Generation and Manipulation of Rat Intestinal Organoids
Summary
Here, we present a protocol to generate rat intestinal organoids and use them in several downstream applications. Rats are often a preferred preclinical model, and the robust intestinal organoid system fills the need for an in vitro system to accompany in vivo studies.
Abstract
When using organoids to assess physiology and cell fate decisions, it is important to use a model that closely recapitulates in vivo contexts. Accordingly, patient-derived organoids are used for disease modeling, drug discovery, and personalized treatment screening. Mouse intestinal organoids are commonly utilized to understand aspects of both intestinal function/physiology and stem cell dynamics/fate decisions. However, in many disease contexts, rats are often preferred over mice as a model due to their greater physiological similarity to humans in terms of disease pathophysiology. The rat model has been limited by a lack of genetic tools available in vivo, and rat intestinal organoids have proven fragile and difficult to culture long-term. Here, we build upon previously published protocols to robustly generate rat intestinal organoids from the duodenum and jejunum. We provide an overview of several downstream applications utilizing rat intestinal organoids, including functional swelling assays, whole mount staining, the generation of 2D enteroid monolayers, and lentiviral transduction. The rat organoid model provides a practical solution to the need of the field for an in vitro model which retains physiological relevance to humans, can be quickly genetically manipulated, and is easily obtained without the barriers involved in procuring human intestinal organoids.
Introduction
The human small intestinal epithelial architecture and cellular composition are complex, reflecting their physiological functions. The primary role of the small intestine is to absorb nutrients from food passing through its lumen1. To maximize this function, the intestinal surface is organized into finger-like protrusions called villi, which increase the absorptive surface area, and cup-like invaginations called crypts, which house and insulate the stem cells. Within the epithelium, various specialized absorptive and secretory cell types are generated to perform distinct functions1. Because of this complexity, it has been difficult to model tissues like the intestine in high passage-transformed immortalized cell lines. However, the study of stem cells, especially adult stem cells and their differentiation mechanisms, has allowed for the development of 3D intestinal organoid cultures. The use of organoid models has transformed the field, in part due to their recapitulation of some architectural components and cell type heterogeneity found in the intact intestine. Intestinal organoids can be cultured long-term in vitro due to the maintenance of the active stem cell population2.
Intestinal organoids have rapidly become an adaptable model to study stem cell biology, cell physiology, genetic disease, and nutrition3,4, as well as a tool to develop novel drug delivery methods5. In addition, patient-derived organoids are being utilized for disease modeling, drug discovery, and personalized treatment screening, among others6,7,8,9. However, human intestinal organoids still present challenges. Tissue availability, requirements for Institutional Review Board approval, and ethical issues limit the widespread use of human samples. Additionally, human intestinal organoids generated from intestinal crypts require two distinct culture conditions for the maintenance of undifferentiated stem cells or to induce the differentiation of mature cell types10. This contrasts with in vivo, where stem cells and mature differentiated cell types are simultaneously present and continuously generated/maintained1. On the other hand, mouse intestinal organoids, which are grown in a less complex cocktail of growth factors, do not require this switch in media composition and can maintain stem cells and differentiated cells in the same media context2,11. However, key differences in mouse intestine when compared to humans can make mouse organoids a suboptimal model in many instances. Overall, many intestinal organoids from larger mammals, including horses, pigs, sheep, cows, dogs, and cats, have been successfully generated in culture conditions more closely aligned with mouse intestinal organoids than the culture conditions of human intestinal organoids12. The differences in growth factor conditions between mouse and human organoids likely reflect differences in stem cell niche composition and differing requirements for stem cell survival, proliferation, and maintenance. Therefore, there is a need for an easily accessible model organoid system that 1) closely resembles human intestinal cell composition, 2) contains stem cells with growth factor requirements like those of human intestinal organoids, and 3) is capable of continuously maintaining undifferentiated and differentiated compartments. Ideally, the system would be from a commonly used preclinical animal model, such that in vivo and in vitro experiments can be correlated and used in tandem.
Rats are a commonly used preclinical model for intestinal physiology and pharmacology studies due to their very similar intestinal physiology and biochemistry to humans13, particularly with regards to intestinal permeability14. Their relatively larger size compared to mice makes them more amenable to surgical procedures. While large animal models, including pigs, are sometimes used, rats are a more affordable model, require less space for husbandry, and have readily commercially available standard strains15. A drawback of using rat models is that the genetic toolkit for in vivo studies is not well developed compared to mice, and the generation of novel rat lines, including knockouts, knock-ins, and transgenics, is often cost-prohibitive. The development and optimization of a robust rat intestinal organoid model would allow for genetic manipulation, pharmacological treatments, and higher throughput studies in an accessible model that retains key physiological relevance to humans. However, the advantages of one rodent organoid model versus another is highly dependent on the particular process or gene being studied; certain genes found in humans may be pseudogenes in mice but not rats16,17. Additionally, species-specific cell subtypes are increasingly being unveiled by single-cell RNAseq18,19,20. Finally, rat and mouse intestinal disease models often display considerable variations in phenotypes21,22 such that the model that more closely recapitulates the symptoms and disease process seen in humans must be selected for downstream work. The generation of a rat intestinal organoid model provides additional flexibility and choice for researchers in selecting a model system most appropriate for their circumstances. Here, existing protocols23,24 are expanded upon for the generation of rat intestinal organoids and a protocol is outlined for the generation and maintenance of rat intestinal organoids from the duodenum or jejunum. In addition, several downstream applications, including lentiviral infection, whole mount staining, and forskolin swelling assays, are described.
Protocol
NOTE: All cell culture should be handled using proper aseptic technique in a tissue culture hood. All animal work in this study was approved by Yale's Institutional Animal Care and Use Committee (IACUC).
1. Preparation of cell culture reagents
- Prepare R-spondin 1 conditioned media following the manufacturer's instructions. Prepare Wnt3a conditioned media following the manufacturer's instructions. Prepare AdDMEM+ as described in Table 1.
- Resuspend gastrin in sterile dH2O to prepare a 100 µM stock. Aliquot and store at -80 °C. Resuspend N-acetylcysteine in sterile water to prepare a 100 mM stock. Aliquot and store at -20 °C for up to 1 month.
- Resuspend recombinant human Noggin in phosphate buffered saline (PBS) + 0.1% bovine serum albumin (BSA) to prepare a 250 µg/mL stock. Aliquot and store at -80 °C. Resuspend recombinant mouse epidermal growth factor (EGF) in PBS + 0.1% BSA to prepare a 100 µg/mL stock. Aliquot and store at -80 °C.
- Dilute recombinant human IGF-1 in PBS + 0.1% BSA to prepare a 100 µg/mL stock. Aliquot and store at -80 °C. Resuspend recombinant human FGF-2 in 5 mM Tris, pH 7.6, to prepare a 100 µg/mL stock. Aliquot and store at -80 °C.
NOTE: For all the growth factors, use an intermediate dilution in PBS + 0.1% BSA to 100x the final concentration in the culture media. Store at -20 °C.
2. Establishment of rat small intestine organoids
NOTE: This protocol was modified from two previously published protocols for rat intestinal organoids23,24.
- Prepare rat intestinal organoid media (rIOM) as per Table 2 and set a water bath at 37 °C. This complete medium is stable for 5 days at 4 °C.
- Prepare 10 mL of 3 mM ethylenediaminetetraacetic acid (EDTA) in PBS in a 15 mL conical tube and keep on ice. Thaw 250 µL of extracellular matrix extract (EME) on ice.
- Fast a rat overnight with access to water ad libitum. Euthanize the rat according to the IACUC-approved protocol. In this protocol, adult male Sprague Dawley rats (weighing ~200 g) were euthanized via CO2 inhalation (Table of Materials). Cervical dislocation was used as a secondary method of euthanasia.
- Use sterile autoclaved forceps and dissection scissors for dissection. Place the euthanized rat onto the dissection surface ventral side up. Pinch the skin layer with forceps; the following cuts should be done at the surface level, so they are only cutting through this skin layer and not deep enough to damage internal organs.
- To open the abdominal cavity, using large, sharp dissection scissors, cut through the skin layer in a large, longitudinal, surface-level cut in the center of the abdomen. Next, stemming from that cut, make two shorter horizontal cuts, one on each side. Use forceps to peel the skin away to expose the abdominal cavity. Cut through the peritoneal membrane to fully expose the internal organs in the abdominal cavity with ready access to the intestine.
- Using scissors and forceps, locate the stomach and identify the duodenum approximately 2-3 cm distal to it, which appears as a yellowish segment. The proximal jejunum is located approximately 4-5 cm distal to the ligament of Treitz, which serves as a landmark between the duodenum and jejunum.
- Place the isolated intestinal fragment into a 10 cm Petri dish. Clean the desired intestinal segment of mesentery as much as possible. Flush with 10 mL of ice-cold PBS until cleared of luminal contents. On a paper towel, cut the intestinal segment into ~2 cm long pieces. Open each intestinal piece longitudinally to expose the epithelium.
- Using a glass microscope slide, scrape the exposed luminal surface to remove villi. Place intestinal pieces into the prepared EDTA solution on ice. Rotate at 4 °C for 30 min on a tube revolver set to 10 rpm.
- At the dissecting microscope, pour the contents of the conical tube into a 10 cm Petri dish. Add in an additional ~5 mL of ice-cold PBS.
- Using fine forceps, hold an intestinal segment and shake vigorously. It will be possible to see the epithelium release into the PBS. At first, the PBS will primarily contain villi.
- Continue to shake. Periodically discard the PBS containing villi and add 10 mL of fresh ice-cold PBS to the intestinal fragments. Continue to shake the fragments and repeat this washing step until villi are no longer released into the PBS, but instead the PBS primarily contains crypts. Do not exceed 15 min for crypt isolation, so that cell viability is not compromised.
- Discard the remaining intestinal fragments. Enrich the remaining PBS in the Petri dish for intestinal crypts. In a tissue culture hood, collect the PBS-containing crypts and filter through a 70 µm cell strainer (Table of Materials).
- Centrifuge at 250 x g for 5 min. Remove the supernatant and resuspend the pellet in 5 mL of AdDMEM+. Centrifuge again at 250 x g for 5 min.
NOTE: It is recommended to use a centrifuge with a swing bucket rotor. - Remove the supernatant, leaving ~50 µL of media with the pellet. Resuspend the pellet in the remaining media and add to the aliquot of the EME on ice. Gently pipette up and down to suspend the crypts evenly throughout the EME. Avoid introducing bubbles.
- Plate 50 µL of EME domes into a 35 mm tissue culture dish and incubate for 20 min in a 37 °C, 5% CO2 tissue culture incubator.
- Add 2 mL of rIOM (Table 2) containing 10 µM Y27632 and 10 µM CHIR99021 (Table 3). After passaging, Y27632 and CHIR99021 can be discontinued.
- Change the rIOM every 2-3 days. Passage as needed, usually between 3-7 days, depending on the initial number of organoids, size, and growth rate.
3. Passaging rat intestinal organoids
- Thaw a 250 µL aliquot of EME on ice and prewarm the rIOM to 37 ˚C.
- Aspirate the medium from a 35 mm plate containing organoids and add 1 mL of dissociation reagent to immediately release the organoids from the EME domes.
- Immediately transfer dissociation reagent with fragmented organoids to a 15 mL conical tube. Wash the culture plate with 2 mL of AdDMEM+ and add to the 15 mL conical tube containing fragmented organoids.
- With a glass Pasteur pipette, gently pipette up and down 15-20 times to fragment the organoids. Centrifuge at 350 x g for 2 min.
- Add ~50 µL of solution from the bottom of the conical tube into the EME. Gently pipette up and down to mix. Avoid making bubbles.
- Plate 50 µL of EME domes into a 35 mm dish and incubate for 20 min in a 37 °C, 5% CO2 tissue culture incubator.
- Add 2 mL of rIOM with 10 µM Y27632 and 10 µM CHIR99021. Change the growth media every 2-3 days. When changing media, Y27632 and CHIR99021 can be omitted.
4. Cryopreservation and thawing of rat intestinal organoids
- Cryopreserving rat intestinal organoids
NOTE: The cryopreservation protocol was modified from a previous protocol for human and mouse organoids25. Prior to freezing, the organoids must have at least two passages in primary culture. It is advisable to grow organoids to the point of large spheroids or slightly budded organoids prior to cryopreservation, as this will result in a higher yield of viable organoids post-thawing. This can be accomplished by increasing the R-spondin conditioned media to 15% and/or by adding 10 mM nicotinamide (Table 3) to the organoid culture.- Using a microscope, count the number of organoids on the 35 mm dish. Label the cryovials such that 200 organoids will be aliquoted into each cryovial.
- Remove the rIOM from the 35 mm dish. Replace with 2 mL of cold tissue culture grade PBS.
- Using a P1000 tip, release the EME domes from the bottom of the culture dish into the PBS by pipetting up and down. Continue to pipette up and down ~20 times to break up the EME and release the organoids. Collect the organoids and PBS into a 15 mL conical tube.
- Add 2 mL of cold PBS to the culture dish and pipette up and down to release any remaining organoids into the PBS. Transfer the PBS to the 15 mL conical tube.
- Pellet the organoids by centrifuging at 290 x g for 5 min. Remove and discard the supernatant without disrupting the organoid pellet.
- Wash the pellet by gently resuspending in 5 mL of cold AdDMEM+. Centrifuge at 200 x g for 4 min. Carefully remove and discard the supernatant.
- Resuspend the organoid pellet in 1 mL of cold freezing medium per 200 organoids. Aliquot 1 mL of organoids in freezing medium per labeled cryovial. Place the cryovials in a freezing container.
- Store the organoids in a freezing container at -80 °C for 24 h, then transfer the cryovials to liquid nitrogen for long-term storage.
- Thawing rat intestinal organoids
NOTE: This protocol was modified from a previous protocol for thawing human and mouse intestinal organoids26.- Thaw a 250 µL aliquot of EME on ice. Prepare rIOM supplemented with 15% R-spondin conditioned media, 10 µM Y27632, and 10 µM CHIR99021. Warm at 37 °C.
- Add 2 mL of defrosting medium (Table 3) to a 15 mL conical tube at room temperature.
- Retrieve and thaw a vial of organoids from liquid nitrogen by placing the vial in a 37 °C water bath until the vial is almost completely thawed.
- Add 1 mL of defrosting medium to the vial and transfer all of the contents to the conical tube containing defrosting medium. Wash the vial twice with 1 mL of defrosting medium and transfer to the conical tube.
- Centrifuge at 200 x g for 5 min. Aspirate the media, leaving ~50 µL of medium with the organoids. Transfer the media containing organoids to 250 µL of EME.
- Evenly distribute the organoids through the EME by pipetting up and down, avoiding bubbles. Pipette six 50 µL domes into a 35 mm tissue culture dish.
- Incubate in the tissue culture incubator for 15-20 min to allow the EME to polymerize. Add 2 mL of the prepared rIOM to the dish.
- After 2 days, replace the media with rIOM. The use of Y27632 and CHIR99021 can be suspended. Organoid growth may be slow in the first passage after thawing. It is advised to passage the organoids twice after thawing before beginning experiments.
5. Generation of rat intestinal 2D monolayers from 3D organoids
NOTE: The following protocol describes the volumes required to generate 24 wells of a 48-well plate coated with EME, starting with six domes of 50 µL (35 mm dish) containing ~300 intestinal organoids/dome (scale: one dome generates four wells), but can be scaled up or down as needed. As written, this protocol achieves ~80% confluence in 4-5 days. At higher confluency, the cells begin to acquire 3D organoid structures again. At low confluence (≤40%) , the cells remain as monolayers and are viable for ~14 days. If the purpose of the study is to use 2D monolayers, scale down so that one dome generates eight wells of a 24-well plate. The wells can also be coated with collagen I to form monolayers.
- Preparation of coated surfaces
- To coat the plate with EME, dilute EME 1:20 in cold AdDMEM+ (Table 1). For coating with collagen, prepare collagen I according to the manufacturer's instructions. Dilute 5 mg/mL collagen I in AdDMEM+ to 100 µg/mL (1:50 in this case).
- Coat the plate with 200 µL of diluted EME or collagen to cover the well surface entirely. Incubate for 1-2 h at 37 °C in a tissue culture incubator. Prepare rat intestinal organoid medium for 2D monolayer culture (rIOM2D) as per Table 4.
- Generation of monolayers
- Aspirate the media from a 35 mm plate containing organoids. Add 1 mL of PBS.
- Disrupt the EME in the wells by scratching with a P1000 tip. Pipette up and down approximately 20 times to loosen all the EME. Transfer everything to a 15 mL conical tube.
- Add 1 mL of PBS to the 35 mm plate to recover any additional organoids and transfer to the same 15 mL conical tube.
- Centrifuge at 350 x g for 2 min and aspirate the supernatant, including EME residue. Add 1 mL of trypsin solution to the organoid pellet and incubate at 37 °C for 2 min.
- Pipette up and down 10 times using a P1000 tip and add 2 mL of AdDMEM+ to neutralize trypsin.
- Centrifuge at 350 x g for 5 min. Aspirate the supernatant and add 4.8 mL of rIOM2D (Table 4). Resuspend the cell pellet.
- Before placing the organoids, take out any excess EME or collagen in AdDMEM+ from the wells. Then, add 200 µL of organoids in rIOM2D and 10 µM Y27632 to each precoated well.
- After 4-16 h, collect the media and centrifuge at 1,000 x g for 1 min. Transfer the supernatant to a new 15 mL conical tube and discard the pellet.
- Wash each well with 300 µL of PBS and add 200 µL of the centrifuged rIOM2D to each well. Change the rIOM2D every 2-3 days, suspending the use of Y27632.
- Reforming 3D organoids from 2D monolayers
NOTE: Monolayers grown on EME can be induced to reform 3D organoids, while monolayers grown on collagen I do not efficiently return to 3D organoids. Usually, 2-3 wells of organoids generated from monolayers can be used to make one dome of 50 µL of EME at day 5.- Dilute the EME 1:4 in rIOM2D. When the monolayers reach ~80% confluency, carefully aspirate the media and add 100 µL of diluted EME to the wells containing monolayers.
- Incubate at 37 °C, 5% CO2 in a tissue culture incubator for 20 min. Afterward, add 100 µL of rIOM2D and return to the tissue culture incubator. 3D organoids will be generated within 5 days of adding diluted EME.
- After small organoids have reformed (~day 5), prepare rIOM. Collect all the well contents, pipetting up and down to disrupt the EME, and transfer to a 15 mL conical tube.
- Centrifuge at 350 x g for 2 min. Aspirate the supernatant and EME residue.
- Add 1 mL of cold AdDMEM+ and centrifuge at 350 x g for 2 min. Remove the supernatant, leaving ~50 µL of media.
- Transfer the 50 µL of media and organoids to a 250 µL EME aliquot. Pipette up and down to distribute the organoids throughout the EME.
- Pipette 50 µL of EME domes into a 35 mm dish and incubate for 20 min in a tissue culture incubator.
- Add 2 mL of rIOM plus 10 µM Y27632. Change the growth media every 2-3 days. When changing the media, Y27632 can be omitted.
6. Genetic manipulation
- Transient transfection of 2D monolayers
CAUTION: Prepare rat intestinal epithelial monolayers following section 5.1 in a 48-well plate. Transfection must be performed at 70%-80% confluency. Always replace the media in the wells with 200 µL of fresh rIOM2D before transfecting. Calculate 260/280 nm and 260/230 nm absorbance ratios of the plasmid DNA; these must be above 1.8 to ensure good transfection results.
NOTE: Use a plasmid control to calculate the efficiency of transfection. A plasmid that encodes a fluorescent protein is recommended for ease. In this protocol, pLJM1-EGFP27 was used.- Prepare 1 mg/mL of 20 kDa polyethylenimine (PEI; Table 3). For each well, prepare tube A (0.6 µg of plasmid + 50 µL of reduced serum medium) and tube B (1.8 µL of PEI + 50 µL of reduced serum medium). Maintain a DNA:PEI ratio of 1:3.
- Vortex both tubes for 30 s. Combine tube A and tube B, and vortex again for 30 s. If necessary, use a centrifuge to spin down. Incubate for 20 min at room temperature.
- Gently add the DNA/PEI complex drop-wise to 2D monolayer. Gently swirl the plate to mix. Incubate at 37 °C, 5% CO2. The expression can usually be detected after 24 h.
NOTE: If it is necessary to return to 3D organoid structures, wait for 48 h after transfection for adding diluted EME.
- Lentiviral transduction of rat intestinal organoids
NOTE: Before working with lentiviruses, obtain proper authorization and specialized training from the institution. Always wear appropriate personal protective equipment (PPE) when handling lentiviruses. While not outlined here, high-quality, concentrated lentivirus is essential for the successful infection of organoids. This protocol utilized empty pLJM1-EGFP vector27 to express soluble GFP. This protocol is modified from a previously published protocol28. The transduction efficiency depends on the quality and concentration of viral particles, efficient dissociation of organoids into small cell clusters, and the gene being expressed. When calculated 5 days after infection, the mean transduction efficiency prior to selection was 19.4% (± 6.5% standard deviation).- At 2 days prior to lentiviral infection, plan to passage (section 3) two dense wells of a 24-well plate for each lentivirus that will be transduced. Each well of a 24-well plate can accommodate one 50 µL dome of EME.
- Once the EME solidifies, add 0.5 mL of rIOM supplemented with 10 mM nicotinamide, 10 µM Y27632, and 2.5 µM CHIR99021. This induces large spheroid morphologies, which is favorable for the efficiency of lentiviral transduction. Within 2 days of plating, rat organoids should be large spheroids. If significant differentiation (i.e., budding) is observed, repassage the organoids.
- Thaw the concentrated virus on ice. Prepare fresh transduction media, as per Table 3.
- Using a P1000 tip, release the EME domes from the bottom of the cell culture dish into the media by pipetting up and down. Continue to pipette up and down 20 times to break up the EME and release the organoids.
- Pipette the organoids and media into a 15 mL conical tube. All the wells can be pooled together, provided the organoids have the same genotype and are from the same line.
- Wash each well twice with 1 mL of cold AdDMEM+. Collect the AdDMEM+ and transfer it to the same 15 mL conical tube.
- Using a glass Pasteur pipette, mechanically disrupt the rat organoids. This is a critical step, as the aim is small multicellular clusters of cells. Pipette up and down ~30 times. Check for efficiency of the disruption under the microscope using a 4x objective. Continue this process until the cell suspension is primarily comprised of cell clusters with few organoids remaining.
NOTE: Alternatively, organoids can be centrifuged at 200 x g for 5 min at room temperature, the supernatant carefully removed, and resuspended in 1 mL of recombinant enzyme replacing conventional trypsin/EDTA, prewarmed to 37 °C in the tissue culture incubator. Incubate the organoids in trypsin replacement for 2 min in a 37 °C water bath, with regular vortexing to promote dissociation. Avoid prolonged incubation with trypsin replacement, as this can promote cell death. Regularly check for efficiency of disruption under the microscope using a 4x objective. When the suspension is composed of primarily single cells with few cell cultures, dilute the trypsin replacement by adding 4 mL of AdDMEM+ and proceed with the following step. - Spin the 15 mL conical tube containing the cell clusters at 200 x g for 5 min at 4 °C. Carefully remove and discard the supernatant, being careful not to disturb the cell pellet.
- Resuspend the cell clusters in 230 µL of transduction medium per well to be infected. For the infection, use one well of a 48-well plate for each lentivirus.
- Plate 230 µL of cell suspension into each well of a labeled 48-well plate. Add 20 µL of concentrated virus to each well. Use a P1000 tip to mix the virus/cell solution in each well and seal the plate with a transparent film.
- Perform spinoculation: centrifuge the plate at 600 x g for 1 h at 32 °C. Unseal the plate and incubate at 37 °C, 5% CO2 for 6 h.
- Preheat a 24-well plate in the 37 °C incubator.
- Following incubation, pipette each well up and down and transfer the contents to a labeled 1.5 mL tube. Wash each well with 750 µL of AdDMEM+ and transfer to the tube. Spin the tubes at 600 x g for 5 min at 4 °C.
- Remove the tubes from the centrifuge and store on ice. Use a P1000 tip to carefully remove and properly dispose of the supernatant.
- Resuspend the cell pellet in EME and plate 50 µL domes into the preheated 24-well plate. Incubate for 15-20 min at 37 °C until the EME is polymerized.
- To each well, add 500 µL of rIOM supplemented with 10 mM nicotinamide, 10 µM Y27632, and 2.5 µM CHIR99021.
- Replace the media with rIOM plus 10 µM Y27632 1 day after infection. Change the media every 2-3 days. If selection will be performed, add selection 48-72 h after infection. For puromycin selection, use 2 µg/mL puromycin.
7. Immunofluorescence whole mount staining of organoids
- Aspirate the media and add 4% paraformaldehyde (PFA) in PBS-Tween 20 (PBS-T) (Table 3) to the organoids in a cell culture dish. Incubate at room temperature for 10 min.
- Release the organoids from the EME by pipetting up and down. Collect the organoids into a 0.75 mL tube. The organoids will settle to the bottom of the tube by gravity within a few minutes. If needed, centrifuge at 100 x g for 1 min.
- Using a transfer pipette, remove the PFA and resuspend the organoids in PBS-T. Let the organoids settle to bottom of the tube. Remove the PBS-T and resuspend in 200 µL of block solution (Table 3).
- Incubate at room temperature on a rocker or nutator for 45 min. Organoids can clump and settle to the bottom of the tube. Periodically finger flick the tube to resuspend and disperse throughout the solution.
- Let the organoids settle to the bottom of the tube. If needed, centrifuge at 100 x g for 1 min. Remove block solution.
- Add primary antibody diluted in 100 µL of block solution. Incubate at room temperature on a nutating mixer for 45 min at 24 rpm. Antibody concentrations vary depending on the antibody used.
NOTE: This step can be extended based on the primary antibody, up to overnight at 4 °C. - Let the organoids settle to the bottom of the tube. If needed, centrifuge at 100 x g for 1 min.
- Wash in PBS-T five times using a transfer pipette. Incubate at room temperature for 5 min on nutating mixer at 24 rpm. Repeat this step twice.
- Add secondary antibody diluted 1:200 and 50 µg/mL 4′,6-diamidino-2-phenylindole (DAPI) diluted in 100 µL of block solution. Incubate at room temperature on a nutating mixer for 30 min at 24 rpm.
- Let the organoids settle to the bottom of the tube. If needed, centrifuge at 100 x g for 1 min. Remove secondary antibody. Wash five times in PBS-T using a transfer pipette.
- Incubate at room temperature for 5 min on a nutating mixer at 24 rpm. Repeat the washing step twice.
- While the organoids are washing, heat an aliquot of VALAP sealant (Table 3) at 40-50 °C to liquify. Using a paintbrush, paint a thin square of VALAP on a microscope slide approximately the size of a coverslip. Use a 22 mm x 22 mm No. 1.5 coverslip .
- Using scissors, cut the end of a P200 pipette tip. Transfer organoids to the VALAP well on the slide. Using a tissue wipe, carefully wick away the PBS-T. Do not let the organoids dry out.
- Flood the VALAP well with an antifade mounting medium (Table 3). For a ~22 mm x 22 mm square, this will require 100-150 µL of antifade.
- The organoids can cluster; swirl the antifade with a pipette tip to redistribute the organoids, if necessary. Mount with 22 mm x 22 mm No. 1.5 coverslip, avoiding air bubbles. Seal the coverslip by painting a thin layer of VALAP onto the edges.
NOTE: For inverted microscopes, the organoids can also be mounted in a 35 mm glass-bottom dish.
8. Forskolin-induced swelling of rat intestinal organoids
- Grow the organoids for 3-5 days after passaging in rIOM. It is advisable to grow organoids in a 24-well plate to ensure that the same region can be easily reimaged. Capture images before the addition of forskolin (T0).
- Add forskolin (Table 3) directly to the organoid media to a final concentration of 10 µM. Add the same volume of dimethyl sulfoxide (DMSO) to the control wells.
- Image the control and forskolin-treated wells at regular intervals, every 15-30 min. When not imaging, keep organoids in the incubator or use a controlled imaging system with incubation. Maximum swelling should be observed by 120 min.
- Follow standard protocols to calculate relative swelling from the acquired images29.
Representative Results
Rat duodenal and jejunal organoids were generated using the protocol outlined in section 2. It is very important during the crypt isolation steps that villi are efficiently depleted from the PBS. If too many villi are plated in the EME with crypts, it can cause death of the entire culture and failure to establish an organoid line. Because of this, it is useful to isolate crypts under a dissecting scope, allowing for visual confirmation of villar depletion. Figure 1 depicts representative villar fragments and crypts (Figure 1A). Note the significantly smaller size of crypts compared to villi (Figure 1B). After plating, the crypts will expand into spheroids over the next few days and will begin to bud and differentiate by day 4-7 (Figure 2). Once the organoids reach an extensively budded stage, they should be passaged. During passaging, it is important to disrupt the organoids enough to split the crypt buds apart, so that organoid numbers can be expanded (Figure 3B).
The successful recovery of organoids after freezing is highly dependent on the state at which they are frozen. Organoids in a highly proliferative undifferentiated state recover with the highest efficiency. Therefore, we recommend inducing them to be spherical and cystic instead of budded and differentiated. To achieve this, Wnt can be hyperactivated by increasing the amount of the Wnt ligand R-spondin in the media and by including nicotinamide in the media, which has been shown to support organoid formation and cell survival in several culture systems30,31. Figure 3A demonstrates a healthy organoid culture just 2 days after thawing. Including BSA in the media during thawing has also helped in the survival of rat intestinal organoid cultures, which have proven to be more delicate than mouse intestinal organoids.
While 3D organoid culture is often preferred because it recapitulates some of the normal intestinal architecture, it makes other approaches, including live imaging, transfections, and lentiviral transductions, more technically challenging. The use of 2D monolayers generated from 3D organoids32 (Figure 4) allows for higher efficiency introduction of the plasmids. While 3D intestinal organoids are traditionally resistant to transient transfections, plasmids encoding for EGFP can be successfully introduced using lipid-based transfection methods. The most cost-effective approach using PEI is outlined in step 6.1 (Figure 5), but electroporation and commercially available transfection reagents have also yielded comparable results (data not shown). Future studies will be focused on whether these approaches can be used for introducing CRISPR constructs into monolayers.
It was important to be able to reform 3D organoids from 2D monolayers after transfection so that they could be maintained as a passageable line with 3D architectural components of crypts. Interestingly, 2D monolayers plated on the EME readily reformed into small spheroids when EME was added back to the top of the cells, whereas a collagen I substrate was not sufficient for the reformation of 3D structures (Figure 6).
While transient transfections are useful for many studies, the formation of stable lines are often more useful, requiring the introduction of lentivirus into the cells. Rat intestinal organoids were infected with lentivirus by modifying previously published protocols (Figure 7). A key step in the protocol is the disruption of organoids into small aggregates or cell clusters. If cultures are not efficiently disrupted and organoids remain intact, the lentiviral particles will not get into the cells. After infection, organoids must recover and regrow. The protocol outlined here allows for the uptake of viral particles by 10%-48% (mean: 19.4% ± 6.5%) of cells before selection.
Whole mount staining of organoids can prove difficult due to incomplete removal of EME residue or incomplete antibody penetrance. The protocol outlined here allows for the robust staining of organoids. The visualization of organoids on a confocal microscope can also prove difficult if they are too far away from the coverslip. By using VALAP, a well with some height is created such that organoids are not crushed by the coverslip, but are still allowed to settle close to the coverslip for ease of imaging. Representative staining against the apical anion channel cystic fibrosis transmembrane conductance regulator (CFTR) and phalloidin to label F-actin is shown in Figure 8.
Finally, organoids have utility in functional assays. Patient-derived organoids from cystic fibrosis patients have been used to screen CFTR function, as treatment with the cAMP agonist forskolin induces robust CFTR-mediated fluid secretion, causing organoid swelling29,33–37. One goal of this work was to identify and develop an organoid model that can be used in parallel to in vivo preclinical studies. Therefore, we aimed to determine whether rat intestinal organoids undergo forskolin-induced swelling. Indeed, within 30 min of forskolin treatment, rat organoids swelled, with a maximal effect observed by 120 min (Figure 9).
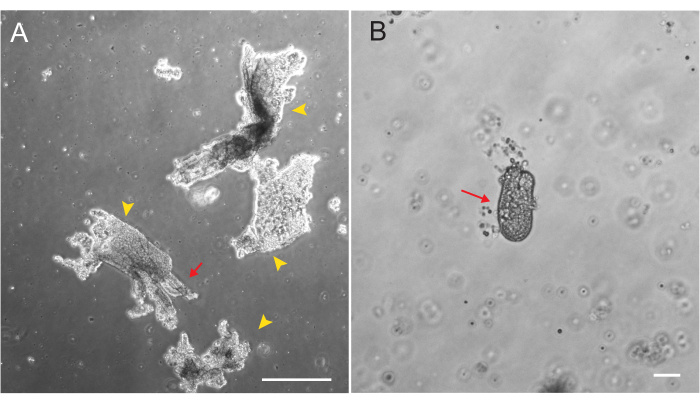
Figure 1: Villar fragments and crypts during epithelial isolation. (A) Representative image of villar fragments in EDTA solution during the crypt isolation protocol. Yellow arrowheads mark villar fragments. Red arrows depict crypts attached to a villar fragment. Note the difference in relative sizes. (B) Higher magnification image of a single crypt (red arrow) so that morphology can be visualized. Scale bars: 100 µm. Please click here to view a larger version of this figure.
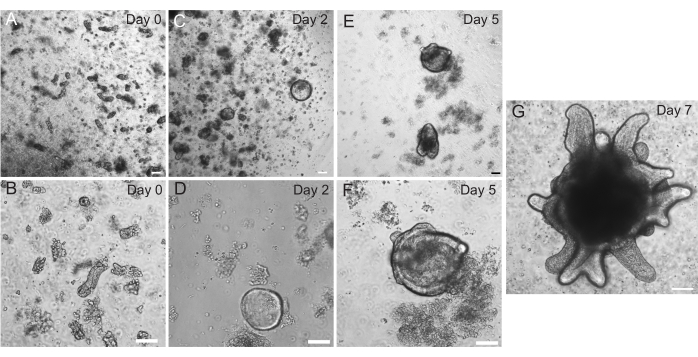
Figure 2: Rat intestinal organoid progression. Rat jejunum crypts were plated in EME immediately after isolation (A,B). Within 2 days, the crypts became spheroids (C,D). By Day 5, they began to initiate crypt buds (E,F), which elaborated and grew by Day 7 (G). Scale bars: 100 µm. Please click here to view a larger version of this figure.
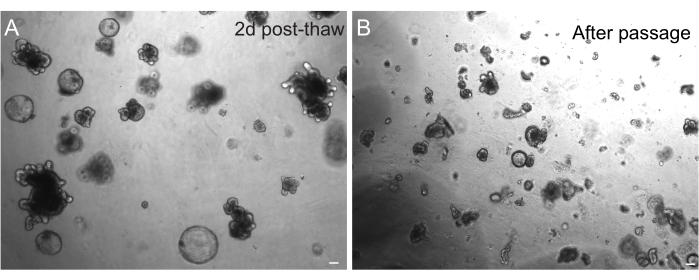
Figure 3: Organoids after thawing and passaging. (A) Rat jejunal organoids were thawed following the outlined protocols after cryopreservation. Note the presence of both spheroids and budded organoids just 2 days after thawing. (B) The same organoid line depicted in A immediately after passaging following the outlined protocol. Note the relative size difference between structures in A and B and the presence of single crypt-like domains in B. Scale bars: 100 µm. Please click here to view a larger version of this figure.
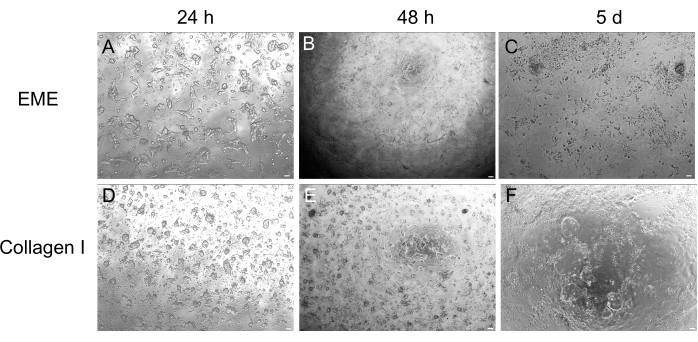
Figure 4: 2D monolayer formation from 3D organoids. (A–C) 2D monolayer progression on EME. (D–F) 2D monolayer progression on collagen I. By day 5, each condition yielded ~80% confluency. Scale bars: 100 µm. Please click here to view a larger version of this figure.
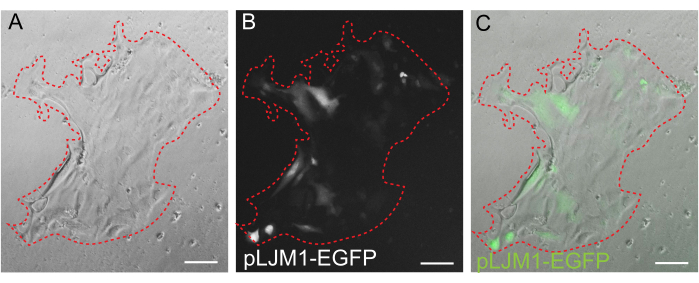
Figure 5: Transient transfection of a 2D monolayer. Representative image of a 2D monolayer grown on EME transiently transfected with pLJM1-EGFP plasmid using PEI. (A) Brightfield, (B) fluorescence (GFP), (C) overlay. The dotted red line marks the monolayer boundary. Scale bars: 100 µm. Please click here to view a larger version of this figure.
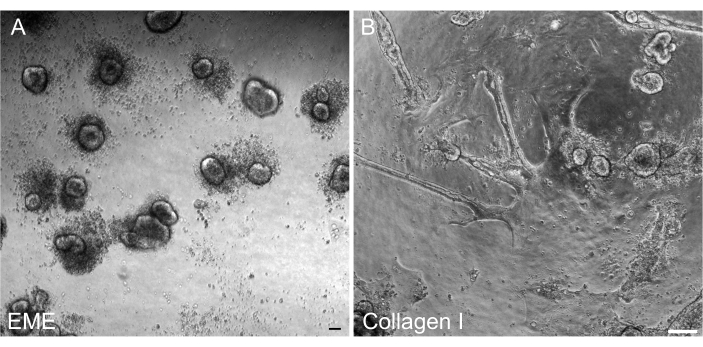
Figure 6: Reformation of 3D organoids from 2D monolayers on EME. (A) Formation of 3D organoids from 2D monolayers grown on EME. Organoids efficiently formed by 5 days after adding EME to the apical surface of the monolayer. Note the abundance of dead cells surrounding the small 3D spheroids. (B) Persistence of 2D monolayers 5 days after collagen I was added to the apical surface of 2D monolayers grown on collagen I. Scale bars: 100 µm. Please click here to view a larger version of this figure.
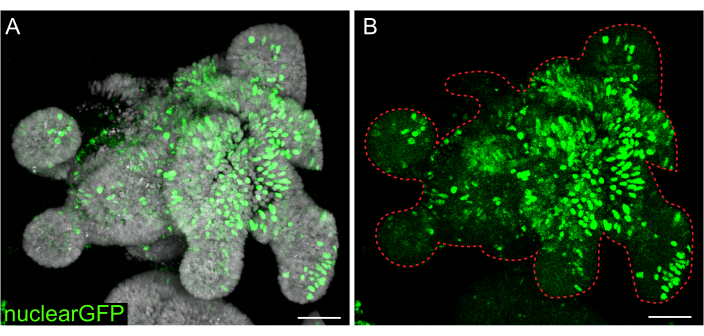
Figure 7: Lentiviral infection of 3D organoids. Rat jejunum organoids were infected with nuclear GFP lentiviral particles using the outlined protocol. After recovery and growth for 5 days, organoids were fixed and counterstained with DAPI. (A) DAPI: gray; nuclearGFP: green. (B) nuclearGFP:green. The dotted red line marks the organoid boundary. Scale bars: 50 µm. Please click here to view a larger version of this figure.
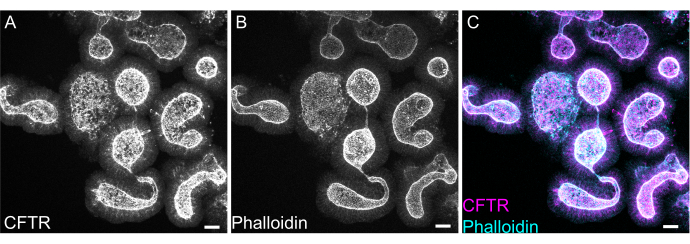
Figure 8: Whole mount immunofluorescence of rat intestinal organoids. (A) CFTR, (B) phalloidin, and (C) merged whole mount immunofluorescence of rat jejunal organoids. Note the apical enrichment of CFTR staining in organoids (gray in A, magenta in C). Phalloidin marks F- actin and prominently labels the apical brush border (gray in B, cyan in C). Scale bars: 25 µm. Please click here to view a larger version of this figure.
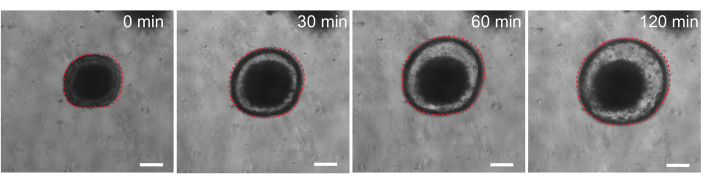
Figure 9: Rat intestinal organoids swell upon forskolin stimulation. Representative time course of rat intestinal organoid swelling after addition of the cAMP agonist forskolin. The 0 min time represents the time point immediately prior to the addition of 10 µM forskolin. Images show the same organoid at 30 min time intervals. Maximal swelling was observed at 120 min after forskolin addition. The dotted red line outlines the organoid boundary. The dark material in the middle of the organoid lumen is comprised of dead cells. Scale bars: 100 µm. Please click here to view a larger version of this figure.
Table 1: AdDMEM+ recipe. Ingredients to make the standard AdDMEM+ media, which is the base media throughout the methods shown here. Please click here to download this Table.
Table 2: Rat intestinal organoid media (rIOM) recipe. Detailed recipe of the standard rat intestinal organoid media, including solvent and storage conditions for recombinant proteins. Please click here to download this Table.
Table 3: Solutions. Recipes and instructions to make other solutions used throughout the protocol. Please click here to download this Table.
Table 4. Rat intestinal organoid medium for 2D monolayer culture (rIOM2D). Modified recipe of organoid culture media optimized for the 2D growth of monolayers. Please click here to download this Table.
Discussion
The development of a rat intestinal organoid model preserves important functional characteristics found in the organ in vivo and is a promising tool for preclinical testing, drug screening, and functional assays. This in vitro model can be used in parallel to in vivo preclinical gastroenterology studies, for which rats are often a preferred model due to their larger intestinal size, shared physiological aspects with humans, and in some cases being better disease models38. Here, a robust step-by-step protocol for the isolation of rat intestinal crypts, generation and long-term culture of rat intestinal organoids, as well as downstream applications including functional forskolin swelling assays, whole mount immunofluorescence, 2D monolayer culture, and lentiviral genetic manipulation is outlined. Rat intestinal organoids are likely to be relevant in many disease contexts where the pathophysiology of mouse models is inappropriate and may provide a better model for human intestinal physiology compared to mouse intestinal organoids.
To establish long-lived organoid cultures that can be passaged and expanded, it is essential to identify the key growth factors required for maintaining intestinal epithelial proliferation. Mouse organoids are most often grown in a simple cocktail of EGF, R-spondin, and Noggin, though it has been reported that Noggin is not necessary for intestinal organoid culture39. Conditioned media can replace recombinant growth factors and the most commonly used cell lines are L-WRN, which secretes Wnt3a, Rspondin-3, and Noggin39, L-Wnt3a, and HA-Rspondin1-Fc 293T cells40. L-WRN conditioned media is sufficient to support not only mouse intestinal39 organoid growth, but the growth of intestinal organoids from several farm animals and companion animals, including dogs, cats, chickens, horses, cows, sheep, and pigs12. However, human intestinal organoids are very different in their growth factor requirements, as they require distinct media formulations for their expansion growth phase (i.e., the progression from small to large spheroids) versus their differentiation phase (i.e., the generation and maturation of differentiated cell types)10. The media requirements of rat intestinal organoids closely mirror those of the expansion growth media for human intestinal organoids, yet, notably, rat organoids are capable of both growth and differentiation in this media environment, considerably simplifying their culture requirements. While our initial attempts focused on establishing and growing rat intestinal organoids in L-WRN-conditioned media, long-term culture was tenuous, and rat intestinal organoid lines suffered from a lack of robustness (data not shown). This may be because L-WRN cell lines are engineered to secrete R-spondin 3, while the 293T-Rspo1 cell line recommended here is engineered to secrete R-spondin 1. It is possible that rat and human organoids prefer R-spondin 1, potentially accounting for the failure of rat organoid lines in L-WRN conditioned media.
To most closely recapitulate the in vivo setting, it is important to develop organoid culture conditions that allow for stem cell survival, maintenance, and proliferation, and that can maintain cellular turnover and simultaneous differentiation events into discrete cell types. Therefore, the concentrations of recombinant proteins and/or proteins in conditioned media need to be tightly titrated and controlled to strike this perfect balance. In particular, optimal Wnt levels are essential to avoid the loss of intestinal organoid cultures. Too little Wnt in conditioned media will be incapable of supporting growth, leading to a loss of stem cells and subsequent organoid death; overactivation of Wnt will cause organoids to be cystic and undifferentiated10. While not detailed here, it is strongly recommended to test each batch of L-Wnt3a and 293T-Rspo1 conditioned media using a Wnt reporter luciferase assay, such as a Topflash cell line41. Previous studies have described that an optimal batch of L-Wnt3a media should result in a 15-fold signal increase at 12.5% and a 300-fold signal increase at 50%, compared to 1% L-Wnt3a10. As rat organoids are more sensitive than mouse organoids to culture requirements, particularly Wnt activation levels, these additional quality control steps greatly assist in facilitating the robustness and reliability of rat organoid cultures. Because a similar reporter line is not available for testing Bmp activity and relative Noggin concentrations in Noggin conditioned media, it is advisable to use recombinant Noggin when possible to precisely control Noggin levels. While mouse intestinal organoids can be grown and maintained in the absence of Noggin39, this has not been attempted for rat intestinal organoid cultures.
Beyond cell culture requirements, the successful initial establishment of a rat organoid line depends critically on the efficient depletion of differentiated villi during crypt isolation. High levels of villar contamination cause crypt death, presumably due to either signals from the dying cells or sequestration of essential factors. To deplete these differentiated villi from epithelial preparations precisely and consistently, it is recommended to perform epithelial isolations with the aid of a stereoscope. Visual examination of the epithelium being released provides a clear cue when to discard the PBS and replace it (Figure 1). Crypts should not be collected until there is sufficient depletion of villi. Villar cells are terminally differentiated and cannot generate organoids in culture. Additionally, subsequent passaging of rat intestinal organoids and their use for any downstream application requires delicate care. Incubation in dissociation reagents for longer periods of time (10 min) result in significant cell death and loss of the organoid line.
Here, a simple and fast protocol for generating intestinal monolayers from rat organoids is described. EME and collagen I substrates have different effects on the epithelium, that can be leveraged depending on the purpose of the study. EME allows for the rapid and efficient adhesion of single cells and the formation of cell projections. In contrast, coating the surface with collagen I delays these processes. Once monolayers reach approximately 80% confluency, cells grown on EME begin to generate 3D organoid structures again. However, they lack sufficient physical and chemical support for continued growth. This reversion back to the organoid state can be prevented by maintaining monolayers in EME at a confluency of 50%-80%. The addition of diluted EME to the apical surface of monolayers promotes the rapid recovery and formation of de novo organoids, generating regions of convergence more quickly and readily. On a collagen I surface, cells can form a uniform monolayer and generate small clusters. However, the addition of collagen I on top of monolayers is not sufficient to induce organoid formation. EME must be diluted when adding to the monolayer surface, as there will be a stronger mechanical resistance for the nascent organoid to overcome. However, this diluted EME does not allow for the robust formation of large organoids. Any de novo generated rat organoids that naturally detach from the surface must be immediately removed and transferred to undiluted EME so that structural support and growth can be restored. Due to the small size of the organoids in this step, the passage of organoids is not recommended until robust growth has been established. The underlying biological significance of why EME can support the reformation of organoids, but whether collagen I can or cannot do this, is not clear. However, there have been reports that cells grown in 3D collagen cannot form budded organoids42,43 or support long-term maintenance. Commercially available EME products are heterogeneous mixtures of extracellular proteins, primarily laminin and collagen IV44. Therefore, the distinct composition of proteins and the ability of an epithelial cell to engage with the extracellular matrix using different cellular complexes could allow for remodeling in EME but not collagen I. Whether collagen I-derived monolayers can be put into EME to support organoid formation and growth has not been tested.
Genetic manipulation of the rat intestinal organoid model is described here, and protocols for lentiviral transduction of 3D organoids and transient transfection of 2D monolayers are outlined. To overcome the low efficiency of lentiviral organoid transduction, a protocol was developed for the transient transfection of 2D monolayers. The flat morphology and exposed apical domains of monolayers provide easier access to viruses and DNA-containing complexes. The expression of an EGFP reporter using the pLJM1-EGFP vector was used for the validation of this technique. GFP reporter expression was observed after 24 h, and was maintained for 5-6 days in monolayers. Future studies focusing on lentiviral transduction of monolayers are likely to have higher efficiency than 3D organoid transduction. Using the above protocols, 3D organoids can be reformed from infected 2D monolayers to facilitate the creation of stable lines. With care, rat intestinal organoids lines can be successfully maintained for over a year, remaining stable over many passages, cryopreserved, successfully thawed, and genetically modified using lentiviral transduction, thereby addressing the need for an accessible and tractable in vitro intestinal organoid model that retains physiological relevance to humans.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank members of the Sumigray and Ameen laboratories for their thoughtful discussions. This work was supported by a Charles H. Hood Foundation Child Health Grant and a Cystic Fibrosis Foundation grant (004741P222) to KS and by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health to NA under award number 2R01DK077065-12.
Materials
| 3-D Culture Matrix Rat Collagen I | Cultrex/R&D Systems | 3447-020-01 | |
| 70 µm cell strainer | Corning/Falcon | 352350 | |
| Advanced DMEM/F12 | Gibco/Thermo Fisher | 12634010 | |
| Amphotericin B | Sigma Aldrich | A2942-20ML | |
| B-27 Supplement (50X), serum free | Thermo Fisher | 17504044 | |
| CHIR99021 | Cayman Chemical | 13122 | |
| CryoStor | Stem Cell Technologies | 100-1061 | |
| Cultrex HA-Rspondin1-Fc 293T cells | R & D Systems | 3710-001-01 | |
| DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) | Molecular Probes/Thermo Fisher | D1306 | |
| FBS | Gibco/Thermo Fisher | 16-000-044 | |
| Gastrin I (human) | Sigma Aldrich | G9145 | |
| Gentle Cell Dissociation Reagent | Stem Cell Technologies | 100-0485 | |
| Glutamax | Thermo Fisher | 35-050-061 | |
| Growth factor-reduced Matrigel, phenol red-free | Corning | 356231 | |
| HEPES | AmericanBio | AB06021 | |
| Lanolin | Beantown Chemical | 144255-250G | |
| L-glutamine | Gibco/Thermo Fisher | A2916801 | |
| L-Wnt3a cells | ATCC | CRL-2647 | |
| N-2 Supplement (100X) | Thermo Fisher | 17502-048 | |
| N-acetylcysteine | Sigma Aldrich | A9165-5G | |
| Nicotinamide | Sigma Aldrich | N0636 | |
| Opti-MEM I Reduced Serum Medium | Gibco/Thermo Fisher | 31985070 | |
| Paraffin | Fisher Scientific | P31-500 | |
| Parafilm | Sigma Aldrich | P7793 | transparent film |
| PBS | Thermo Fisher | 10010023 | |
| Penicillin/Streptomycin | Gibco/Thermo Fisher | 15140122 | |
| pLJM1-EGFP | Addgene | 19319 | |
| Polybrene | Millipore | TR-1003-G | |
| Polyethylenimine hydrochloride (PEI) | Sigma Aldrich | 764965 | |
| p-phenylenediamine | Acros Organics/Thermo Fisher | 417481000 | |
| Puromycin | VWR | J593-25mg | |
| Recombinant human FGF2 protein | Peprotech | 100-18B-250ug | |
| Recombinant human IGF-1 protein | Biolegend | B356441 | |
| Recombinant human Noggin protein | R & D Systems | 6057-NG-100 | |
| Recombinant mouse EGF protein | Thermo Fisher | PMG8041 | |
| Sprague Dawley rat | Charles River Laboratories | Strain 001 | |
| Triton X-100 | American Bioanalytical | AB02025-00500 | |
| TrypLE Express Enzyme | Gibco/Thermo Fisher | 12604013 | |
| Y27632 dihydrochloride | Sigma Aldrich | Y0503 |
References
- Beumer, J., Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nature Reviews. Molecular Cell BiologyI. 22 (1), 39-53 (2021).
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262-265 (2009).
- Yin, Y. -. B., de Jonge, H. R., Wu, X., Yin, Y. -. L. Enteroids for nutritional studies. Molecular Nutrition & Food Research. 63 (16), 1801143 (2019).
- Cai, T., et al. Effects of six common dietary nutrients on murine intestinal organoid growth. PLoS One. 13 (2), e0191517 (2018).
- Davoudi, Z., et al. Gut Organoid as a new platform to study alginate and chitosan mediated PLGA nanoparticles for drug delivery. Marine Drugs. 19 (5), 282 (2021).
- Vlachogiannis, G., et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 359 (6378), 920-926 (2018).
- Yin, Y. B., de Jonge, H. R., Wu, X., Yin, Y. L. Mini-gut: a promising model for drug development. Drug Discovery Today. 24 (9), 1784-1794 (2019).
- Zietek, T., et al. Organoids to study intestinal nutrient transport, drug uptake and metabolism – update to the human model and expansion of applications. Frontiers in Bioengineering and Biotechnology. 8, 577656 (2020).
- Gunther, C., Winner, B., Neurath, M. F., Stappenbeck, T. S. Organoids in gastrointestinal diseases: from experimental models to clinical translation. Gut. 71 (9), 1892-1908 (2022).
- Pleguezuelos-Manzano, C., et al. Establishment and culture of human intestinal organoids derived from adult stem cells. Current Protocols in Immunology. 130 (1), 106 (2020).
- Zhao, Z., et al. Organoids. Nature Reviews Methods Primers. 2 (1), 94 (2022).
- Powell, R. H., Behnke, M. S. WRN conditioned media is sufficient for in vitro propagation of intestinal organoids from large farm and small companion animals. Biology Open. 6 (5), 698-705 (2017).
- Fagerholm, U., Johansson, M., Lennernas, H. Comparison between permeability coefficients in rat and human jejunum. Pharmaceutical Research. 13 (9), 1336-1342 (1996).
- Dubbelboer, I. R., Dahlgren, D., Sjogren, E., Lennernas, H. Rat intestinal drug permeability: A status report and summary of repeated determinations. European Journal of Pharmaceutics and Biopharmaceutics. 142, 364-376 (2019).
- Bryda, E. C. The Mighty Mouse: the impact of rodents on advances in biomedical research. Missouri Medicine. 110 (3), 207-211 (2013).
- Zhang, Z., Carriero, N., Gerstein, M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends in Genetics. 20 (2), 62-67 (2004).
- Lambracht-Washington, D., Fischer Lindahl, K. Active MHC class Ib genes in rat are pseudogenes in the mouse. Immunogenetics. 56 (2), 118-121 (2004).
- Busslinger, G. A., et al. Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Reports. 34 (10), 108819 (2021).
- Burclaff, J., et al. A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics. Cellular and Molecular Gastroenterology and Hepatology. 13 (5), 1554-1589 (2022).
- Haber, A. L., et al. A single-cell survey of the small intestinal epithelium. Nature. 551 (7680), 333-339 (2017).
- Olivier, A. K., Gibson-Corley, K. N., Meyerholz, D. K. Animal models of gastrointestinal and liver diseases. Animal models of cystic fibrosis: gastrointestinal, pancreatic, and hepatobiliary disease and pathophysiology. American Journal of Physiology. Gastrointestinal and Liver Physiology. 308 (6), G459-G471 (2015).
- Lu, P., et al. Animal models of gastrointestinal and liver diseases. Animal models of necrotizing enterocolitis: pathophysiology, translational relevance, and challenges. American Journal of Physiology. Gastrointestinal and Liver Physiology. 306 (11), G917-G928 (2014).
- Fujii, M., et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 23 (6), 787-793 (2018).
- Hedrich, W. D., et al. Development and characterization of rat duodenal organoids for ADME and toxicology applications. Toxicology. 446, 152614 (2020).
- How to Cryopreserve Intestinal Organoids. STEMCELL Available from: https://www.stemcell.com/technical-resources/area-of-interest/organoid-research/intestinal-research/tech-tips-protocols/how-to-cryopreserve-intestinal-organoids.html (2023)
- How to Thaw Intestinal Organoids. STEMCELL Available from: https://www.stemcell.com/technical-resources/educational-materials/protocols/how-to-thaw-intestinal-organoids.html (2023)
- Sancak, Y., et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320 (5882), 1496-1501 (2008).
- Van Lidth de Jeude, J. F., Vermeulen, J. L., Montenegro-Miranda, P. S., Vanden Brink, G. R., Heijmans, J. A protocol for lentiviral transduction and downstream analysis of intestinal organoids. Journal of Visualized Experiments. (98), e52531 (2015).
- Boj, S. F., et al. Forskolin-induced swelling in intestinal organoids: an in vitro assay for assessing drug response in cystic fibrosis patients. Journal of Visualized Experiments. (120), e55159 (2017).
- Regent, F., et al. Nicotinamide promotes formation of retinal organoids from human pluripotent stem cells via enhanced neural cell fate commitment. Frontiers in Cellular Neuroscience. 16, 878351 (2022).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Breau, K. A., et al. Efficient transgenesis and homology-directed gene targeting in monolayers of primary human small intestinal and colonic epithelial stem cells. Stem Cell Reports. 17 (6), 1493-1506 (2022).
- Berkers, G., et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Reports. 26 (7), 1701-1708 (2019).
- deWinter-de Groot, K. M., et al. Forskolin-induced swelling of intestinal organoids correlates with disease severity in adults with cystic fibrosis and homozygous F508del mutations. Journal of Cystic Fibrosis. 19 (4), 614-619 (2020).
- Dekkers, J. F., vander Ent, C. K., Beekman, J. M. Novel opportunities for CFTR-targeting drug development using organoids. Rare Diseases. 1, 27112 (2013).
- Dekkers, J. F., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature Medicine. 19 (7), 939-945 (2013).
- van Mourik, P., Beekman, J. M., vander Ent, C. K. Intestinal organoids to model cystic fibrosis. European Respiratory Journal. 54 (1), 1802379 (2019).
- Tong, T., et al. Transport of artificial virus-like nanocarriers through intestinal monolayers via microfold cells. Nanoscale. 12 (30), 16339-16347 (2020).
- Miyoshi, H., Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nature Protocols. 8 (12), 2471-2482 (2013).
- Ootani, A., et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nature Medicine. 15 (6), 701-706 (2009).
- Xu, Q., et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 116 (6), 883-895 (2004).
- Jabaji, Z., et al. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS One. 9 (9), e107814 (2014).
- Sachs, N., Tsukamoto, Y., Kujala, P., Peters, P. J., Clevers, H. Intestinal epithelial organoids fuse to form self-organizing tubes in floating collagen gels. Development. 144 (6), 1107-1112 (2017).
- Aisenbrey, E. A., Murphy, W. L. Synthetic alternatives to Matrigel. Nature Reviews Materials. 5 (7), 539-551 (2020).

