cross-section of the DONA. Please click here to view a larger version of this figure.
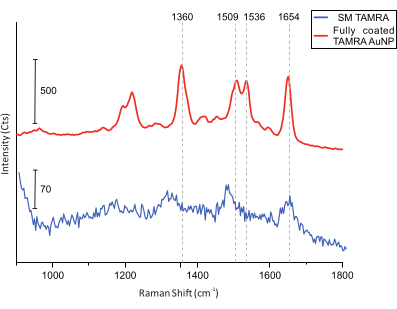
Figure 6: SERS spectra of DONAs equipped with fully coated TAMRA AuNPs and with a single TAMRA molecule. The vertical gridlines indicate the main TAMRA peaks. The main TAMRA peaks are visible in the fully coated TAMRA AuNP. Although the signal-to-noise ratio for the SM TAMRA spectra is lower, the main peaks are identifiable: 1,360 cm-1: C-C stretching; 1,509 cm-1: C=C stretching; 1,536 cm-1: C=C stretching; 1,654 cm-1: C=O stretching. Please click here to view a larger version of this figure.
Table 1: List of DNA origami fork staples. Please click here to download this Table.
Table 2: List of modified DNA strands. Please click here to download this Table.
Discussion
SM SERS is a powerful tool that allows researchers to study the behavior and interactions of individual molecules within a sample1. Such a technique allows the analysis of systems at an unprecedented level of sensitivity, providing new insights into the fundamental behavior of single molecules and the distribution of chemical or physical properties over an ensemble of molecules, and helps to identify relevant intermediates in chemical processes. However, placing a single molecule in the hot spot while also making sure that the hot spot has enough surface enhancement can be quite challenging27. The described DONAs in this protocol can precisely place a single molecule in the hot spot between two gold nanoparticles while ensuring a 1011-fold surface enhancement is reached.
The gap between the nanoparticles is critical for the DONA assembly to reach the required surface enhancement for SM SERS studies. The DONAs are optimized for spherical nanoparticles with sizes between 60 and 80 nm. Furthermore, the quality of the nanoparticles can significantly impact the hybridization of the nanoparticles with the DNA origami forks; when the nanoparticles that are used in the coating step are older than 6 months, the efficiency of hybridization begins to decline.
Another critical aspect of the protocol is that the steps that require an exact ratio between components must be followed precisely, or the DONAs will not be formed correctly. The DNA origami fork is extremely sensitive to the temperature ramping protocol, with changes affecting the structure's integrity or preventing forks from forming.
Amorphous carbon generation is a significant issue during SERS measurements because its peaks are typically in the same range as the fingerprint area for many molecules (1,200-1,700 cm-1). While the formation is not yet fully understood, it is usually associated with high laser power or long integration times28. As a precaution, the lowest laser power and the shortest integration time possible should be used. However, this is not easily accomplished because a balance must be achieved between obtaining the desired SERS signal and avoiding the generation of amorphous carbon.
The DONAs are very versatile as an SM system regarding the different types and shapes of nanoparticles that could be used, such as silver spheres, gold flowers, or stars. Additionally, the molecule under investigation can be easily replaced by changing only the DNA strand in the middle of the bridge, with no changes to the procedure. Switching to proteins like cytochrome C could be done by having a pyridine-modified DNA capture strand in the bridge, which would bind the cytochrome C and ensure that it is in the hot spot for SM SERS measurements22. This also translates to flexibly choosing the laser for irradiation, potentially using a laser that gives maximum enhancement.
In summary, this method is reliable for assembling DONA structures and using them for single-molecule surface-enhanced Raman spectroscopy measurements.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research was supported by the European Research Council (ERC; consolidator Grant No. 772752).
Materials
| 100 kDa MWCO Amicon filters, 0.5 mL | Merck | UFC5100BK | |
| 10x TAE buffer (0.4 M Tris, 0.2 M acetic acid, 0.01 M EDTA) | SIGMA Aldrich | T9650 | |
| 60 nm goldspheres, bare (citrate) | NanoComposix | AUCN60 | |
| ACCESS-NC-A AFM probes | SCHAEFER-TEC | ||
| Agarose powder | SIGMA Aldrich | 9012-36-6 | |
| AuNP DNA coating strands | IDT | ||
| AuNP DNA coating strands (TAMRA) | SIGMA Aldrich | ||
| Glycerol | SIGMA Aldrich | 56-81-5 | |
| Heraeus Fresco 17 centrifuge | Thermo Fisher Scientific | ||
| HORIBA OmegaScope with a LabRAM HR evolution | HORIBA | ||
| Magnesium chloride | SIGMA Aldrich | 7786-30-3 | |
| Nanofork DNA bridge strand (TAMRA) | Metabion | ||
| Nanofork DNA staple strands | SIGMA Aldrich | ||
| ParafilmM | Carl Roth | CNP8.1 | |
| Primus 25 Thermocycler | Peqlab/VWR | ||
| Silicon wafer | Siegert wafer | BW14076 | |
| Single-stranded scaffold DNA, type p7249 (M13mp18) | Tilibit nanosystems | ||
| TCEP solution | SIGMA Aldrich | 51805-45-9 |
References
- Heck, C., Kanehira, Y., Kneipp, J., Bald, I. Placement of single proteins within the SERS hot spots of self-assembled silver nanolenses. Angewandte Chemie. 57 (25), 7444-7447 (2018).
- Dey, S., et al. DNA origami. Nature Reviews Methods Primers. 1 (1), 13 (2021).
- Tasciotti, E. Smart cancer therapy with DNA origami. Nature Biotechnology. 36 (3), 234-235 (2018).
- Ijäs, H., et al. Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Research. 49 (6), 3048-3062 (2021).
- Wang, S., et al. DNA origami-enabled biosensors. Sensors. 20 (23), 6899 (2020).
- Kabusure, K. M., et al. Optical characterization of DNA origami-shaped silver nanoparticles created through biotemplated lithography. Nanoscale. 14 (27), 9648-9654 (2022).
- Kershner, R. J., et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nature Nanotechnology. 4 (9), 557-561 (2009).
- Dutta, A., et al. Molecular states and spin crossover of hemin studied by DNA origami enabled single-molecule surface-enhanced Raman scattering. Nanoscale. 14 (44), 16467-16478 (2022).
- Langer, J., et al. Present and future of surface-enhanced Raman scattering. ACS Nano. 14 (1), 28-117 (2020).
- Jones, R. R., Hooper, D. C., Zhang, L., Wolverson, D., Valev, V. K. Raman techniques: fundamentals and frontiers. Nanoscale Research Letters. 14 (1), 231 (2019).
- Blackie, E. J., Le Ru, E. C., Etchegoin, P. G. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. Journal of the American Chemical Society. 131 (40), 14466-14472 (2009).
- Sepúlveda, B., Angelomé, P. C., Lechuga, L. M., Liz-Marzán, L. M. LSPR-based nanobiosensors. Nano Today. 4 (3), 244-251 (2009).
- Kelly, K. L., Coronado, E., Zhao, L. L., Schatz, G. C. The optical properties of metal nanoparticles: The influence of size, shape, and dielectric environment. The Journal of Physical Chemistry B. 107 (3), 668-677 (2003).
- Funston, A. M., Novo, C., Davis, T. J., Mulvaney, P. Plasmon coupling of gold nanorods at short distances and in different geometries. Nano Letters. 9 (4), 1651-1658 (2009).
- Niu, R., et al. DNA origami-based nanoprinting for the assembly of plasmonic nanostructures with single-molecule surface-enhanced Raman scattering. Angewandte Chemie. 60 (21), 11695-11701 (2021).
- Fang, W., et al. Quantizing single-molecule surface-enhanced Raman scattering with DNA origami metamolecules. Science Advances. 5 (9), (2019).
- Zhan, P., et al. DNA origami directed assembly of gold bowtie nanoantennas for single-molecule surface-enhanced Raman scattering. Angewandte Chemie. 57 (11), 2846-2850 (2018).
- Choi, H. K., et al. Single-molecule surface-enhanced Raman scattering as a probe of single-molecule surface reactions: promises and current challenges. Accounts of Chemical Research. 52 (11), 3008-3017 (2019).
- Liu, N., Liedl, T. DNA-assembled advanced plasmonic architectures. Chemical Reviews. 118 (6), 3032-3053 (2018).
- Acuna, G. P., et al. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science. 338 (6106), 506-510 (2012).
- Shen, B., Kostiainen, M. A., Linko, V. DNA origami nanophotonics and plasmonics at interfaces. Langmuir. 34 (49), 14911-14920 (2018).
- Tapio, K., et al. A versatile DNA origami-based plasmonic nanoantenna for label-free single-molecule surface-enhanced Raman spectroscopy. ACS Nano. 15 (4), 7065-7077 (2021).
- Liu, B., Liu, J. Freezing-driven DNA adsorption on gold nanoparticles: tolerating extremely low salt concentration but requiring high DNA concentration. Langmuir. 35 (19), 6476-6482 (2019).
- Aliano, A., et al. AFM, tapping mode. Encyclopedia of Nanotechnology. , 99 (2012).
- Recording Raman spectral images and profiles. Horiba Scientific Available from: https://www.horiba.com/int/scientific/technologies/raman-imaging-and-spcetroscopy/recording-spectral-images-and=profiles/ (2023)
- Raman Images Explained. Renishaw Available from: https://www.renishaw.com/en/raman-images-explained-25810 (2023)
- Kogikoski, S., Tapio, K., von Zander, R. E., Saalfrank, P., Bald, I. Raman enhancement of nanoparticle dimers self-assembled using DNA origami nanotriangles. Molecules. 26 (6), 1684 (2021).
- Heck, C., Kanehira, Y., Kneipp, J., Bald, I. Amorphous carbon generation as a photocatalytic reaction on DNA-assembled gold and silver nanostructures. Molecules. 24 (12), 2324 (2019).

